Characteristics of Oil Body Development and the Cloning and Expression Analysis of PDAT Genes in Eucommia ulmoides
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Stress Treatment Methods
2.2. Determination of E. ulmoides Oil
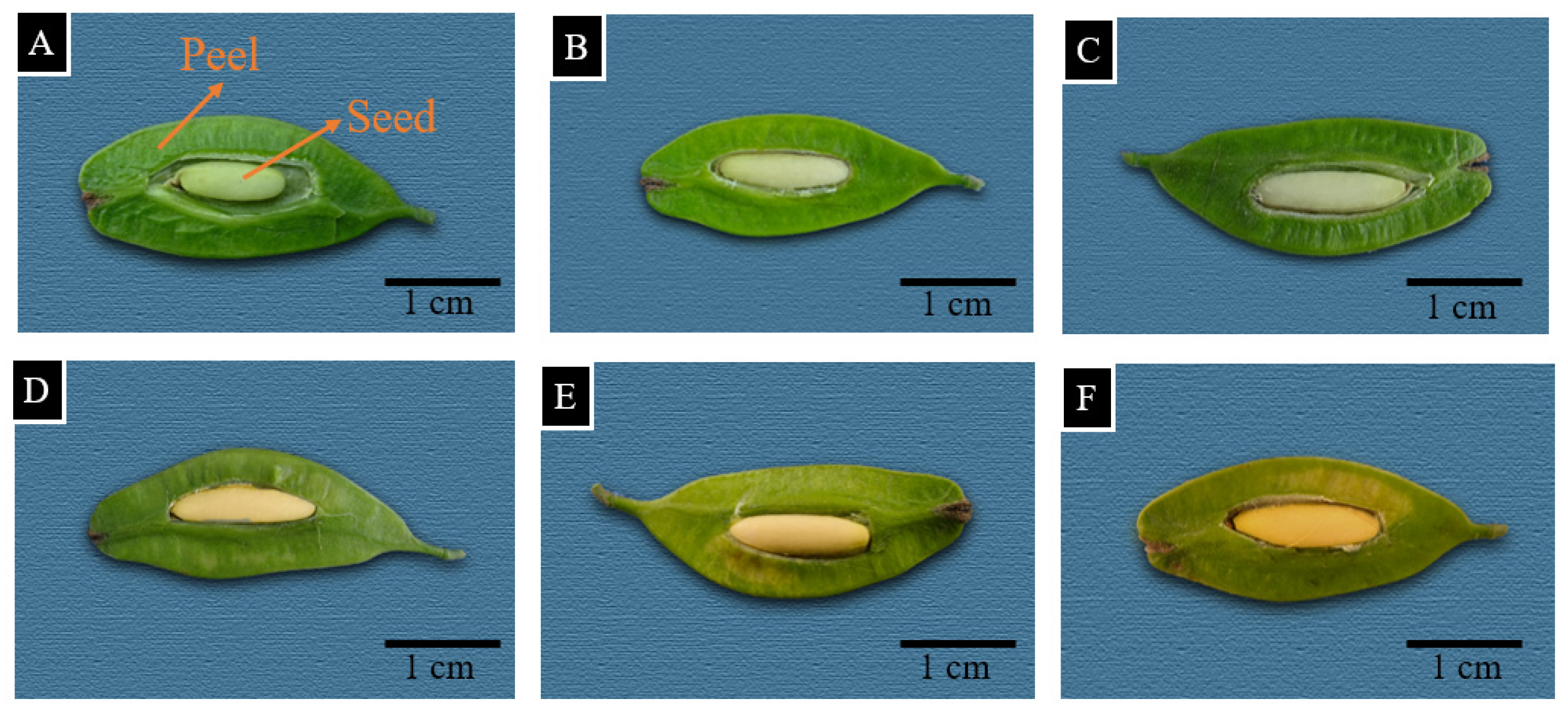
2.3. Observation of E. ulmoides Seed Oil Bodies
2.4. Identification of the PDAT Gene Family in E. ulmoides
2.5. Bioinformatics Analysis of PDAT Protein
2.6. RNA Extraction and Quantitative Reverse Transcription PCR Assay
3. Results
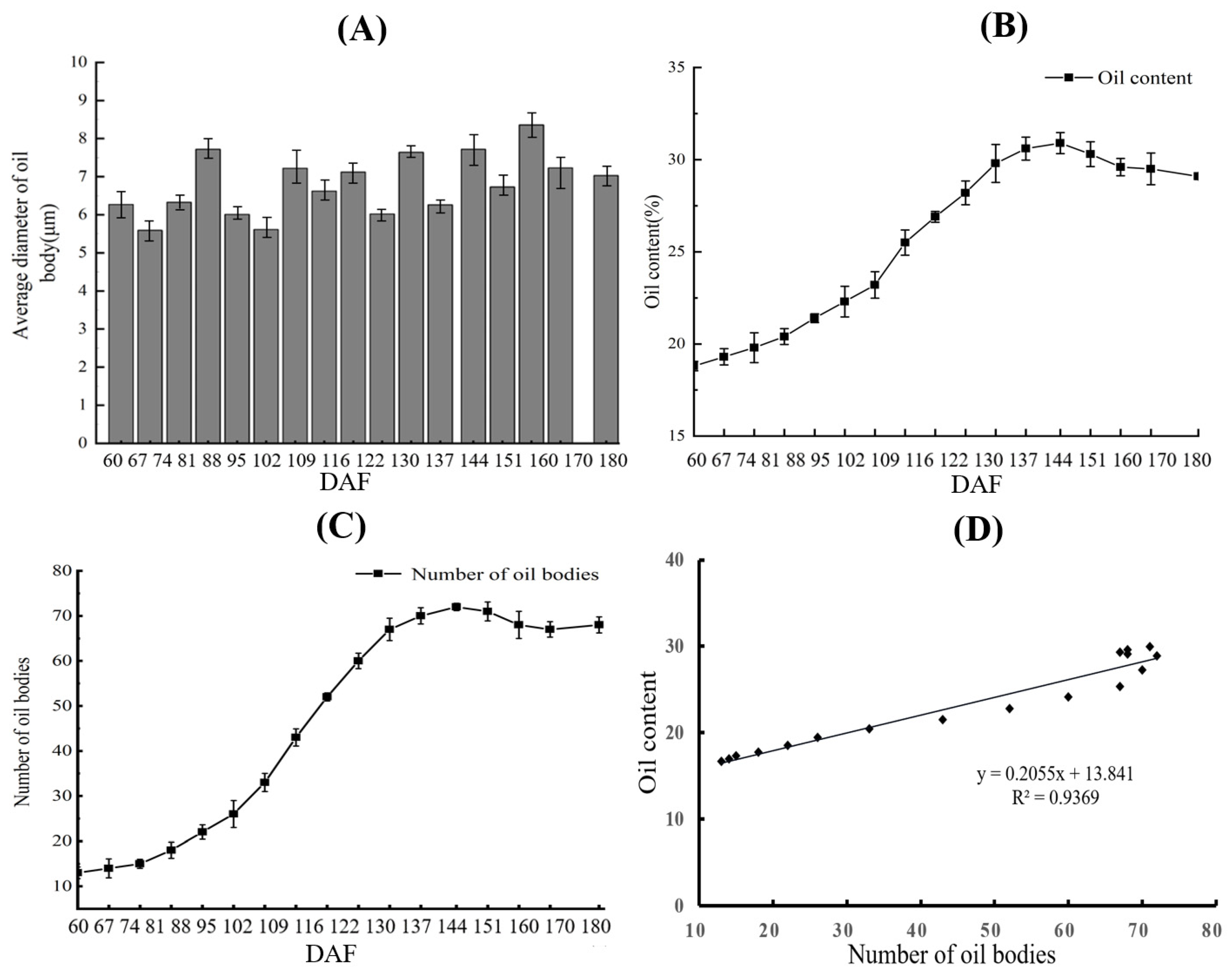
3.1. Correlation Analysis between Oil Bodies and Oil Content
3.2. Identification and Analysis of the Physicochemical Properties of EuPDAT Proteins in E. ulmoides
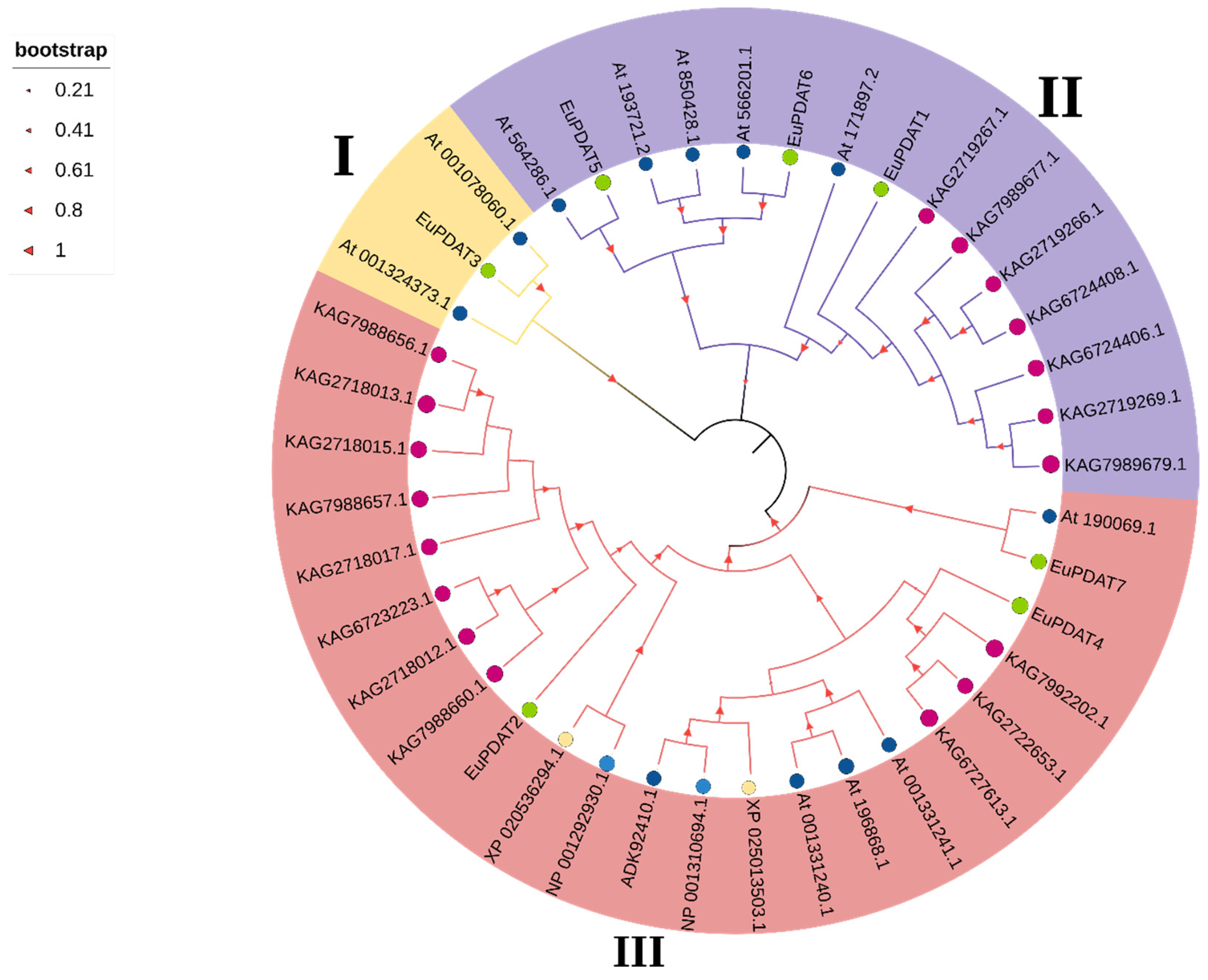
3.3. Phylogenetic Tree, Gene Structure, and Conserved Motif Analysis of the EuPDAT Genes in E. ulmoides
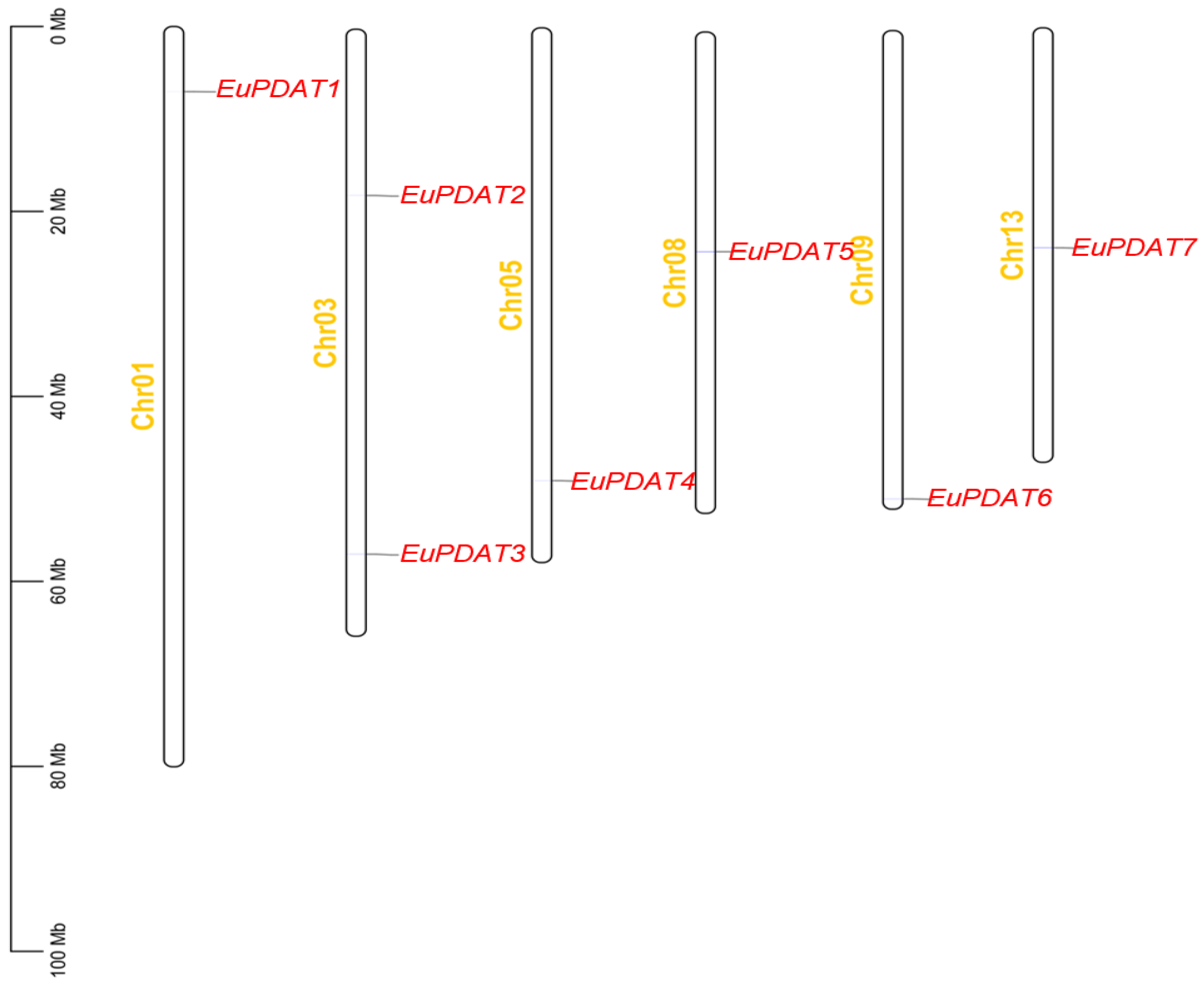
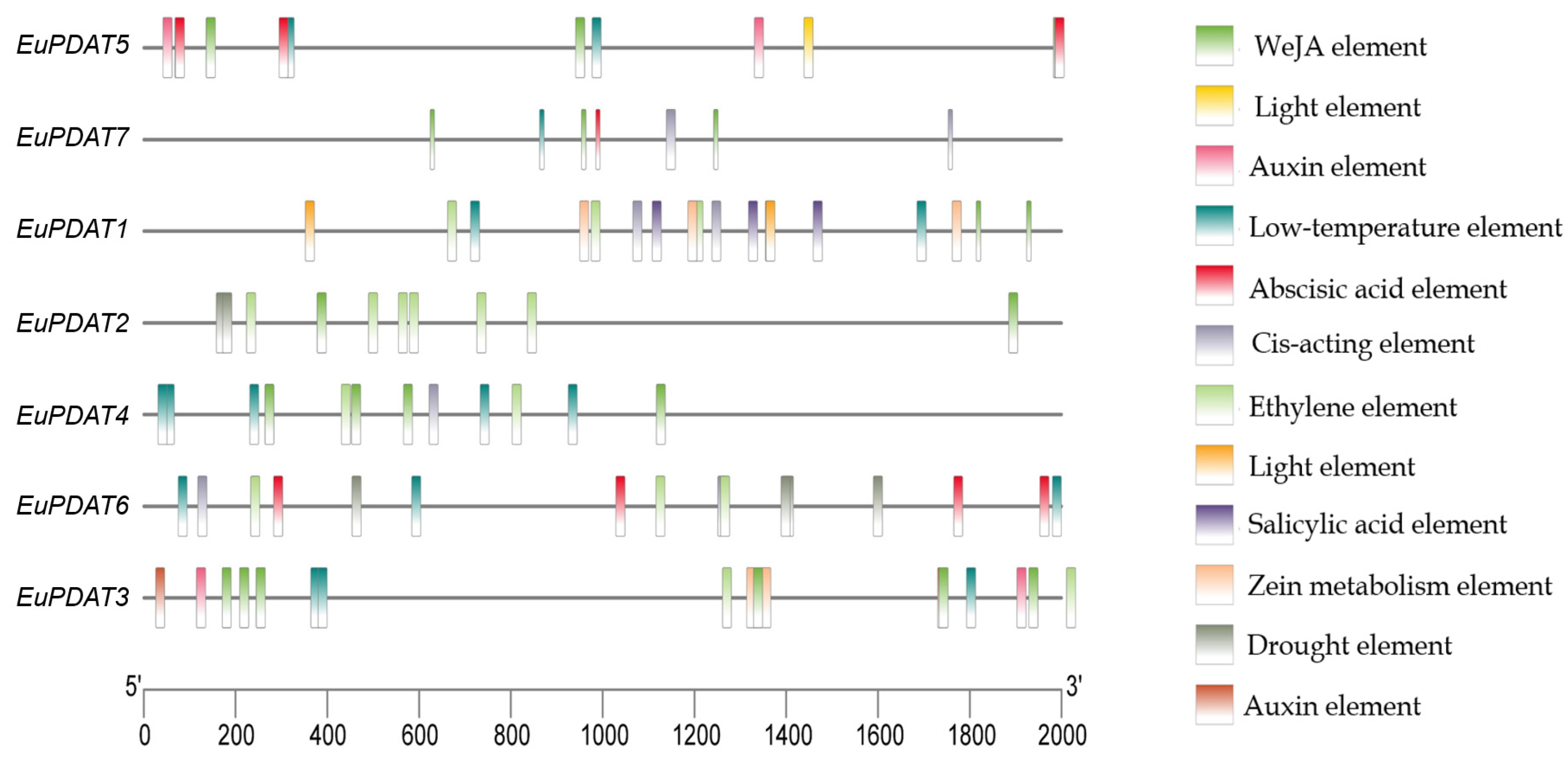
3.4. Chromosomal Locations and Putative Cis-Element Analysis of the EuPDAT Genes in E. ulmoides
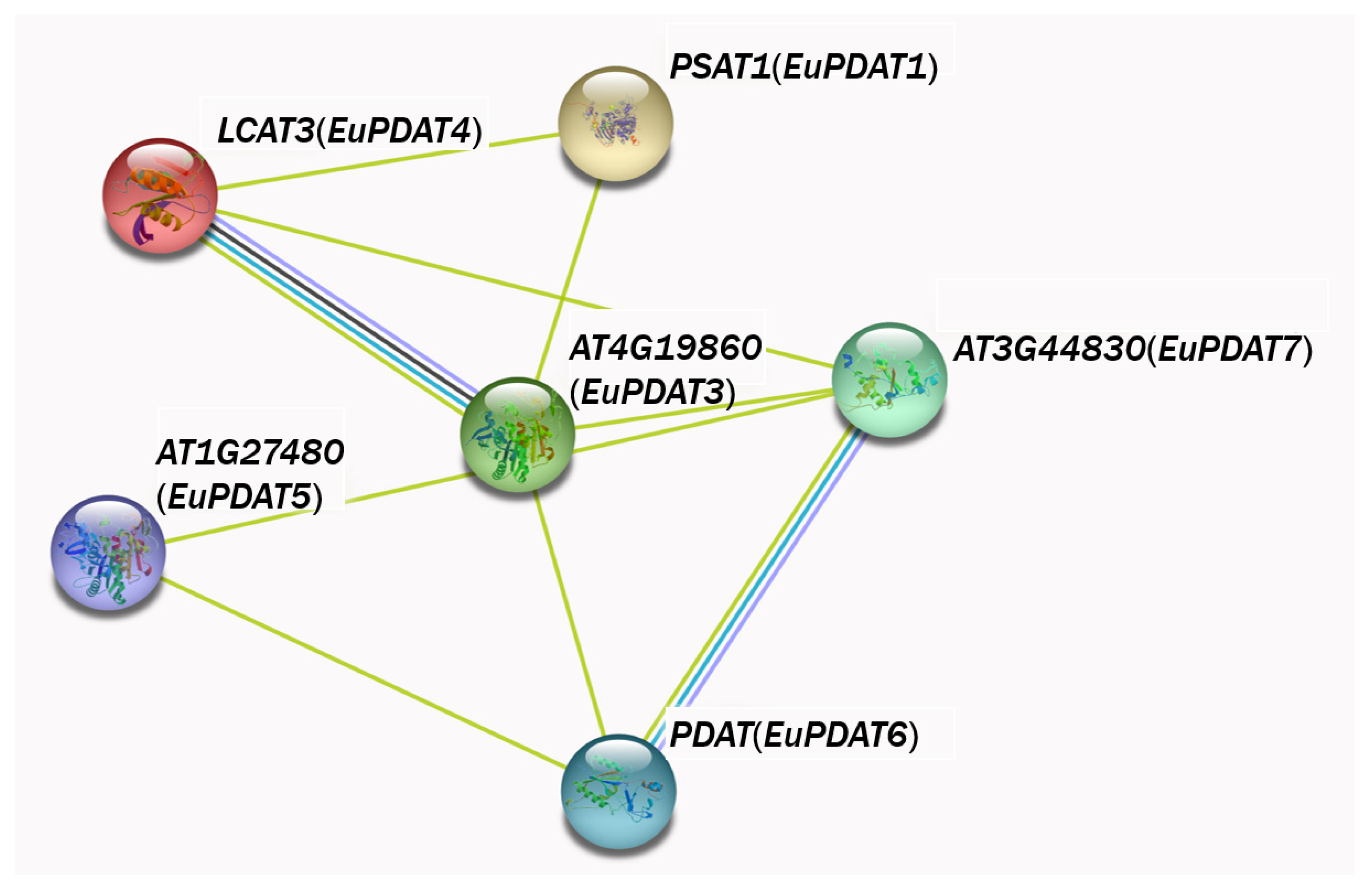
3.5. PDAT Protein Interaction Network Analysis of E. ulmoides
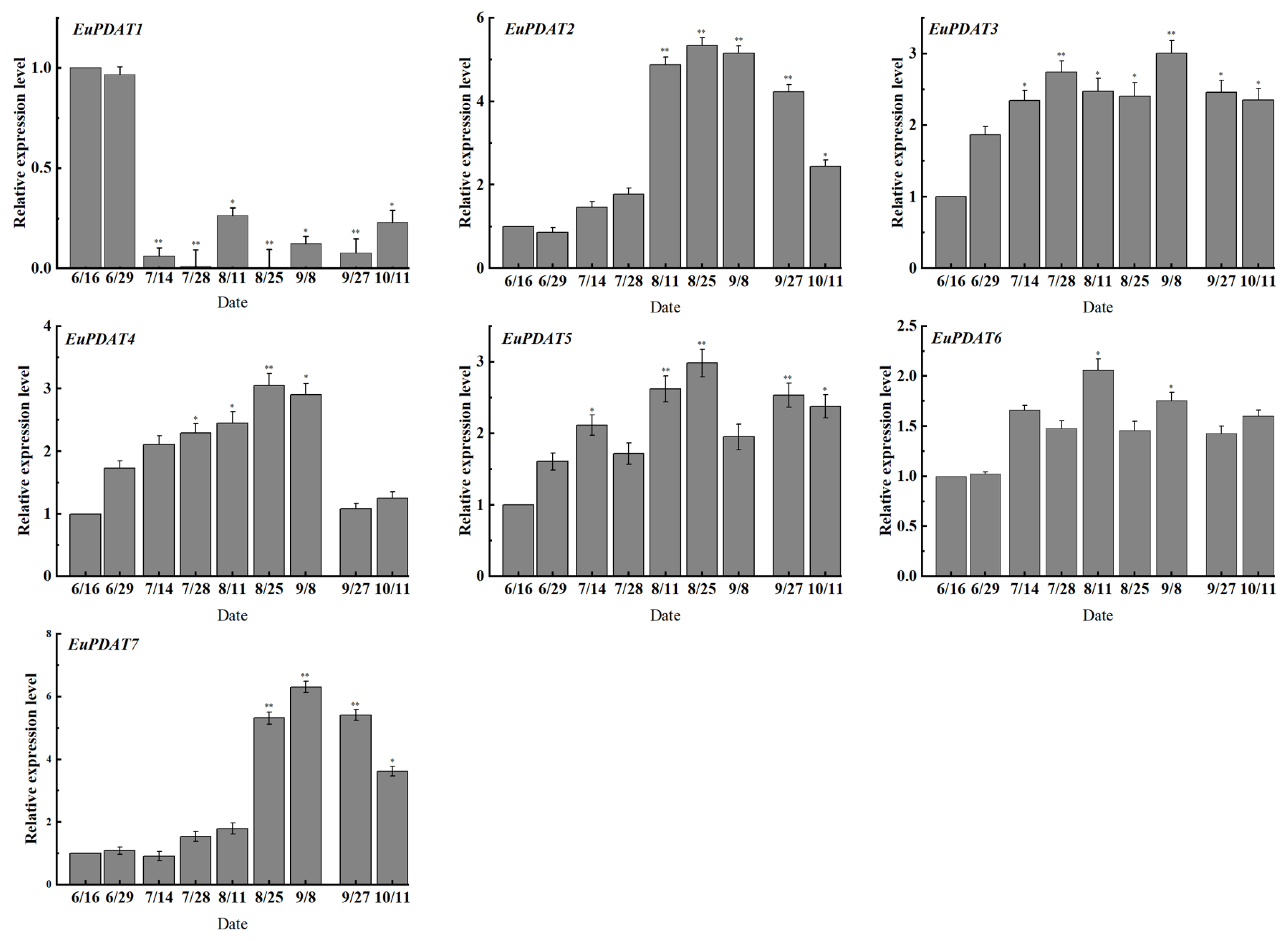
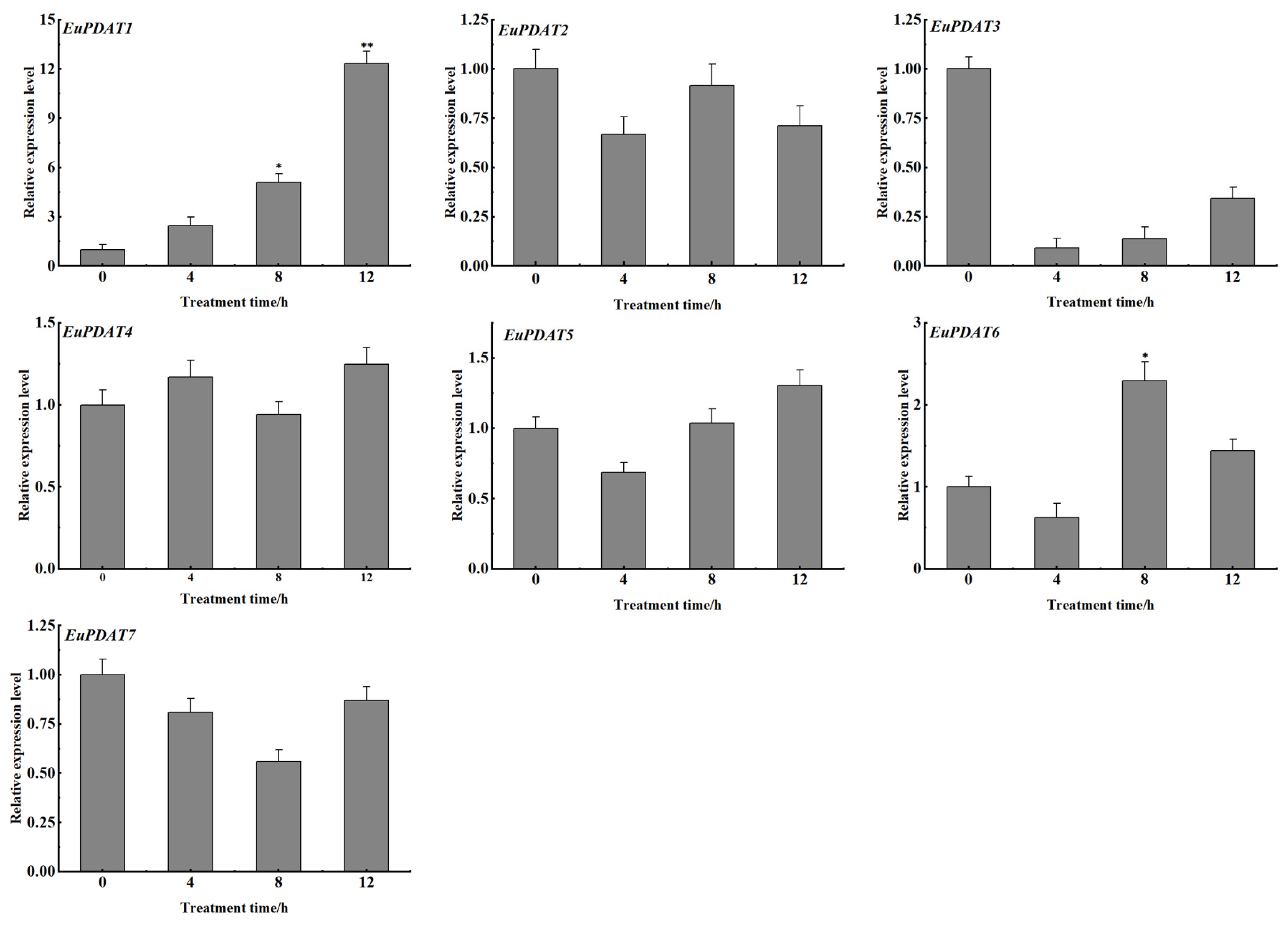
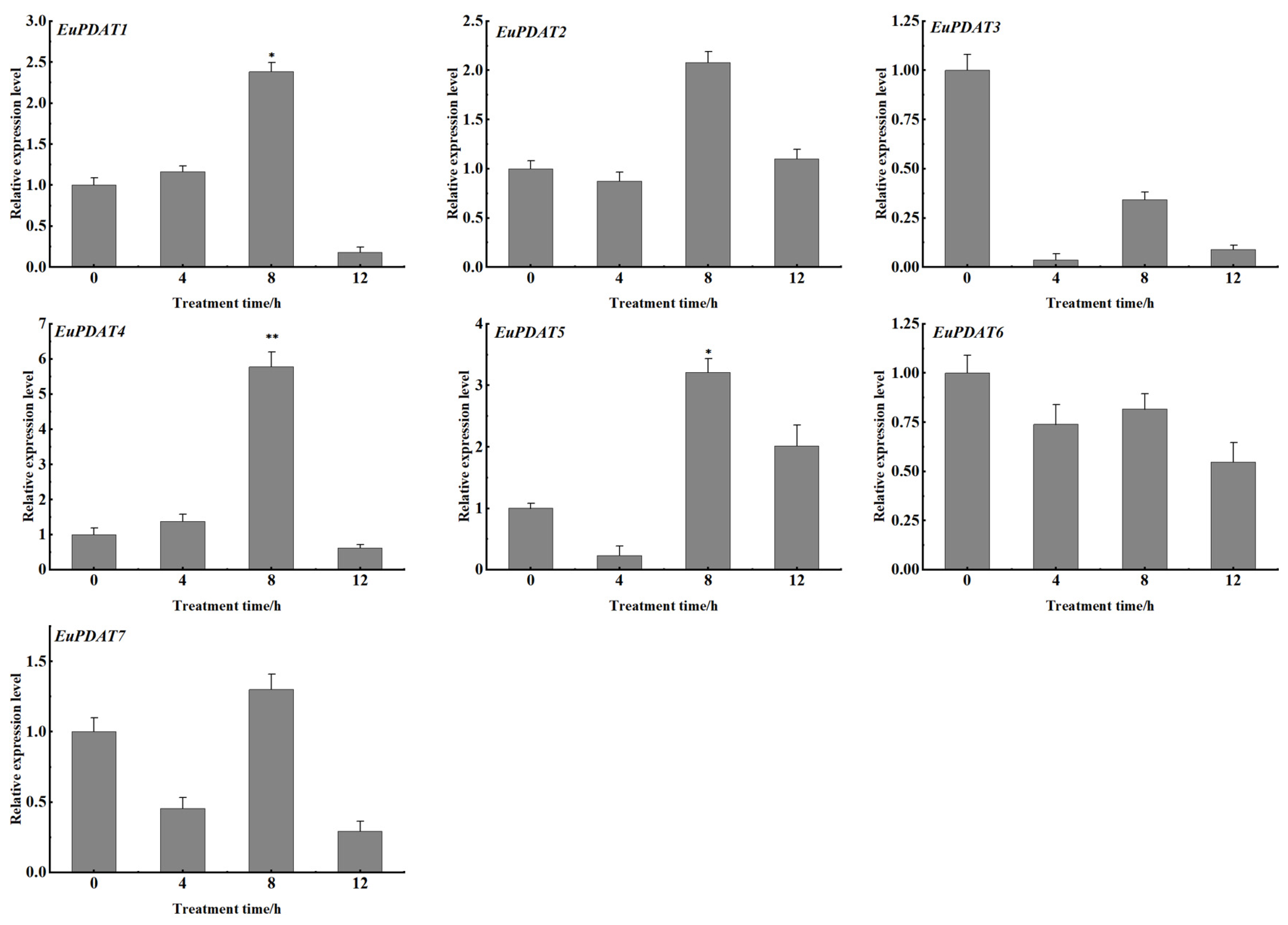
3.6. Expression Profiles of EuPDAT Genes at Different Stages and Treatments
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wang, X.Q.; Liang, X.Q.; Zhao, J.; Huang, B.B. Cultivar Characterization of Tea Seed Oils by Their Active Components and Antioxidant Capacity. J. Am. Oil Chem. Soc. 2014, 91, 629–639. [Google Scholar] [CrossRef]
- Frandsen, G.I.; Mundy, J.; Tzen, J. Oil bodies and their associated proteins, oleosin and caleosin. Physiol. Plant. 2010, 112, 301–307. [Google Scholar] [CrossRef]
- Song, Y.; Wang, X.D.; Rose, R.J. Oil body biogenesis and biotechnology in legume seeds. Plant Cell Rep. 2017, 36, 1519–1532. [Google Scholar] [CrossRef]
- Siloto, R.M.P.; Findlay, K.; Lopez-Villalobos, A.; Yeung, E.C.; Nykiforuk, C.L.; Moloney, M.M. The Accumulation of Oleosins Determines the Size of Seed Oilbodies in Arabidopsis. Plant Cell 2006, 18, 1961–1974. [Google Scholar] [CrossRef]
- Tzen, J.T.; Huang, A.H. Surface structure and properties of plant seed oil bodies. J. Cell Biol. 1992, 117, 327–335. [Google Scholar] [CrossRef]
- Dong, J.S.; Shi, D.Q.; Gao, J.Q.; Li, C.L.; Liu, J.; Yang, W.C. Correlation Between the Quantity and the Sum of Areas of Oil Bodies and Oil Content in Rapeseed (Brassica napus). Chin. Bull. Bot. 2009, 44, 79–85. [Google Scholar]
- Kennedy, E.P. Biosynthesis of complex lipids. Fed. Proc. 1961, 20, 934–940. [Google Scholar]
- Liu, X.; Ouyang, L.; Zhou, Z. Phospholipid: Diacylglycerol acyltransferase contributes to the conversion of membrane lipids into triacylglycerol in Myrmecia incisa during the nitrogen starvation stress. Sci. Rep. 2016, 6, 26610. [Google Scholar] [CrossRef]
- Fan, J.; Yan, C.; Xu, C. Phospholipid:diacylglycerol acyltransferase-mediated triacylglycerol biosynthesis is crucial for protection against fatty acid-induced cell death in growing tissues of Arabidopsis. Plant J. 2013, 76, 930–942. [Google Scholar] [CrossRef] [PubMed]
- Ståhl, U.; Carlsson, A.S.; Lenman, M.; Dahlqvist, A.; Huang, B.; Banaś, W.; Banaś, A.; Stymne, S. Cloning and Functional Characterization of a Phospholipid:Diacylglycerol Acyltransferase from Arabidopsis. Plant Physiol. 2004, 135, 1324–1335. [Google Scholar] [CrossRef]
- Zheng, L.; Shockey, J.; Guo, F.; Shi, L.; Li, X.; Shan, L.; Wan, S.; Peng, Z. Discovery of a new mechanism for regulation of plant triacylglycerol metabolism: The peanut diacylglycerol acyltransferase-1 gene family transcriptome is highly enriched in alternative splicing variants. J. Plant Physiol. 2017, 219, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Peng, F.Y.; Weselake, R.J. Genome-Wide Analysis of PHOSPHOLIPID:DIACYLGLYCEROL ACYLTRANSFERASE (PDAT) Genes in Plants Reveals the Eudicot-Wide PDAT Gene Expansion and Altered Selective Pressures Acting on the Core Eudicot PDAT Paralogs. Plant Physiol. 2015, 167, 887–904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fenyk, S.; Woodfield, H.K.; Romsdahl, T.B.; Wallington, E.J.; Bates, R.E.; Fell, D.A.; Chapman, K.D.; Fawcett, T.; Harwood, J.L. Overexpression of phospholipid: Diacylglycerol acyltransferase inBrassica napus results in changes in lipid metabolism and oil accumulation. Biochem. J. 2022, 479, 805–823. [Google Scholar] [CrossRef] [PubMed]
- Voelker, T.A.; Worrell, A.C.; Anderson, L.; Bleibaum, J.; Fan, C.; Hawkins, D.J.; Radke, S.E.; Davies, H.M. Fatty acid biosynthesis redirected to medium chains in transgenic oilseed plants. Science 1992, 257, 72–74. [Google Scholar] [CrossRef]
- Simpson, J.P.; Ohlrogge, J.B. A Novel Pathway for Triacylglycerol Biosynthesis Is Responsible for the Accumulation of Massive Quantities of Glycerolipids in the Surface Wax of Bayberry (Myrica pensylvanica) Fruit. Plant Cell 2016, 28, 248–264. [Google Scholar] [CrossRef]
- Banaś, W.; Sanchez Garcia, A.; Banaś, A.; Stymne, S. Activities of acyl-CoA:diacylglycerol acyltransferase (DGAT) and phospholipid:diacylglycerol acyltransferase (PDAT) in microsomal preparations of developing sunflower and safflower seeds. Planta 2013, 237, 1627–1636. [Google Scholar] [CrossRef]
- Kroon, J.T.M.; Wei, W.; Simon, W.J.; Slabas, A.R. Identification and functional expression of a type 2 acyl-CoA:diacylglycerol acyltransferase (DGAT2) in developing castor bean seeds which has high homology to the major triglyceride biosynthetic enzyme of fungi and animals. Phytochemistry 2006, 67, 2541–2549. [Google Scholar] [CrossRef]
- Kim, H.U.; Lee, K.; Go, Y.S.; Jung, J.H.; Suh, M.; Kim, J.B. Endoplasmic Reticulum-Located PDAT1-2 from Castor Bean Enhances Hydroxy Fatty Acid Accumulation in Transgenic Plants. Plant Cell Physiol. 2011, 52, 983–993. [Google Scholar] [CrossRef]
- Chapman, K.D.; Ohlrogge, J.B. Compartmentation of Triacylglycerol Accumulation in Plants. J. Biol. Chem. 2012, 287, 2288–2294. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Siloto, R.M.; Wickramarathna, A.D.; Mietkiewska, E.; Weselake, R.J. Identification of a pair of phospholipid:diacylglycerol acyltransferases from developing flax (Linum usitatissimum L.) seed catalyzing the selective production of trilinolenin. J. Biol. Chem. 2013, 288, 24173–24188. [Google Scholar] [CrossRef]
- Furmanek, T.; Demski, K.; Banaś, W.; Haslam, R.; Napier, J. The Utilization of the Acyl-CoA and the Involvement PDAT and DGAT in the Biosynthesis of Erucic Acid-Rich Triacylglycerols in Crambe Seed Oil. Lipids 2014, 49, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Caldo, K.; Nath, D.P.; Ozga, J.; Lemieux, M.J.; Weselake, R.J.; Chen, G. Properties and Biotechnological Applications of Acyl-CoA:diacylglycerol Acyltransferase and Phospholipid:diacylglycerol Acyltransferase from Terrestrial Plants and Microalgae. Lipids 2018, 53, 663–688. [Google Scholar]
- Du, H.Y.; Du, Q.X. Foundation, problems and countermeasures of Eucommia ulmoides industry high-quality development in China. Nonwood For. Res. 2020, 38, 1–10. [Google Scholar]
- Tu, T.H.; Kim, H.; Yang, S.; Kim, J.K.; Kim, J.G. Linoleic acid rescues microglia inflammation triggered by saturated fatty acid. Biochem. Biophys. Res. Commun. 2019, 513, 201–206. [Google Scholar] [CrossRef]
- Inaba, Y.; Nakahigashi, K.; Ito, T.; Tomita, M. Alteration of fatty acid chain length of Chlamydomonas reinhardtii by simultaneous expression of medium-chain-specific thioesterase and acyl carrier protein. Phycol. Res. 2017, 65, 94–99. [Google Scholar] [CrossRef]
- Asefy, Z.; Tanomand, A.; Hoseinnejhad, S.; Ceferov, Z.; Oshaghi, E.A.; Rashidi, M. Unsaturated fatty acids as a co-therapeutic agents in cancer treatment. Mol. Biol. Rep. 2021, 48, 2909–2916. [Google Scholar] [CrossRef]
- Feng, Y.; Wang, L.; Fu, J.; Wuyun, T.; Du, H.; Tan, X.; Zou, F.; Li, F. Transcriptome sequencing discovers genes related to fatty acid biosynthesis in the seeds of Eucommia ulmoides. Genes Genom. 2016, 38, 275–283. [Google Scholar] [CrossRef]
- Kaseke, T.; Opara, U.L.; Fawole, O.A. Fatty acid composition, bioactive phytochemicals, antioxidant properties and oxidative stability of edible fruit seed oil: Effect of preharvest and processing factors. Heliyon 2020, 6, e4962. [Google Scholar] [CrossRef]
- Were, B.A.; Onkware, A.O.; Gudu, S.; Welander, M.; Carlsson, A.S. Seed oil content and fatty acid composition in East African sesame (Sesamum indicum L.) accessions evaluated over 3 years. Field Crops Res. 2006, 97, 254–260. [Google Scholar] [CrossRef]
- Heil, C.S.; Wehrheim, S.S.; Paithankar, K.S.; Grininger, M. Fatty Acid Biosynthesis: Chain-Length Regulation and Control. Chembiochem 2019, 20, 2298–2321. [Google Scholar] [CrossRef]
- Qing, J.; Du, Q.; Meng, Y.; Liu, P.; Du, H.; Wang, L. Genome-wide identification and expression pattern analysis of the ribonuclease T2 family in Eucommia ulmoides. Sci. Rep. 2021, 11, 6900. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.P.; Xiao, P.; Du, S.G.; Jiang, X.M.; Fu, Y.X.; Guo, X.L. Oil Bodies Observation and the Correlation Between the Oil Bodies and Oil Content in Camellia oleifera Seeds. J. Chin. Cereals Oils Assoc. 2014, 29, 82–85. [Google Scholar]
- Chen, H.; Pan, C.D.; Wang, B.; Xiao, Z.Z.; Hu, Y.; Hu, G.J. Oil Body Observation in Seed Development and Its Analysis in Seed of Juglans regia ‘Wen185’ andJ. regia ‘Xinxin2’ in Period of Seed Maturity. Sci. Agric. Sin. 2015, 48, 3899–3909. [Google Scholar]
- Zhang, Q.; Chen, T.; Wang, X.; Wang, J.; Gu, K.; Yu, J.; Hu, D.; Hao, Y. Genome-wide identification and expression analyses of homeodomain-leucine zipper family genes reveal their involvement in stress response in apple (Malus × domestica). Hortic. Plant J. 2022, 8, 261–278. [Google Scholar] [CrossRef]
- Zhang, T.; Li, X.; Zhao, Q.; Shi, Y.; Hao, Y.; You, C. Genome-wide identification and functional characterization of the MdCLE peptide family in apple (Malus × domestica). Hortic. Plant J. 2022, 8, 279–288. [Google Scholar] [CrossRef]
- Xie, T.; Chen, C.; Li, C.; Liu, J.; Liu, C.; He, Y. Genome-wide investigation of WRKY gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genom. 2018, 19, 490. [Google Scholar] [CrossRef] [Green Version]
- Lin, T.K.; Zhong, L.; Santiago, J.L. Anti-Inflammatory and Skin Barrier Repair Effects of Topical Application of Some Plant Oils. Int. J. Mol. Sci. 2018, 19, 70. [Google Scholar] [CrossRef]
- Marmon, S.; Sturtevant, D.; Herrfurth, C.; Chapman, K.; Stymne, S.; Feussner, I. Two Acyltransferases Contribute Differently to Linolenic Acid Levels in Seed Oil. Plant Physiol. 2017, 173, 2081–2095. [Google Scholar] [CrossRef]
- Tzen, J.T.C. Integral Proteins in Plant Oil Bodies. ISRN Bot. 2012, 2012, 173954. [Google Scholar] [CrossRef]
- Kodad, O.; Rafel Socias I Company. Variability of Oil Content and of Major Fatty Acid Composition in Almond (Prunus amygdalus Batsch) and Its Relationship with Kernel Quality. J. Agric. Food Chem. 2008, 56, 4096–4101. [Google Scholar] [CrossRef]
- Hu, Z.; Wang, X.; Zhan, G.; Liu, G.; Hua, W.; Wang, H. Unusually large oilbodies are highly correlated with lower oil content in Brassica napus. Plant Cell Rep. 2009, 28, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Heneen, W.K.; Karlsson, G.; Brismar, K.; Gummeson, P.; Marttila, S.; Leonova, S.; Carlsson, A.S.; Bafor, M.; Banas, A.; Mattsson, B.; et al. Fusion of oil bodies in endosperm of oat grains. Planta 2008, 228, 589–599. [Google Scholar] [CrossRef] [PubMed]
- Ghosal, A.; Banas, A.; St Hl, U.; Dahlqvist, A.; Lindqvist, Y.; Stymne, S. Saccharomyces cerevisiae phospholipid:diacylglycerol acyl transferase (PDAT) devoid of its membrane anchor region is a soluble and active enzyme retaining its substrate specificities. BBA Mol. Cell Biol. Lipids 2007, 1771, 1457–1463. [Google Scholar] [CrossRef]
- Dahlqvist, A.; Stahl, U.; Lenman, M.; Banas, A.; Lee, M.; Sandager, L.; Ronne, H.; Stymne, S. Phospholipid:diacylglycerol acyltransferase: An enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. USA 2000, 97, 6487–6492. [Google Scholar] [CrossRef]
- Yoon, K.; Han, D.; Li, Y.; Sommerfeld, M.; Hu, Q. Phospholipid:Diacylglycerol Acyltransferase Is a Multifunctional Enzyme Involved in Membrane Lipid Turnover and Degradation While Synthesizing Triacylglycerol in the Unicellular Green MicroalgaChlamydomonas reinhardtii. Plant Cell 2012, 24, 3708–3724. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Carlsson, A.S.; Francis, T.; Zhang, M.; Hoffman, T.; Giblin, M.E.; Taylor, D.C. Triacylglycerol synthesis by PDAT1 in the absence of DGAT1 activity is dependent on re-acylation of LPC by LPCAT2. BMC Plant Biol. 2012, 12, 4. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Zhang, J.; Gai, J.; Chen, S. Cloning and comparative analysis of the gene encoding diacylglycerol acyltransferase from wild type and cultivated soybean. Theor. Appl. Genet. 2006, 112, 1086–1097. [Google Scholar] [CrossRef]
- Mueller, S.P.; Unger, M.; Guender, L.; Fekete, A.; Mueller, M.J. Phospholipid:Diacylglycerol Acyltransferase-Mediated Triacylglyerol Synthesis Augments Basal Thermotolerance. Plant Physiol. 2017, 175, 486–497. [Google Scholar] [CrossRef]
- Liu, P.F.; Wang, L.; Du, Q.X.; Du, L.Y. Estimation of potential suitable distribution area and the ecological characteristics of Eucommia ulmoides Oliv. in China. Acta Ecol. Sin. 2020, 40, 5674–5684. [Google Scholar]
- Li, R. Urgent Need to Improve Professional Cooperation in Eucommia Seed Oil Industry. China Forestry Industry. 2016, 8, 63–64. [Google Scholar]
- Alrashidi, M.; Derawi, D.; Salimon, J.; Firdaus Yusoff, M. An investigation of physicochemical properties of Nigella sativa L. Seed oil from Al-Qassim by different extraction methods. J. King Saud Univ. Sci. 2020, 32, 3337–3342. [Google Scholar] [CrossRef]




| Name | Primer Sequence |
|---|---|
| EuPDAT1-F | ATGGGAATCAAAGTGGCAGTC |
| EuPDAT1-R | TCACCGTGAAACGTTAATCTGAG |
| EuPDAT2-F | ATGGCTTCTTCCTCTAAAAATTCTG |
| EuPDAT2-R | TTACACATAATGCAAGAGGAGATCA |
| EuPDAT3-F | ATGGCTTCTTCAGTTCTTCGGTT |
| EuPDAT3-R | TCACAACTGAATATTCAATCTCTCCG |
| EuPDAT4-F | ATGTCGATGTTGAGGCGGAG |
| EuPDAT4-R | TCAAGCTTTCCCCATACTATTTACA |
| EuPDAT5-F | ATGCTCGGGGGATGCTGTT |
| EuPDAT5-R | TTAGAGAACTCGGGAGCTTTCCTC |
| EuPDAT6-F | ATGGCGCTGATTCGAAGAAG |
| EuPDAT6-R | CTAAAAGTCTCTTCCTTTGTACTGCA |
| EuPDAT7-F | ATGGACCTTGATCTATGGATGTTG |
| EuPDAT7-R | TCATATGATAGATGGTCCCTTACCC |
| Name | Primer Sequence |
|---|---|
| EuPDAT1-F | AGACCTCCGACGGTTTTTCC |
| EuPDAT1-R | GGTTCCTTTGGAGGACCGAG |
| EuPDAT2-F | GTGAGTTGCCTTGCGGATTC |
| EuPDAT2-R | CAACCTGCAGCCGTAGTACA |
| EuPDAT3-F | GAGAACACCAAAGCGGAGGA |
| EuPDAT3-R | GCGCCACGAATTTCATCTCC |
| EuPDAT4-F | GGACTCCGCAAAGAAAGGGA |
| EuPDAT4-R | GTCTCGGTCAGCTCGGTAAG |
| EuPDAT5-F | TTCCCGCATATCTGGAACCG |
| EuPDAT5-R | TAACCAGGCCTCCACCTTCT |
| EuPDAT6-F | CGCTATGGATCCCACACCTC |
| EuPDAT6-R | CCGAGAGCGTCGATGAGAAA |
| EuPDAT7-F | AAAACGACTCCGATGCGACT |
| EuPDAT7-R | TCAGTGGCAGTGGACCAATC |
| Actin-F | TTGTTAGCAACTGGGATGATATGG |
| Actin-R | CAGGGTGTTCTTCAGGAGCAA |
| Gene Name | Number of Amino Acids | Molecular Weight (kDa) | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Subcellular Location |
|---|---|---|---|---|---|---|---|
| EuPDAT 1 | 75 | 37.8 | 5.12 | 41.07 | 91.64 | 0.634 | Plasma membrane |
| EuPDAT 2 | 392 | 95.4 | 8.29 | 45.40 | 80.55 | 0.721 | Endoplasmic reticulum |
| EuPDAT 3 | 313 | 76.5 | 5.95 | 43.05 | 81.01 | 0.737 | Plasma membrane |
| EuPDAT 4 | 96 | 86.8 | 7.56 | 37.02 | 77.94 | 0.846 | Endoplasmic reticulum |
| EuPDAT 5 | 182 | 65.7 | 5.91 | 44.77 | 85.98 | 0.959 | Plasma membrane |
| EuPDAT 6 | 138 | 81.4 | 5.59 | 49.23 | 70.47 | 0.781 | Nucleus |
| EuPDAT 7 | 178 | 78.6 | 7.09 | 31.28 | 97.07 | 0.813 | Endoplasmic reticulum |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, J.; Qing, J.; Liu, C.; Wang, Q.; Du, H.; Liu, P.; Du, L.; Wang, L.; Du, Q. Characteristics of Oil Body Development and the Cloning and Expression Analysis of PDAT Genes in Eucommia ulmoides. Agronomy 2022, 12, 2197. https://doi.org/10.3390/agronomy12092197
Zhong J, Qing J, Liu C, Wang Q, Du H, Liu P, Du L, Wang L, Du Q. Characteristics of Oil Body Development and the Cloning and Expression Analysis of PDAT Genes in Eucommia ulmoides. Agronomy. 2022; 12(9):2197. https://doi.org/10.3390/agronomy12092197
Chicago/Turabian StyleZhong, Jian, Jun Qing, Chenlu Liu, Qi Wang, Hongyan Du, Panfeng Liu, Lanying Du, Lu Wang, and Qingxin Du. 2022. "Characteristics of Oil Body Development and the Cloning and Expression Analysis of PDAT Genes in Eucommia ulmoides" Agronomy 12, no. 9: 2197. https://doi.org/10.3390/agronomy12092197
APA StyleZhong, J., Qing, J., Liu, C., Wang, Q., Du, H., Liu, P., Du, L., Wang, L., & Du, Q. (2022). Characteristics of Oil Body Development and the Cloning and Expression Analysis of PDAT Genes in Eucommia ulmoides. Agronomy, 12(9), 2197. https://doi.org/10.3390/agronomy12092197






