Breeding of the Long-Grain Restorer of Indica-Japonica Hybrid Rice by Using the Genetic Effects of Grain Shape QTLs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Validation of Grain Size Alleles Variations
2.3. Development of the Grain Size Allele-Specific Marker
2.4. DNA Extraction and PCR
2.5. Examination of Grain Size-Related Traits
3. Results
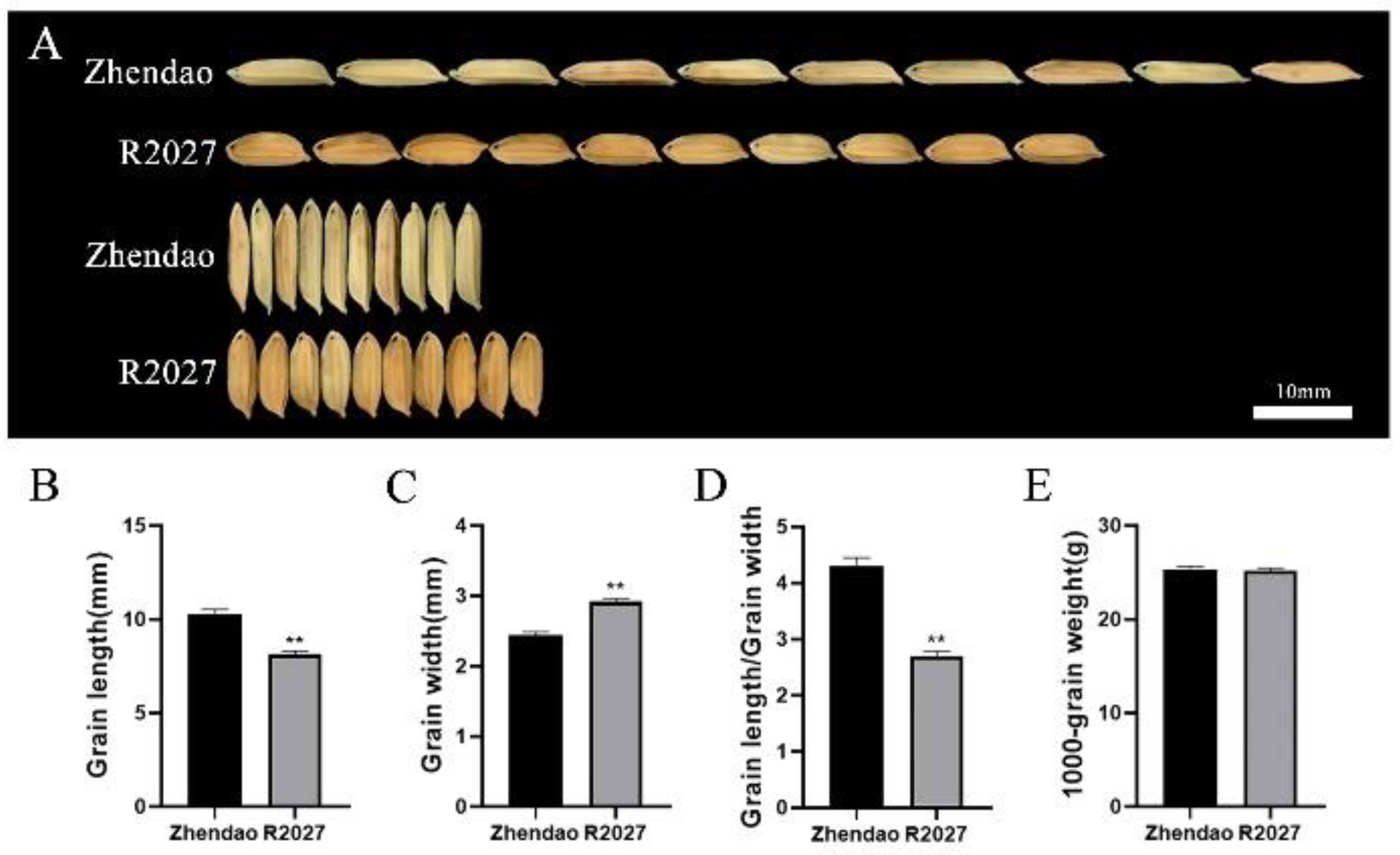
3.1. Phenotype of Grain Size in Zhendao and R2027
3.2. Study of the Candidate Functional Variants of Five Grain-Size Genes
3.3. Functional Marker Development of Five Size Genes
3.4. Detection of Restorer Genes in the Zhendao and R2027 Recombinant Inbred Lines
3.5. Detection of Major Restorer Genes in TF Families
3.6. Effects of the Aggregation of Different Grain-Shape Genes on the Grain Shape in TF Families
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Crawford, G.W.; Shen, C. The origins of rice agriculture: Recent progress in East Asia. Antiquity 1998, 72, 858. [Google Scholar] [CrossRef]
- Sweeney, M.; McCouch, S. The complex history of the domestication of rice. Ann. Bot. 2007, 100, 951–957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, Y.; Zhang, Q. Genetic and Molecular Bases of Rice Yield. Annu. Rev. Plant Biol. 2010, 61, 421–442. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.; Tian, Z.; Rao, Y.; Dong, G.; Yang, Y.; Huang, L.; Leng, Y.; Xu, J.; Sun, C.; Zhang, G.; et al. Rational design of high-yield and superior-quality rice. Nat. Plants 2017, 3, 17031. [Google Scholar] [CrossRef]
- Jiang, H.; Zhang, A.; Liu, X.; Chen, J. Grain Size Associated Genes and the Molecular Regulatory Mechanism in Rice. Int. J. Mol. Sci. 2022, 23, 3169. [Google Scholar] [CrossRef]
- Lin, H.-X.; Song, X.-J.; Huang, W.; Shi, M.; Zhu, M.-Z. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 2007, 39, 623–630. [Google Scholar] [CrossRef]
- Hu, J.; Wang, Y.; Fang, Y.; Zeng, L.; Xu, J.; Yu, H.; Shi, Z.; Pan, J.; Zhang, D.; Kang, S.; et al. A Rare Allele of GS2 Enhances Grain Size and Grain Yield in Rice. Mol. Plant 2015, 8, 1455–1465. [Google Scholar] [CrossRef] [Green Version]
- Fan, C.; Xing, Y.; Mao, H.; Lu, T.; Han, B.; Xu, C.; Li, X.; Zhang, Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006, 112, 1164–1171. [Google Scholar] [CrossRef]
- Li, Y.; Fan, C.; Xing, Y.; Jiang, Y.; Luo, L.; Sun, L.; Shao, D.; Xu, C.; Li, X.; Xiao, J.; et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 2011, 43, 1266–1269. [Google Scholar] [CrossRef]
- Duan, P.; Xu, J.; Zeng, D.; Zhang, B.; Geng, M.; Zhang, G.; Huang, K.; Huang, L.; Xu, R.; Ge, S.; et al. Natural Variation in the Promoter of GSE5 Contributes to Grain Size Diversity in Rice. Mol. Plant 2017, 10, 685–694. [Google Scholar] [CrossRef]
- Tong, H.; Jin, Y.; Liu, W.; Li, F.; Fang, J.; Yin, Y.; Qian, Q.; Zhu, L.; Chu, C. DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 2009, 58, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Song, X.J.; Kuroha, T.; Ayano, M.; Furuta, T.; Nagai, K.; Komeda, N.; Segami, S.; Miura, K.; Ogawa, D.; Kamura, T.; et al. Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proc. Natl. Acad. Sci. USA 2015, 112, 76–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Xiong, G.; Hu, J.; Jiang, L.; Yu, H.; Xu, J.; Fang, Y.; Zeng, L.; Xu, E.; Xu, J.; et al. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 2015, 47, 944–948. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, K.; Yuan, Q.; Liu, X.; Liu, Z.; Lin, X.; Zeng, R.; Zhu, H.; Dong, G.; Qian, Q.; et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 2012, 44, 950–954. [Google Scholar] [CrossRef]
- Wang, S.; Li, S.; Liu, Q.; Wu, K.; Zhang, J.; Wang, S.; Wang, Y.; Chen, X.; Zhang, Y.; Gao, C.; et al. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 2015, 47, 949–954. [Google Scholar] [CrossRef]
- Liu, J.; Chen, J.; Zheng, X.; Wu, F.; Lin, Q.; Heng, Y.; Tian, P.; Cheng, Z.; Yu, X.; Zhou, K.; et al. GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants 2017, 3, 17043. [Google Scholar] [CrossRef]
- Yan, S.; Zou, G.; Li, S.; Wang, H.; Liu, H.; Zhai, G.; Guo, P.; Song, H.; Yan, C.; Tao, Y. Seed size is determined by the combinations of the genes controlling different seed characteristics in rice. Theor. Appl. Genet. 2011, 123, 1173–1181. [Google Scholar] [CrossRef]
- Shomura, A.; Izawa, T.; Ebana, K.; Ebitani, T.; Kanegae, H.; Konishi, S.; Yano, M. Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 2008, 40, 1023–1028. [Google Scholar] [CrossRef]
- Sun, L.; Li, X.; Fu, Y.; Zhu, Z.; Tan, L.; Liu, F.; Sun, X.; Sun, X.; Sun, C. GS6, A Member of the GRAS Gene Family, Negatively Regulates Grain Size in Rice. J. Integr. Plant Biol. 2013, 55, 938–949. [Google Scholar] [CrossRef]
- Ishimaru, K.; Hirotsu, N.; Madoka, Y.; Murakami, N.; Hara, N.; Onodera, H.; Kashiwagi, T.; Ujiie, K.; Shimizu, B.; Onishi, A.; et al. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 2013, 45, 707–711. [Google Scholar] [CrossRef]
- Liu, Q.; Han, R.; Wu, K.; Zhang, J.; Ye, Y.; Wang, S.; Chen, J.; Pan, Y.; Li, Q.; Xu, X.; et al. G-protein betagamma subunits determine grain size through interaction with MADS-domain transcription factors in rice. Nat. Commun. 2018, 9, 852. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Chen, S.; Yu, S. Functional markers developed from multiple loci in GS3 for fine marker-assisted selection of grain length in rice. Theor. Appl. Genet. 2011, 122, 905–913. [Google Scholar] [CrossRef]
- Xia, D.; Zhou, H.; Liu, R.; Dan, W.; Li, P.; Wu, B.; Chen, J.; Wang, L.; Gao, G.; Zhang, Q.; et al. GL3.3, a Novel QTL Encoding a GSK3/SHAGGY-like Kinase, Epistatically Interacts with GS3 to Produce Extra-long Grains in Rice. Mol. Plant 2018, 11, 754–756. [Google Scholar] [CrossRef] [Green Version]
- Takano-Kai, N.; Jiang, H.; Powell, A.; McCouch, S.; Takamure, I.; Furuya, N.; Doi, K.; Yoshimura, A. Multiple and independent origins of short seeded alleles of GS3 in rice. Breed Sci. 2013, 63, 77–85. [Google Scholar] [CrossRef] [Green Version]
- Mao, H.; Sun, S.; Yao, J.; Wang, C.; Yu, S.; Xu, C.; Li, X.; Zhang, Q. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 19579–19584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Ma, B.; Bian, Z.; Li, X.; Zhang, C.; Liu, J.; Li, Q.; Liu, Q.; He, Z. Grain Size Selection Using Novel Functional Markers Targeting 14 Genes in Rice. Rice 2020, 13, 63. [Google Scholar] [CrossRef]
- Xu, Y.H.; Zhu, Y.Y.; Zhou, H.C.; Li, Q.; Sun, Z.X.; Liu, Y.G.; Lin, H.X.; He, Z.H. Identification of a 98-kb DNA segment containing the rice Eui gene controlling uppermost internode elongation, and construction of a TAC transgene sublibrary. Mol. Genet. Genom. 2004, 272, 149–155. [Google Scholar] [CrossRef]
- Akagi, H.; Nakamura, A.; Yokozeki-Misono, Y.; Inagaki, A.; Takahashi, H.; Mori, K.; Fujimura, T. Positional cloning of the rice Rf-1 gene, a restorer of BT-type cytoplasmic male sterility that encodes a mitochondria-targeting PPR protein. Theor. Appl. Genet. 2004, 108, 1449–1457. [Google Scholar] [CrossRef]
- Wang, Z.; Zou, Y.; Li, X.; Zhang, Q.; Chen, L.; Wu, H.; Su, D.; Chen, Y.; Guo, J.; Luo, D.; et al. Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell 2006, 18, 676–687. [Google Scholar] [CrossRef] [Green Version]
- Tang, H.; Luo, D.; Zhou, D.; Zhang, Q.; Tian, D.; Zheng, X.; Chen, L.; Liu, Y.G. The rice restorer Rf4 for wild-abortive cytoplasmic male sterility encodes a mitochondrial-localized PPR protein that functions in reduction of WA352 transcripts. Mol. Plant 2014, 7, 1497–1500. [Google Scholar] [CrossRef]
- Kazama, T.; Toriyama, K. A fertility restorer gene, Rf4, widely used for hybrid rice breeding encodes a pentatricopeptide repeat protein. Rice 2014, 7, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; He, Z.; Appels, R.; Xia, X. Functional markers in wheat: Current status and future prospects. Theor. Appl. Genet. 2012, 125, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Salgotra, R.K.; Stewart, C.N., Jr. Functional Markers for Precision Plant Breeding. Int. J. Mol. Sci. 2020, 21, 4792. [Google Scholar] [CrossRef] [PubMed]
- Nan, J.; Feng, X.; Wang, C.; Zhang, X.; Wang, R.; Liu, J.; Yuan, Q.; Jiang, G.; Lin, S. Improving rice grain length through updating the GS3 locus of an elite variety Kongyu 131. Rice 2018, 11, 21–29. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Li, J.; Qu, J.; Yan, S. Development of Genome-Wide Insertion and Deletion Polymorphism Markers from Next-Generation Sequencing Data in Rice. Rice 2015, 8, 63. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Chen, L.; Liu, Y.; Dai, H.; He, J.; Kang, H.; Pan, G.; Huang, J.; Qiu, Z.; Wang, Q.; et al. Marker assisted pyramiding of two brown planthopper resistance genes, Bph3 and Bph27 (t), into elite rice Cultivars. Rice 2016, 9, 27. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Ye, S.; Mou, T. Molecular Breeding of Rice Restorer Lines and Hybrids for Brown Planthopper (BPH) Resistance Using the Bph14 and Bph15 Genes. Rice 2016, 9, 53. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Liu, K.; Zha, W.; Zhou, L.; Li, M.; Xu, H.; Li, P.; Chen, Z.; Yang, G.; Chen, P.; et al. Identification and verification of grain shape QTLs by SNP array in rice. PLoS ONE 2021, 16, e0260133. [Google Scholar] [CrossRef]
- Zhao, D.S.; Li, Q.F.; Zhang, C.Q.; Zhang, C.; Yang, Q.Q.; Pan, L.X.; Ren, X.Y.; Lu, J.; Gu, M.H.; Liu, Q.Q. GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat. Commun. 2018, 9, 1240. [Google Scholar] [CrossRef] [Green Version]
- Cerioli, T.; Hernandez, C.O.; Angira, B.; McCouch, S.R.; Robbins, K.R.; Famoso, A.N. Development and validation of an optimized marker set for genomic selection in southern U.S. rice breeding programs. Plant Genome 2022, 15, e20219. [Google Scholar] [CrossRef]
- Moonsap, P.; Laksanavilat, N.; Sinumporn, S.; Tasanasuwan, P.; Kate-Ngam, S.; Jantasuriyarat, C. Genetic diversity of Indo-China rice varieties using ISSR, SRAP and InDel markers. J. Genet. 2019, 98, 80. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Cui, X.; Li, R.; Huang, P.; Zong, J.; Yao, D.; Li, G.; Zhang, D.; Yuan, Z. Development of genome-wide insertion/deletion markers in rice based on graphic pipeline platform. J. Integr. Plant Biol. 2015, 57, 980–991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohata, R.; Koitabashi, K.; Kitashiba, H.; Nishio, T. Sensitive mutant detection by concentrating mutant DNA with allele-specific capture and its application to analysis of contaminated grains in rice. Plant Cell Rep. 2018, 37, 865–872. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Ma, Z.; Zhao, M.; Xiao, M.; Zhao, J.; Wang, C.; Gao, H.; Bai, Y.; Wang, H.; Sui, G. Research and Development Strategies for Hybrid japonica Rice. Rice 2020, 13, 36. [Google Scholar] [CrossRef] [PubMed]
- Melonek, J.; Duarte, J.; Martin, J.; Beuf, L.; Murigneux, A.; Varenne, P.; Comadran, J.; Specel, S.; Levadoux, S.; Bernath-Levin, K.; et al. The genetic basis of cytoplasmic male sterility and fertility restoration in wheat. Nat. Commun. 2021, 12, 1036. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, X.; Xu, Z.; Zhao, X.; Wan, Z.; Cheng, X.; Liu, Q.; Gu, M.; Tang, S. The Effects of Rf4 and the Genetic Mechanism Behind Fertility Restoration of Wild Abortive Cytoplasmic Male Sterility (WA-CMS) in Japonica Rice (Oryza sativa ssp. Japonica). Rice 2022, 15, 59. [Google Scholar] [CrossRef] [PubMed]




| Primer Names | Primer Sequence (5′-3′) | Enzyme Site | Product Size (bp) |
|---|---|---|---|
| GW6a_InDel-1F | GACTTATCAGCCGCACTG | 206/200 | |
| GW6a_InDel-1R GW6a_InDel-2F GW6a_InDel-2R | CTCTTGACCCACCTTGAATA ATGTTCGTTCTGGTCTTGA GCTGCCAATTCACATTACT | 216/191 | |
| GS6_InDel-F | GCGATGGAGATGGAGATG | 149/161 | |
| GS6_InDel-R | AGAGTGAGAGCAGAGACC | ||
| GW5_InDel-F | GGACTAATTACAGCGATAACC | 1627/415 | |
| GW5_InDel-F | GAACGGCAGAATGAGGAG | ||
| GL7_InDel-1F | CTCACGCACATCCAACTG | 106/117 | |
| GL7_InDel-1R | ATACCACATCTCATCTCAC | ||
| GL7_InDel-2F GL7_InDel-2R TGW6_SNP-F TGW6_SNP-R | GTGAGATGAGATGTGGTAT TGAAATAAGCGGGAGGGA CCGATAGCAGCATGAACTA GGTCAATGCAACGATCAGAT | Sac I | 134/116 167/137 |
| Cultivar | Grain Morphology | Genetype | |||||||
|---|---|---|---|---|---|---|---|---|---|
| GL | GW | GL/GW | TGW | GL7 | GW6a | GS6 | GW5 | TGW6 | |
| Zhendao R2027 | 10.20 ± 0.35 8.10 ± 0.22 | 2.42 ± 0.21 2.97 ± 0.16 | 4.39 ± 0.02 2.76 ± 0.01 | 25.79 ± 0.12 25.14 ± 0.34 | gl7 GL7 | gw6a GW6a | gs6 GS6 | GW5 gw5 | TGW6 tgw6 |
| TF-1 | 9.91 ± 0.30 | 2.40 ± 0.20 | 4.16 ± 0.02 | 22.34 ± 0.25 | gl7 | - | gs6 | - | - |
| TF-2 | 9.87 ± 0.35 | 2.37 ± 0.14 | 4.18 ± 0.02 | 22.90 ± 0.24 | gl7 | - | gs6 | - | - |
| TF-3 | 9.86 ± 0.40 | 2.43 ± 0.19 | 4.07 ± 0.02 | 23.30 ± 0.27 | gl7 | - | gs6 | - | - |
| TF-4 | 9.73 ± 0.38 | 2.38 ± 0.21 | 4.16 ± 0.02 | 23.00 ± 0.22 | gl7 | - | gs6 | - | - |
| TF-5 | 9.70 ± 0.38 | 2.38 ± 0.19 | 4.10 ± 0.02 | 22.17 ± 0.29 | gl7 | - | gs6 | - | - |
| TF-6 | 9.64 ± 0.25 | 2.42 ± 0.20 | 4.00 ± 0.02 | 22.47 ± 0.29 | gl7 | - | gs6 | - | - |
| TF-7 | 9.39 ± 0.39 | 2.37 ± 0.22 | 4.00 ± 0.02 | 19.01 ± 0.16 | gl7 | - | gs6 | - | - |
| TF-8 | 9.39 ± 0.30 | 2.30 ± 0.17 | 4.08 ± 0.01 | 19.78 ± 0.21 | gl7 | - | gs6 | - | - |
| TF-9 | 9.81 ± 0.39 | 2.49 ± 0.17 | 3.95 ± 0.02 | 23.00 ± 0.15 | - | gw6a | gs6 | - | - |
| TF-10 | 9.46 ± 0.39 | 2.40 ± 0.19 | 3.95 ± 0.02 | 22.32 ± 0.18 | - | gw6a | gs6 | - | - |
| TF-11 | 9.42 ± 0.32 | 2.43 ± 0.18 | 3.90 ± 0.02 | 22.56 ± 0.24 | - | gw6a | gs6 | - | - |
| TF-12 | 9.37 ± 0.36 | 2.40 ± 0.26 | 3.93 ± 0.01 | 21.89 ± 0.19 | - | gw6a | gs6 | - | - |
| TF-13 | 9.34 ± 0.39 | 2.39 ± 0.18 | 3.95 ± 0.02 | 21.45 ± 0.38 | - | gw6a | gs6 | - | - |
| TF-14 | 9.19 ± 0.30 | 2.32 ± 0.15 | 3.97 ± 0.02 | 19.23 ± 0.15 | - | gw6a | gs6 | - | - |
| TF-15 | 9.15 ±0.42 | 2.31 ± 0.15 | 3.98 ± 0.01 | 20.75 ± 0.22 | - | gw6a | gs6 | - | - |
| TF-16 | 9.11 ± 0.41 | 2.30 ± 0.16 | 3.98 ± 0.01 | 20.52 ± 0.30 | - | gw6a | gs6 | - | - |
| TF-17 | 9.01 ± 0.35 | 2.39 ± 0.15 | 3.79 ± 0.02 | 20.38 ± 0.22 | - | gw6a | gs6 | - | - |
| TF-18 | 8.68 ± 0.32 | 2.34 ± 0.24 | 3.73 ± 0.01 | 18.52 ± 0.14 | - | gw6a | gs6 | - | - |
| TF-19 | 8.78 ±0.31 | 2.51 ± 0.18 | 3.52 ± 0.01 | 25.18 ± 0.18 | - | - | gs6 | - | - |
| TF-20 | 8.96 ± 0.38 | 2.53 ± 0.19 | 3.53 ± 0.02 | 25.80 ± 0.31 | - | - | gs6 | - | - |
| TF-21 | 8.90 ± 0.32 | 2.57 ± 0.16 | 3.48 ± 0.02 | 24.81 ± 0.28 | - | - | gs6 | - | - |
| TF-22 | 8.85 ±0.16 | 2.56 ± 0.18 | 3.48 ± 0.02 | 25.88 ± 0.31 | - | - | gs6 | - | - |
| TF-23 | 8.96 ± 0.43 | 2.65 ± 0.19 | 3.38 ± 0.02 | 27.88 ± 0.35 | - | - | gs6 | - | - |
| TF-24 | 8.82 ± 0.51 | 2.64 ± 0.15 | 3.35 ± 0.01 | 25.70 ± 0.33 | - | - | gs6 | - | - |
| TF-25 | 8.77 ± 0.28 | 2.66 ± 0.17 | 3.30 ± 0.02 | 27.58 ± 0.22 | - | - | gs6 | - | - |
| TF-26 | 8.77 ± 0.26 | 2.56 ± 0.18 | 3.42 ± 0.01 | 27.27 ± 0.31 | - | - | gs6 | - | - |
| TF-27 | 8.68 ± 0.31 | 2.57 ± 0.18 | 3.37 ± 0.02 | 26.75 ± 0.29 | - | - | - | gw5 | - |
| TF-28 | 8.67 ± 0.21 | 2.60 ± 0.18 | 3.33 ± 0.02 | 25.14 ± 0.28 | - | - | - | gw5 | - |
| TF-29 | 8.64 ± 0.24 | 2.55 ± 0.19 | 3.40 ± 0.01 | 23.95 ± 0.20 | - | - | - | gw5 | - |
| TF-30 | 8.58 ± 0.40 | 2.58 ± 0.17 | 3.34 ± 0.03 | 23.87 ± 0.24 | - | - | - | gw5 | - |
| TF-31 | 8.58 ± 0.20 | 2.60 ± 0.18 | 3.31 ± 0.01 | 23.18 ± 0.18 | - | - | - | gw5 | tgw6 |
| TF-32 | 8.53 ± 0.28 | 2.58 ± 0.16 | 3.30 ± 0.01 | 24.75 ± 0.32 | - | - | - | gw5 | tgw6 |
| TF-33 | 8.49 ± 0.28 | 2.63 ± 0.19 | 3.24 ± 0.02 | 24.30 ± 0.29 | - | - | - | gw5 | tgw6 |
| TF-34 | 8.46 ± 0.31 | 2.59 ± 0.16 | 3.28 ± 0.00 | 23.91 ± 0.31 | - | - | - | - | tgw6 |
| TF-35 | 8.27 ± 0.32 | 2.61 ± 0.16 | 3.17 ± 0.01 | 23.11 ± 0.18 | - | - | - | - | tgw6 |
| TF-36 | 8.16 ± 0.21 | 2.62 ± 0.21 | 3.12 ± 0.01 | 23.72 ± 0.20 | - | - | - | - | tgw6 |
| Trait | Zhendao | R2027 | TF19 | TF20 | TF21 | TF22 | TF23 |
|---|---|---|---|---|---|---|---|
| Plant height (cm) | 132.11 ± 1.02 | 134.26 ± 1.32 | 125.08 ± 1.32 | 132.66 ± 0.68 | 130.70 ± 1.21 | 130.72 ± 0.69 | 128.72 ± 0.69 |
| Tiller number | 6.20 ± 1.50 | 5.80 ± 1.40 | 11.80 ± 1.80 | 6.00 ± 1.20 | 7.00 ± 1.60 | 6.40 ± 1.20 | 6.80 ± 1.30 |
| Yield per plant (g) | 25.23 ± 4.11 | 26.01 ± 3.77 | 27.75 ± 2.73 | 26.66 ± 3.80 | 27.66 ± 3.10 | 27.14 ± 2.55 | 26.44 ± 3.08 |
| 1000-grain weight (g) | 25.79 ± 0.12 | 25.14 ± 0.34 | 25.18 ± 0.18 | 25.80 ± 0.31 | 24.81 ± 0.28 | 25.88 ± 0.31 | 27.88 ± 0.35 |
| Seed-setting rate (%) | 79.82 ± 0.89 | 70.82 ± 0.68 | 83.40 ± 0.45 | 81.70 ± 0.69 | 79.20 ± 1.02 | 80.20 ± 0.79 | 81.10 ± 0.43 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, K.; Peng, Z.; Sun, Z.; Zhou, Z.; Li, Y.; Zhou, R.; He, D.; Huang, C.; Chen, D.; Cheng, S.; et al. Breeding of the Long-Grain Restorer of Indica-Japonica Hybrid Rice by Using the Genetic Effects of Grain Shape QTLs. Agronomy 2023, 13, 107. https://doi.org/10.3390/agronomy13010107
Liu K, Peng Z, Sun Z, Zhou Z, Li Y, Zhou R, He D, Huang C, Chen D, Cheng S, et al. Breeding of the Long-Grain Restorer of Indica-Japonica Hybrid Rice by Using the Genetic Effects of Grain Shape QTLs. Agronomy. 2023; 13(1):107. https://doi.org/10.3390/agronomy13010107
Chicago/Turabian StyleLiu, Keke, Zequn Peng, Zhihao Sun, Zhengping Zhou, Yanhui Li, Ran Zhou, Dengmei He, Chenbo Huang, Daibo Chen, Shihua Cheng, and et al. 2023. "Breeding of the Long-Grain Restorer of Indica-Japonica Hybrid Rice by Using the Genetic Effects of Grain Shape QTLs" Agronomy 13, no. 1: 107. https://doi.org/10.3390/agronomy13010107
APA StyleLiu, K., Peng, Z., Sun, Z., Zhou, Z., Li, Y., Zhou, R., He, D., Huang, C., Chen, D., Cheng, S., Cao, L., Zhan, X., & Sun, L. (2023). Breeding of the Long-Grain Restorer of Indica-Japonica Hybrid Rice by Using the Genetic Effects of Grain Shape QTLs. Agronomy, 13(1), 107. https://doi.org/10.3390/agronomy13010107






