Novel Aspects of Regulation of Nitrogen Responses in the Tea Plant (Camellia sinensis (L.))
Abstract
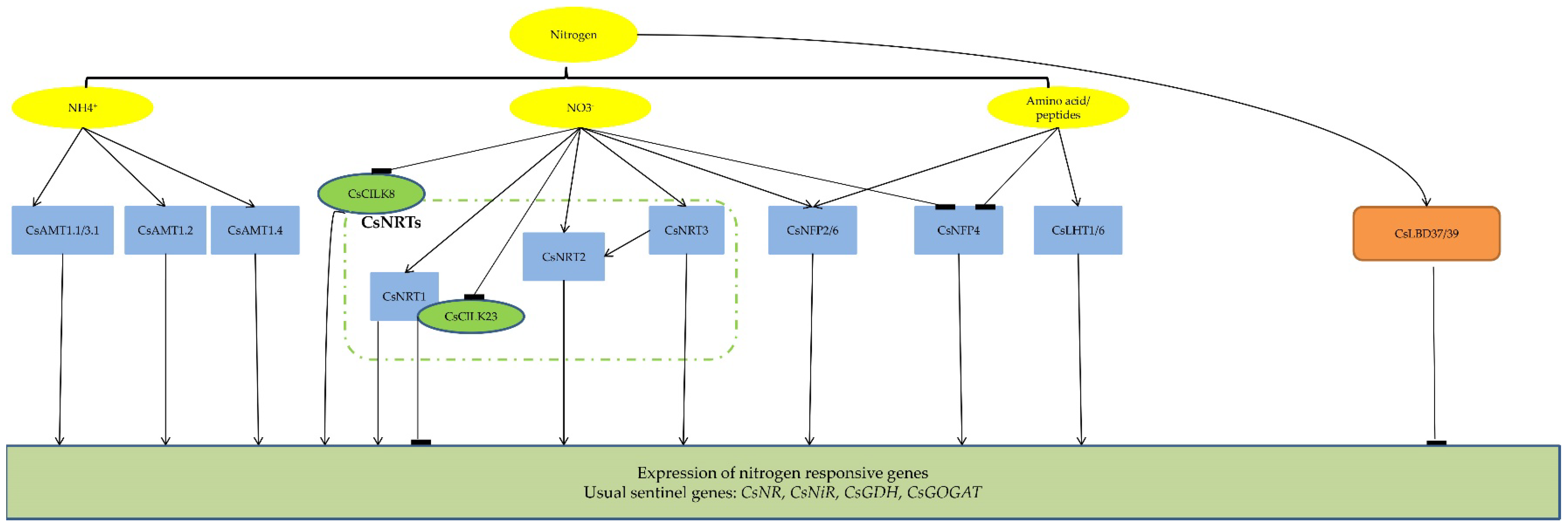
1. Introduction
2. Short-Term Nitrogen Effects: The Genes Involved in Nitrogen Response Process
2.1. Nitrogen Transporters
2.2. Transcription Factors
2.3. Genes Related to N Assimilation
2.4. Protein Kinases
2.5. Amino Acid Transporters CsLHT1 and CsLHT6
3. Long-Term Nitrogen Effects
3.1. Nitrogen and Root System Architecture
3.2. Nitrogen and Leaf Metabolism
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chang, H. Thea, A section of beverageial tea trees of the genus Camellia. Acta Sci. Nat. 1981, 1, 87–99. [Google Scholar]
- Ming, T.L. A revision of Camellia sect. Thea. Acta Bot. Yunnanica 1992, 14, 115–132. [Google Scholar]
- Hirel, B.; Le Gouis, J.; Ney, B.; Gallais, A. The challenge of improving nitrogen use efficiency in crop plants: Towards a more central role for genetic variability and quantitative genetics within integrated approaches. J. Exp. Bot. 2007, 58, 2369–2387. [Google Scholar] [CrossRef] [PubMed]
- Krapp, A.; David, L.C.; Chardin, C.; Girin, T.; Marmagne, A.; Leprince, A.S.; Chaillou, S.; Ferrario-Mery, S.; Meyer, C.; Daniel-Vedele, F. Nitrate transport and signalling in Arabidopsis. J. Exp. Bot. 2014, 65, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Vidal, E.A.; Moyano, T.C.; Canales, J.; Gutierrez, R.A. Nitrogen control of developmental phase transitions in Arabidopsis thaliana. J. Exp. Bot. 2014, 65, 5611–5618. [Google Scholar] [CrossRef]
- Fredes, I.; Moreno, S.; Diaz, F.P.; Gutierrez, R.A. Nitrate signaling and the control of Arabidopsis growth and development. Curr. Opin. Plant Biol. 2019, 47, 112–118. [Google Scholar] [CrossRef]
- Linda, G.; Sandra, J.; Torgny, N. Plant nitrogen status and cooccurrence of organic and inorganic nitrogen sources influence root uptake by scots pine seedlings. Tree Physiol. 2014, 34, 205–213. [Google Scholar]
- Xie, S.; Yang, F.; Feng, H.; Yu, Z.; Wei, X.; Liu, C.; Wei, C. Potential to reduce chemical fertilizer application in tea plantations at various spatial scales. Int. J. Environ. Res. Public. Health 2022, 19, 5243. [Google Scholar] [CrossRef]
- Bertrand, H.; Tétu, T.; Lea, P.J.; Dubois, F. Improving nitrogen use efficiency in crops for sustainable agriculture. Sustainability 2011, 3, 1452–1485. [Google Scholar]
- Cassman, K.G.; Dobermann, A.R.; Walters, D.T. Agroecosystems, nitrogen-use efficiency, and nitrogen management. Ambio 2002, 31, 132–140. [Google Scholar] [CrossRef]
- Galloway, J.N.; Townsend, A.R.; Erisman, J.W.; Bekunda, M.; Cai, Z.C.; Freney, J.R.; Martinelli, L.A.; Seitzinger, S.P.; Sutton, M.A. Transformation of the nitrogen cycle: Recent trends, questions, and potential solutions. Science 2008, 320, 889–892. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Hu, B.; Chu, C. Nitrogen use efficiency in crops: Lessons from Arabidopsis and rice. J. Exp. Bot. 2017, 68, 2477–2488. [Google Scholar] [CrossRef] [PubMed]
- Gruber, N.; Galloway, J.N. An Earth-system perspective of the global nitrogen cycle. Nature 2008, 451, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Tachibana, N.; Yoshikawa, S.; Ikeda, K. Behavior of inorganic nitrogen in tea field soils heavily applied with nitrogen and changes in amino acid content of the first crop accompanying the reduction of nitrogen. Jpn. J. Crop Sci. 1995, 64, 523–528. [Google Scholar] [CrossRef]
- Venkatesan, S.; Ganapathy, M.N.K. Impact of nitrogen and potassium fertilizer application on quality of CTC teas. Food Chem. 2004, 84, 325–328. [Google Scholar] [CrossRef]
- Ishigaki, K. Comparison between ammonium-nitrogen and nitrate-nitrogen on the efiect of tea plant growth. Jpn. Agric. Res. Quart. 1974, 8, 101–105. [Google Scholar]
- Moreno, J.M.; García-Martinez, J.L. Extraction of tracheal sap from citrus and analysis of its nitrogenous compounds. Physiol. Plant. 1980, 50, 298–303. [Google Scholar] [CrossRef]
- Morita, A.; Tuji, M. Nitrate and oxalate contents of tea plants (Camellia sinensis L.) with special reference to types of green tea and effect of shading. Soil Sci. Plant Nutr. 2002, 48, 547–553. [Google Scholar] [CrossRef][Green Version]
- Wang, Y.Y.; Cheng, Y.H.; Chen, K.E.; Tsay, Y.F. Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 2018, 69, 85–122. [Google Scholar] [CrossRef]
- Williams, L.E.; Miller, A.J. Transporters responsible for the uptake and partitioning of nitrogenous solutes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 659. [Google Scholar] [CrossRef]
- Friedel, J.K.; Scheller, E. Composition of hydrolysable amino acids in soil organic matter and soil microbial biomass. Soil Biol. Biochem. 2002, 34, 315–325. [Google Scholar] [CrossRef]
- Yu, Z.; Zhang, Q.; Kraus, T.E.C.; Dahlgren, R.A.; Anastasio, C.; Zasoski, R.J. Contribution of amino compounds to dissolved organic nitrogen in forest soils. Biogeochemistry 2002, 61, 173–198. [Google Scholar] [CrossRef]
- Kranabetter, J.M.; Dawson, C.R.; Dunn, D.E. Indices of dissolved organic nitrogen, ammonium and, nitrate across productivity gradients of boreal forests. Soil Biol. Biochem. 2007, 39, 3147–3158. [Google Scholar] [CrossRef]
- Ruan, J.Y.; Ma, L.F.; Shi, Y.Z.; Han, W.Y. Uptake of fluoride by tea plant (Camellia sinensis L) and the impact of aluminium. J. Sci. Food Agric. 2003, 83, 1342–1348. [Google Scholar] [CrossRef]
- Ruan, J.; Haerdter, R.; Gerendás, J. Impact of nitrogen supply on carbon/nitrogen allocation: A case study on amino acids and catechins in green tea [Camellia sinensis (L.) O. Kuntze] plants. Plant Biol. 2010, 12, 724–734. [Google Scholar] [CrossRef] [PubMed]
- Gansel, X.; Munos, S.; Tillard, P.; Gojon, A. Differential regulation of the NO3− and NH4+ transporter genes AtNrt2.1 and AtAmt1.1. Arabidopsis: Relation with long-distance and local controls by N status of the plant. Plant J. 2001, 26, 143–155. [Google Scholar] [CrossRef]
- Camanes, G.; Bellmunt, E.; Javier, G.A.; Pilar, G.A.; Cerezo, M. Reciprocal regulation between AtNRT2.1 and AtAMT1.1 expression and the kinetics of NH4+ and NO3− influxes. J. Plant Physiol. 2012, 169, 268–274. [Google Scholar] [CrossRef]
- Kronzucker, H.J.; Siddiqi, M.Y.; Glass, A.D.M.; Kirk, G.J.D. Nitrate-ammonium synergism in rice. a subcellular flux analysis. Plant Physiol. 1999, 119, 1041–1046. [Google Scholar] [CrossRef]
- Glass, A.D.M.; Britto, D.T.; Kaiser, B.N.; Kinghorn, J.R.; Kronzucker, H.J.; Kumar, A.; Okamoto, M.; Rawat, S.; Siddiqi, M.Y.; Unkles, S.E.; et al. The regulation of nitrate and ammonium transport systems in plants. J. Exp. Bot. 2002, 53, 855–864. [Google Scholar] [CrossRef]
- Okamoto, M.; Vidmar, J.J.; Glass, A.D.M. Regulation of NRT1 and NRT2 gene families of Arabidopsis thaliana: Responses to nitrate provision. Plant Cell Physiol. 2003, 44, 304–317. [Google Scholar] [CrossRef]
- Yuan, L.; Loqué, D.; Kojima, S.; Rauch, S.; Ishiyama, K.; Inoue, E.; Takahashi, H.; Wirén, N. The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. The Plant Cell 2007, 19, 2636–2652. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Wang, W.; Ou, S.; Tang, J.; Li, H.; Che, R.; Zhang, Z.; Chai, X.; Wang, H.; Wang, Y.; et al. Variation in NRT1.1b contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 2015, 47, 834. [Google Scholar] [CrossRef]
- Ludewig, U.; Neuhauser, B.; Dynowski, M. Molecular mechanisms of ammonium transport and accumulation in plants. FEBS Lett. 2007, 581, 2301–2308. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xiang, F.; Zhong, M.; Zhou, L.; Liu, H.; Li, S.; Wang, X. Transcriptome and metabolite analysis identifies nitrogen utilization genes in tea plant (Camellia sinensis). Sci. Rep. 2017, 7, 1693. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Dong, C.; Yang, T.; Bao, S.; Fang, W.; Lucas, W.J.; Zhang, Z. The tea plant CsLHT1 and CsLHT6 transporters take up amino acids, as a nitrogen source, from the soil of organic tea plantations. Hortic. Res. 2021, 8, 178. [Google Scholar] [CrossRef]
- Hou, L.; Zhang, Z.; Dou, S.; Zhang, Y.; Pang, X.; Li, Y. Genome-wide identification, characterization, and expression analysis of the expansin gene family in Chinese jujube (Ziziphus jujuba Mill.). Planta 2019, 249, 815–829. [Google Scholar] [CrossRef]
- Ruan, J.Y.; Gerendas, J.; Hardter, R.; Sattelmacher, B. Effect of nitrogen form and root-zone pH on growth and nitrogen uptake of tea (Camellia sinensis L.) plants. Ann. Bot. 2008, 99, 301–310. [Google Scholar] [CrossRef]
- Yang, Y.Y.; Li, X.H.; Ratcliffe, R.G.; Ruan, J.Y. Characterization of ammonium and nitrate uptake and assimilation in roots of tea plants. Russ. J. Plant Physiol. 2013, 60, 91–99. [Google Scholar] [CrossRef]
- Fan, K.; Fan, D.; Ding, Z.; Su, Y.; Wang, X. Cs-miR156 is involved in thenitrogen form regulation of catechins accumulation in tea plant (Camellia sinensis L.). Plant Physiol. Bioch. 2015, 97, 350–360. [Google Scholar] [CrossRef]
- Wang, R.C.; Okamoto, M.; Xing, X.J.; Crawford, N.M. Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol. 2003, 132, 556–567. [Google Scholar] [CrossRef]
- Nunes-Nesi, A.; Fernie, A.R.; Stitt, M. Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen Interactions. Mol. Plant. 2010, 3, 973–996. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.H.; Tsay, Y.F. Nitrate, ammonium, and potassium sensing and signaling. Curr. Opin. Plant Biol. 2010, 13, 604–610. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Liu, Y.; Wang, L.; Bai, P.; Ruan, L.; Zhang, C.; Wei, K.; Cheng, H. Molecular cloning and expression analysis of ammonium transporters in tea plants (Camellia sinensis L.) under different nitrogen treatments. Gene 2018, 658, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Graff, L.; Loque, D.; Kojima, S.; Tsuchiya, Y.N.; Takahashi, H.; Wirén, N.v. AtAMT1;4, a pollen-specific high-affinity ammonium transporter of the plasma membrane in Arabidopsis. Plant Cell Physiol. 2009, 50, 13–25. [Google Scholar] [CrossRef]
- Wang, Y.; Xuan, Y.; Wang, S.; Fan, D.; Wang, X.; Zheng, X. Genome-wide identification, characterization, and expression analysis of the ammonium transporter gene family in tea plants (Camellia sinensis L.). Physiol. Plantarum. 2022, 174, 13646. [Google Scholar] [CrossRef]
- Bao, A.; Liang, Z.; Zhao, Z.; Cai, H. Overexpressing of OsAMT1-3, a high affinity ammonium transporter gene, modifies rice growth andcrbon-nitrogen metabolic status. Int. J. Mol. Sci. 2015, 16, 9037–9063. [Google Scholar] [CrossRef]
- Zhang, F.; He, W.; Yuan, Q.; Wei, K.; Ruan, L.; Wang, L.; Cheng, H. Transcriptome analysis identifies CsNRT genes involved in nitrogen uptake in tea plants, with a major role of CsNRT2.4. Plant Physiol. Bioch. 2021, 167, 970–979. [Google Scholar] [CrossRef]
- Hu, Y.; Fernandez, V. Nitrate transporters in leaves and their potential roles in foliar uptake of nitrogen dioxide. Front. Plant Sci. 2014, 5, 360. [Google Scholar] [CrossRef]
- Liu, M.; Burgos, A.; Zhang, Q.; Tang, D.; Shi, Y.; Ma, L.; Yi, X.; Ruan, J. Analyses of transcriptome profiles and selected metabolites unravel the metabolic response to NH4+ and NO3− as signaling molecules in tea plant (Camellia sinensis L.). Sci. Hortic. 2017, 218, 293–303. [Google Scholar] [CrossRef]
- Leran, S.; Varala, K.; Boyer, J.C.; Chiurazzi, M.; Crawford, N.; Daniel-Vedele, F.; David, L.; Dickstein, R.; Fernandez, E.; Forde, B.; et al. A unified nomenclature of nitrate transporter 1/peptide transporter family members in plants. Trends Plant Sci. 2014, 19, 5–9. [Google Scholar] [CrossRef]
- Wang, H.; Wan, Y.; Buchner, P.; King, R.; Ma, H.; Hawkesford, M.J. Phylogeny and gene expression of the complete nitrate transporter 1/peptide transporter family in triticum aestivum. J. Exp. Bot. 2020, 71, 4531–4546. [Google Scholar] [CrossRef] [PubMed]
- Corratge-Faillie, C.; Lacombe, B. Substrate (un) specificity of Arabidopsis NRT1/PTR family (NPF) proteins. J. Exp. Bot. 2017, 68, 3107–3113. [Google Scholar] [CrossRef]
- Wang, Y.; Wei, K.; Ruan, L.; Bai, P.; Wu, L.; Wang, L.; Cheng, H. Systematic investigation and expression profiles of the nitrate transporter 1/peptide transporter family (NPF) in tea Plant (Camellia sinensis L.). Int. J. Mol. Sci. 2022, 23, 6663. [Google Scholar] [CrossRef]
- Shuai, B.; Reynaga-Pena, C.G.; Springer, P.S. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 2002, 129, 747–761. [Google Scholar] [CrossRef] [PubMed]
- Majer, C.; Hochholdinger, F. Defining the boundaries: Structure and function of LOB domain proteins. Trends Plant Sci. 2011, 16, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Xia, E.; Zhang, H.; Sheng, J.; Li, L.; Zhang, Q.; Kim, C.; Zhang, Y.; Liu, Y.; Zhu, T.; Li, W.; et al. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol. Plant. 2017, 10, 866–877. [Google Scholar] [CrossRef]
- Zhang, X.; He, Y.; He, W.; Su, H.; Wang, Y.; Hong, G.; Xu, P. Structural and functional insights into the LBD family involved in abiotic stress and flavonoid synthases in Camellia sinensis. Sci. Rep. 2019, 9, 15651. [Google Scholar] [CrossRef] [PubMed]
- Teng, R.; Yang, N.; Liu, C.; Chen, Y.; Wang, Y.; Zhuang, J. CsLBD37, a LBD/ASL transcription factor, affects nitrate response and flowering of tea plant. Sci. Hortic. 2022, 306, 111457. [Google Scholar] [CrossRef]
- Teng, R.; Yang, N.; Li, J.; Liu, C.; Chen, Y.; Li, T.; Wang, Y.; Xiong, A.; Zhuang, J. Isolation and characterization of an LBD transcription factor CsLBD39 from tea plant (Camellia sinensis L.) and its roles in modulating nitrate content by regulating nitrate-metabolism-related genes. Int. J. Mol. Sci. 2022, 23, 9294. [Google Scholar] [CrossRef] [PubMed]
- Aslam, M.; Huffaker, R.C. Role of nitrate and nitrite in the induction of nitrate reductase in leaves of barley seedlings. Plant Physiol. 1989, 91, 1152–1156. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Liu, M.Y.; Zhang, Q.; Ma, L.; Ruan, J. Preferential assimilation of NH4+ over NO3− in tea plant associated with genes involved in nitrogen transportation, utilization and catechins biosynthesis. Plant Sci. 2019, 291, 110369. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Xie, S.; Wang, L.; Jing, S.; Zhu, X.; Li, X.; Wei, Z.; Yuan, H. Identification of up-regulated genes in tea leaves under mild infestation of green leafhopper. Sci. Hortic. 2011, 130, 476–481. [Google Scholar] [CrossRef]
- Wang, L.; Feng, X.; Yao, L.; Ding, C.; Lei, L.; Wang, X. Characterization of CBL-CIPK signaling complexes and their involvement in cold response in tea plant. Plant Physiol. Bioch. 2020, 154, 195–203. [Google Scholar] [CrossRef]
- Liu, H.; Wang, Y.; Li, H.; Teng, R.; Wang, Y.; Zhuang, J. Genome-wide identification and expression analysis of calcineurin B-like protein and calcineurin B-like protein-interacting protein kinase family genes in tea plant. DNA Cell Biol. 2019, 38, 824–839. [Google Scholar] [CrossRef]
- Hu, H.; Wang, Y.; Tsay, Y.F. AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J. 2009, 57, 264–278. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.H.; Lin, S.H.; Hu, H.C.; Tsay, Y.-F. CHL1 functions as a nitrate sensor in plants. Cell 2009, 138, 1184–1194. [Google Scholar] [CrossRef]
- Fischer, W.N.; Loo, D.; Koch, W.; Ludewig, U.; Boorer, K.; Tegeder, M.; Fronmmer, W.B. Low and high affinity amino acid H+-cotransporters for cellular import of neutral and charged amino acids. Plant J. 2002, 29, 717–731. [Google Scholar] [CrossRef]
- Fischer, W.N.; André, B.; Rentsch, B.; Krolkiewicz, S.; Tegeder, M.; Breitkreuz, K.; Frommer, W. Amino acid transport in plants. Trends Plant Sci. 1998, 3, 188–195. [Google Scholar] [CrossRef]
- Perchlik, M.; Foster, J.; Tegeder, M. Different and overlapping functions of Arabidopsis LHT6 and AAP1 transporters in root amino acid uptake. J. Exp. Bot. 2014, 65, 5193–5204. [Google Scholar] [CrossRef]
- Svennerstam, H.; Ganeteg, U.; Nasholm, T. Root uptake of cationic amino acids by Arabidopsis depends on functional expression of amino acid permease 5. New Phytol. 2008, 180, 620–630. [Google Scholar] [CrossRef]
- Svennerstam, H.; Jämtgård, S.; Ahmad, I.; Huss-Danell, K.; Näsholm, T.; Ganeteg, U. Transporters in Arabidopsis roots mediating uptake of amino acids at naturally occurring concentrations. New Phytol. 2011, 191, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, S.; Gumy, C.; Blatter, E.; Boeffel, S.; Fricke, W.; Rentsch, D. In planta function of compatible solute transporters of the AtProT family. J. Exp. Bot. 2011, 62, 787–796. [Google Scholar] [CrossRef]
- Drew, M.C. Comparison of the effects of a localized supply of phosphate, nitrate, ammonium and potassium on the growth of the seminal root system, and the shoot in barley. New Phytol. 1975, 75, 479–490. [Google Scholar] [CrossRef]
- Hodge, A. The plastic plant: Root responses to heterogeneous supplies of nutrients. New Phytol. 2004, 162, 9–24. [Google Scholar] [CrossRef]
- Hodge, A. Plastic plants and patchy soils. J. Exp. Bot. 2006, 57, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Hodge, A. Root decisions. Plant Cell Environ. 2009, 32, 628–640. [Google Scholar] [CrossRef]
- De, K.H.; Visser, E.J.; Huber, H.; Mommer, L.; Hutchings, M.J. A modular concept of plant foraging behaviour: The interplay between local responses and systemic control. Plant Cell Environ. 2009, 32, 704–712. [Google Scholar]
- Farley, R.A.; Fitter, A.H. The responses of seven co-occurring woodland her baceous perennials to localized nutrient-rich patches. J. Ecol. 1999, 87, 849–859. [Google Scholar] [CrossRef]
- Jagadish, R.; Shanmugaselvan, V.A. Influence of nitrogen and potassium on root nutrient and root CEC of different tea cultivars (Camellia sinensis, C. assamica and C.assamica spp. Lasiocalyx). Rhizosphere 2016, 1, 36–44. [Google Scholar]
- Kaur, G.; Asthir, B.; Bains, N.S.; Farooq, M. Nitrogen nutrition, its assimilation and remobilization in diverse wheat genotypes. Int. J. Agric. Biol. 2015, 17, 531–538. [Google Scholar] [CrossRef]
- Atkinson, J.A.; Rasmussen, A.; Traini, R.; Voß, U.; Sturrock, C.; Mooney, S.J.; Darren, M.; Malcolm, J. Branching out in roots: Uncovering form, function, and regulation. Plant Physiol. 2014, 166, 538–550. [Google Scholar] [CrossRef] [PubMed]
- Giehl, R.F.; Von, W.N. Root nutrient foraging. Plant Physiol. 2014, 166, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.F.; Gu, F.; Ma, H.Y.; Tian, C.Y. Effect of nitrate on root development and nitrogen uptake of Suaeda physophora under NaCl salinity. Pedosphere 2010, 20, 536–544. [Google Scholar] [CrossRef]
- Ruan, L.; Wang, L.; Wei, K.; Cheng, H.; Li, H.; Shao, S.; Wu, L. Comparative analysis of nitrogen spatial heterogeneity responses in low nitrogen susceptible and tolerant tea plants (Camellia sinensis L.). Sci. Hortic. 2019, 246, 182–189. [Google Scholar] [CrossRef]
- Liu, X.; Hu, X.M.; Jin, L.F.; Shi, C.Y.; Liu, Y.Z.; Peng, S.A. Identification and transcript analysis of two glutamate decarboxylase genes, CsGAD1 and CsGAD2, reveal the strong relationship between CsGAD1 and citrate utilization in citrus fruit. Mol. Biol. Rep. 2014, 41, 6253–6262. [Google Scholar] [CrossRef] [PubMed]
- Mei, X.; Chen, Y.; Zhang, L.; Fu, Z.M.; Wei, Q.; Grierson, D.; Zhou, Y.; Huang, Y.; Dong, F.; Yang, Z. Dual mechanisms regulating glutamate decarboxylases and accumulation of gamma-aminobutyric acid in tea (Camellia sinensis L.) leaves exposed to multiple stresses. Sci. Rep. 2016, 6, 23685. [Google Scholar] [CrossRef]
- Shelp, B.J.; Bozzo, G.G.; Zarei, A.; Simpson, J.P.; Trobacher, C.P.; Allan, W.L. Strategies and tools for studying the metabolism and function of γ-aminobutyrate in plants. II. Integrated analysis. Botany 2012, 90, 781–793. [Google Scholar] [CrossRef]
- Fait, A.; Fromm, H.; Walter, D.; Galili, G.; Fernie, A.R. Highway or byway: The metabolic role of the gaba shunt in plants. Trends Plant Sci. 2008, 13, 14–19. [Google Scholar] [CrossRef]
- Shimajiri, Y.; Oonishi, T.; Ozaki, K.; Kainou, K.; Akama, K. Genetic manipulation of the γ-aminobutyric acid (GABA) shunt in rice: Overexpression of truncated glutamate decarboxylase (GAD2) and knockdown of γ-aminobutyric acid transaminase (GABA-T) lead to sustained and high levels of GABA accumulation in rice kernels. Plant Biotechnol. J. 2013, 11, 594–604. [Google Scholar]
- Miyashita, Y.; Dolferus, R.; Ismond, K.P.; Good, A.G. Alanine aminotransferase catalyses the breakdown of alanine after hypoxia in Arabidopsis thaliana. Plant J. 2007, 49, 1108–1121. [Google Scholar] [CrossRef]
- Hu, S.; Zhang, M.; Yang, Y.; Xuan, W.; Zou, Z.; Arkorful, E.; Chen, Y.; Ma, Q.; Jeyaraj, A.; Chen, X.; et al. A novel insight into nitrogen and auxin signaling in lateral root formation in tea plant [Camellia sinensis (L.) O. Kuntze]. BMC Plant Biol. 2020, 20, 232. [Google Scholar] [CrossRef]
- Ruan, L.; Wei, K.; Wang, L.Y.; Cheng, H.; Zhang, F.; Wu, L.Y.; Bai, P.X.; Zhang, C.C. Characteristics of NH4+ and NO3− fluxes in tea (Camellia sinensis L.) roots measured by scanning ion-selective electrode technique. Sci. Rep. 2016, 6, 38370. [Google Scholar] [CrossRef]
- Zhong, R.Q.; Lee, C.; McCarthy, R.L.; Reeves, C.K.; Jones, E.G.; Ye, Z.H. Transcriptional activation of secondary wall biosynthesis by Rice and maize NAC and MYB transcription factors. Plant Cell Physiol. 2011, 52, 1856–1871. [Google Scholar] [CrossRef] [PubMed]
- Bollhoner, B.; Prestele, J.; Tuominen, H. Xylem cell death: Emerging understanding of regulation and function. J. Exp. Bot. 2012, 63, 1081–1094. [Google Scholar] [CrossRef] [PubMed]
- Ratcliffe, O.J.; Nadzan, G.C.; Reuber, T.L.; Riechmann, J.L. Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol. 2001, 126, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Wang, Y.; Cheng, X.; Yang, T.; Su, Y.; Lin, S.; Zhang, S.; Zhang, Z. Nitrogen-regulated theanine and flavonoid biosynthesis in tea plant roots: Protein-level regulation revealed by multiomics analyses. J. Agric. Food Chem. 2021, 69, 10002–10016. [Google Scholar] [CrossRef]
- Jiang, X.; Liu, Y.; Wu, Y.; Tan, H.; Meng, F.; Wang, Y.; Li, M.; Zhao, L.; Liu, L.; Qian, Y.; et al. Analysis of accumulation patterns and preliminary study on the condensation mechanism of proanthocyanidins in the tea plant (Camellia sinensis L.). Sci. Rep. 2015, 5, 8742. [Google Scholar] [CrossRef]
- Buer, C.; Muday, G.; Djordjevic, M. Flavonoids are differentially taken up and transported long distances in Arabidopsis. Plant Physiol. 2007, 145, 478–490. [Google Scholar] [CrossRef]
- Jiang, C.; Ying, Y.; Liu, X.; Wang, Z. Response of flag leaf lipid peroxidation and protective enzyme activity of wheat cultivars with different heat tolerance to high temperature stress after anthesis. Zuo Wu Xue Bao 2007, 33, 143–148. [Google Scholar]
- Liu, W.; Xu, K.; Su, H.; Wang, H. Effect of nitrogen nutrition on physiological index of spinach senescence. Plant Nutr. Fert. Sci. 2007, 13, 1110–1115. [Google Scholar]
- Xiao, F.; Yang, Z.Q.; Huang, H.J.; Yang, F.; Zhu, L.Y.; Han, D. Nitrogen fertilization in soil affects physiological characteristics and quality of green tea leaves. Hortscience 2018, 53, 715–722. [Google Scholar] [CrossRef]
- Zhang, Y.C.; Wang, L.C. Effect of low temperature stress on membrane permeability and protection of activity in tea leaves. North. Hortic. 2010, 9, 38–40. [Google Scholar]
- Yang, Y.Y. Quality-Related Constituents in Tea Leaves as Affected by Nitrogen. Ph.D. Thesis, Nanjing Agricultural University, Nanjing, China, 2011. [Google Scholar]
- Zhao, W.X. Effect of Nitrogen Fertilizer on Yield and Quality of Tea and Ecological Fitness of Toxoptera Aurantii Boyer. Ph.D. Thesis, Fujian Agriculture and Forestry University, Fujian, China, 2007. [Google Scholar]
- Zhang, Y.; Wang, L.; Wei, K.; Ruan, L.; Wu, L.; He, M.; Tong, H.; Cheng, H. Differential regulatory mechanisms of secondary metabolites revealed at different leaf positions in two related tea cultivars. Sci. Hortic. 2020, 272, 109579. [Google Scholar] [CrossRef]
- Coschigano, K.T.; Melo-Oliveira, R.; Lim, J.; Coruzzi, G.M. Arabidopsis gls mutants and distinct Fd-GOGAT genes. Implications for photorespiration and primary nitrogen assimilation. Plant Cell 1998, 10, 741–752. [Google Scholar] [CrossRef]
- Liu, Z.W.; Li, H.; Wang, W.L.; Wu, Z.J.; Cui, X.; Zhuang, J. CsGOGAT is important in dynamic changes of theanine content in postharvest tea plant leaves under different temperature and shading spreadings. J. Agric. Food Chem. 2017, 65, 9693–9702. [Google Scholar] [CrossRef]
- Liu, Z.W.; Li, H.; Liu, J.X.; Wang, Y.; Zhuang, J. Integrative transcriptome, proteome, and microRNA analysis reveals the effects of nitrogen sufficiency and deficiency conditions on theanine metabolism in the tea plant (Camellia sinensis L.). Hortic. Res. 2020, 7, 65. [Google Scholar] [CrossRef]
- Cheng, S.; Fu, X.; Wang, X.; Liao, Y.; Zeng, L.; Dong, F.; Yang, Z. Studies on the biochemical formation pathway of the amino acid L-theanine in tea (Camellia sinensis L.) and other plants. J. Agric. Food Chem. 2017, 65, 7210–7216. [Google Scholar] [CrossRef]
- Dong, C.; Li, F.; Yang, T.; Feng, L.; Zhang, S.; Li, F.; Li, W.; Xu, G.; Bao, S.; Wan, X.; et al. Theanine transporters identified in tea plants (Camellia sinensis L.). Plant J. 2019, 101, 57–70. [Google Scholar] [CrossRef]
- Rana, N.K.; Mohanpuria, P.; Kumar, V.; Yadav, S.K. A CsGS is regulated at transcriptional level during developmental stages and nitrogen utilization in Camellia sinensis L. (L.) O. Kuntze. Mol. Biol. Rep. 2010, 37, 703–710. [Google Scholar] [CrossRef]
- Wan, X.C.; Xia, T. Secondary Metabolism of Tea Plant, 1st ed.; Science Press: Beijing, China, 2015; pp. 50–53. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, X.; Sun, Z.; Zhang, X.; Han, X. Novel Aspects of Regulation of Nitrogen Responses in the Tea Plant (Camellia sinensis (L.)). Agronomy 2023, 13, 144. https://doi.org/10.3390/agronomy13010144
Xie X, Sun Z, Zhang X, Han X. Novel Aspects of Regulation of Nitrogen Responses in the Tea Plant (Camellia sinensis (L.)). Agronomy. 2023; 13(1):144. https://doi.org/10.3390/agronomy13010144
Chicago/Turabian StyleXie, Xueying, Zilin Sun, Xinjian Zhang, and Xiaoyang Han. 2023. "Novel Aspects of Regulation of Nitrogen Responses in the Tea Plant (Camellia sinensis (L.))" Agronomy 13, no. 1: 144. https://doi.org/10.3390/agronomy13010144
APA StyleXie, X., Sun, Z., Zhang, X., & Han, X. (2023). Novel Aspects of Regulation of Nitrogen Responses in the Tea Plant (Camellia sinensis (L.)). Agronomy, 13(1), 144. https://doi.org/10.3390/agronomy13010144






