RicePedigree: Rice Pedigree Database for Documentation and Assistance in Rice Breeding
Abstract
1. Introduction
2. Materials and Methods
2.1. Breeding History Database and the Web Application Construction
2.2. Clustering of NCBI-Deposited Accessions
3. Results
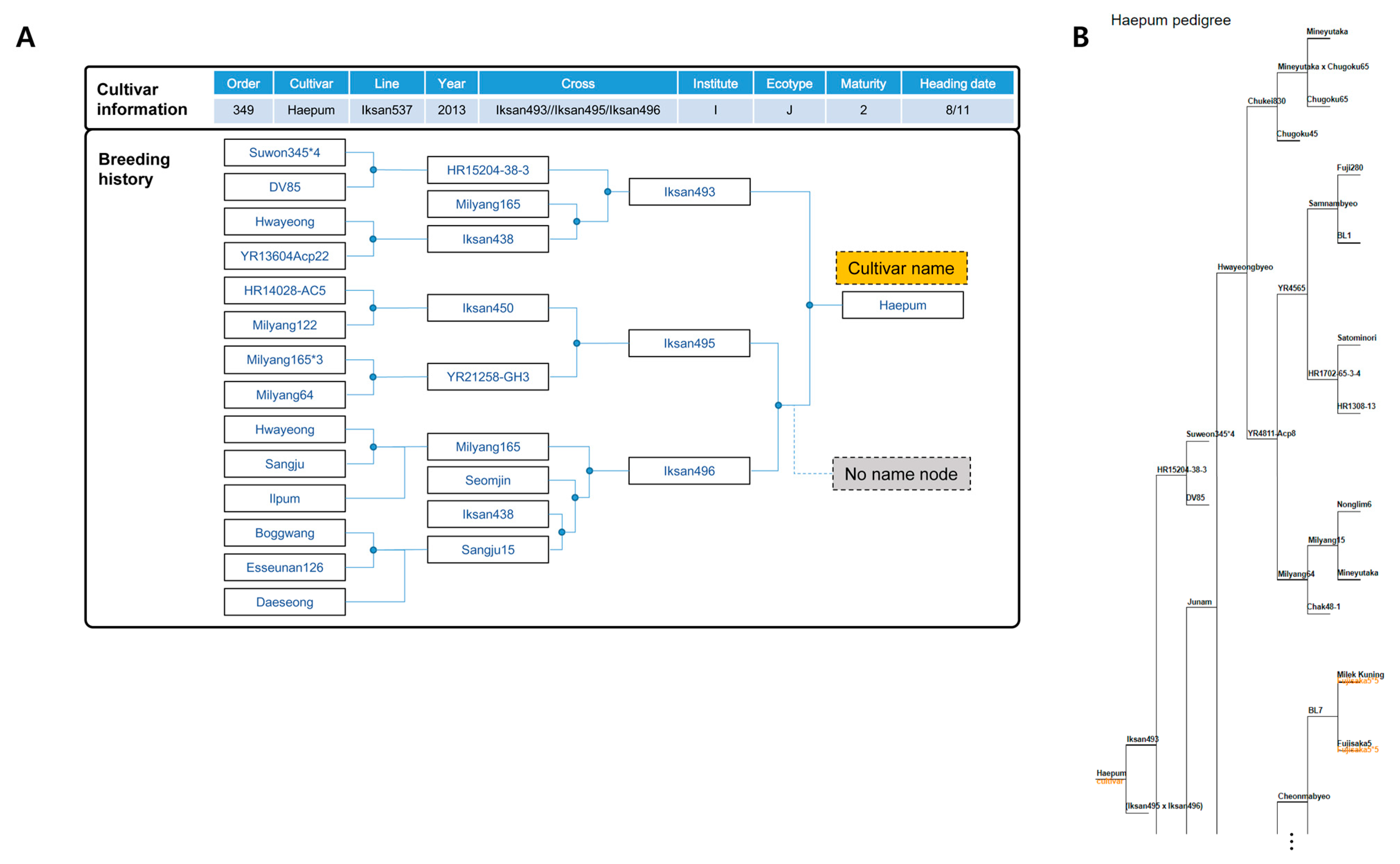
3.1. Systematic Representation of Pedigree
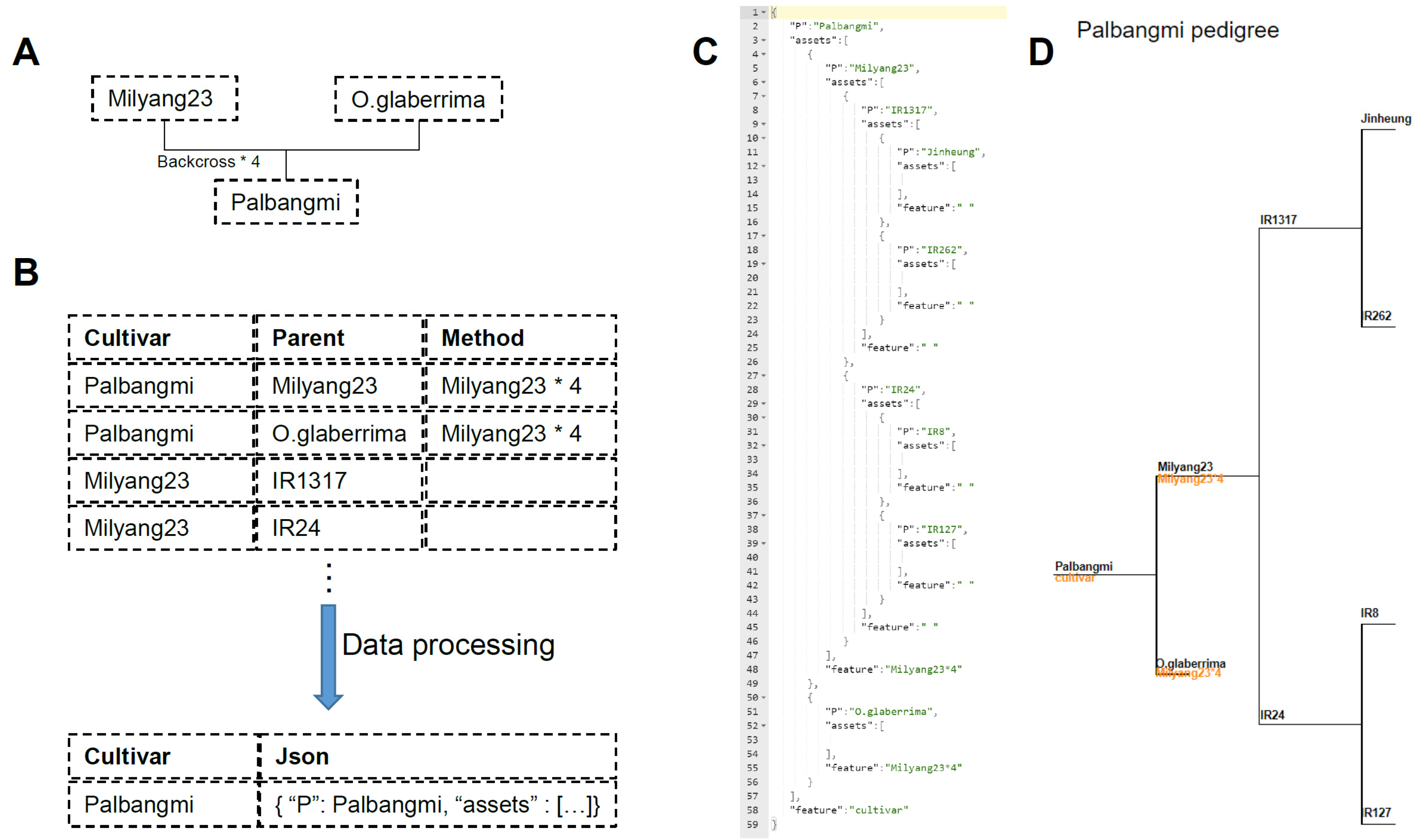
3.2. Data Processing of Pedigree Information
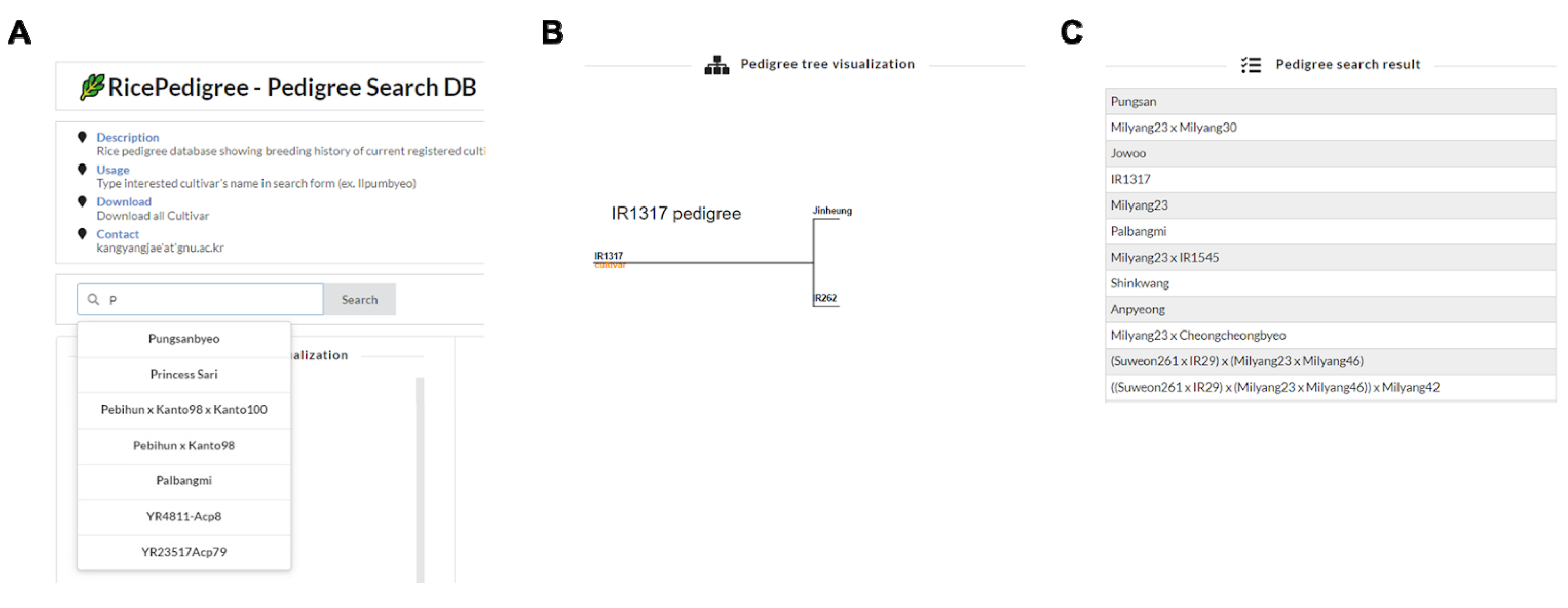
3.3. Database and Web Application Development
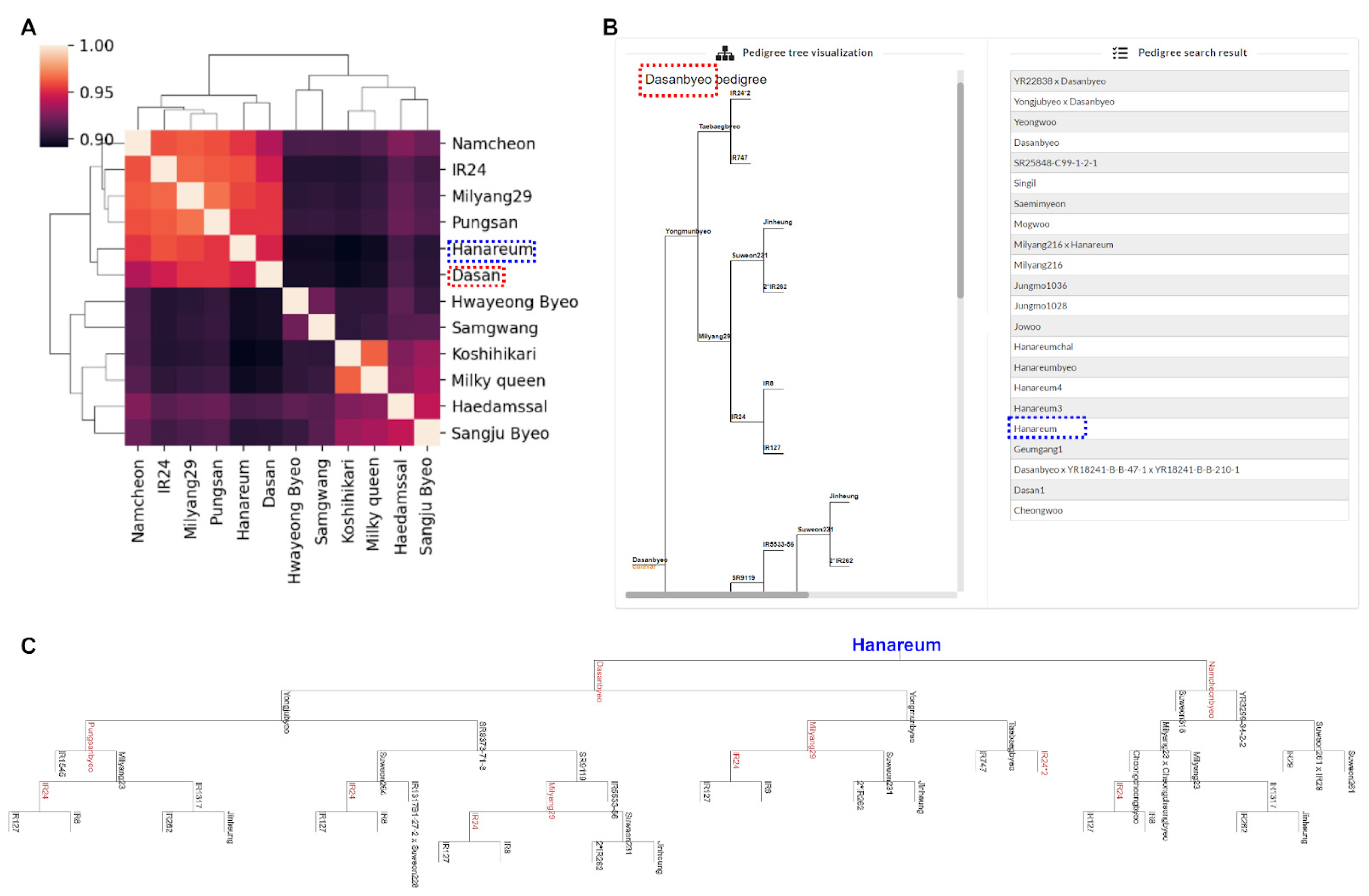
3.4. Case Study: Supporting Evidence for Genomic Similarity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Prohens, J. Plant Breeding: A Success Story to be Continued Thanks to the Advances in Genomics. Front. Plant Sci. 2011, 2, 51. [Google Scholar] [CrossRef] [PubMed]
- Breseghello, F.; Coelho, A.S.G. Traditional and Modern Plant Breeding Methods with Examples in Rice (Oryza sativa L.). J. Agric. Food Chem. 2013, 61, 8277–8286. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.J.; Lee, T.; Lee, J.; Shim, S.; Jeong, H.; Satyawan, D.; Kim, M.Y.; Lee, S. Translational genomics for plant breeding with the genome sequence explosion. Plant Biotechnol. J. 2016, 14, 1057–1069. [Google Scholar] [CrossRef] [PubMed]
- Verdeprado, H.; Kretzschmar, T.; Begum, H.; Raghavan, C.; Joyce, P.; Lakshmanan, P.; Cobb, J.N.; Collard, B.C. Association mapping in rice: Basic concepts and perspectives for molecular breeding. Plant Prod. Sci. 2018, 21, 159–176. [Google Scholar] [CrossRef]
- Spindel, J.; Begum, H.; Akdemir, D.; Virk, P.; Collard, B.; Redoña, E.; Atlin, G.; Jannink, J.-L.; McCouch, S.R. Genomic Selection and Association Mapping in Rice (Oryza Sativa): Effect of Trait Genetic Architecture, Training Population Composition, Marker Number and Statistical Model on Accuracy of Rice Genomic Selection in Elite, Tropical Rice Breeding Lines. PLoS Genet. 2015, 11, e1004982. [Google Scholar]
- Zhou, X.; Huang, X. Genome-Wide Association Studies in Rice: How to Solve the Low Power Problems? Mol. Plant 2019, 12, 10–12. [Google Scholar] [CrossRef] [PubMed]
- Ankamah-Yeboah, T.; Janss, L.L.; Jensen, J.D.; Hjortshøj, R.L.; Rasmussen, S.K. Genomic Selection Using Pedigree and Marker-by-Environment Interaction for Barley Seed Quality Traits From Two Commercial Breeding Programs. Front. Plant Sci. 2020, 11, 539. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.D.; Baldo, A.M.; Mueller, L.A. Ricebase: A Breeding and Genetics Platform for Rice, Integrating Individual Molecular Markers, Pedigrees and Whole-Genome-Based Data. Database 2016, 2016, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E.; Bink, M.C.A.M.; Van De Weg, W.E. Pedimap: Software for the Visualization of Genetic and Phenotypic Data in Pedigrees. J. Hered. 2012, 103, 903–907. [Google Scholar] [CrossRef] [PubMed]
- Song, M.-T.; Lee, J.-K.; Yang, S.-J.; Choi, H.-C.; Hwang, H.-G.; Kim, H.-Y.; Park, K.-G.; Cho, Y.-S.; Moon, H.-P.; Han, W.-S.; et al. KRBIMS (Korean Rice Breeding Information Management System): A Database for Rice Breeding Information Management. Korea J. Breed. Sci. 2002, 34, 111–115. [Google Scholar]
- Leinonen, R.; Sugawara, H.; Shumway, M. International Nucleotide Sequence Database Collaboration The Sequence Read Archive. Nucleic Acids Res. 2011, 39, D19–D21. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza Sativa Nipponbare Reference Genome Using next Generation Sequence and Optical Map Data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Waskom, M.; Botvinnik, O.; O’Kane, D.; Hobson, P.; Ostblom, J.; Lukauskas, S.; Gemperline, D.C.; Augspurger, T.; Halchenko, Y.; Cole, J.B.; et al. seaborn: Statistical data visualization. J. Open Source Softw. 2018, 6, 3021. [Google Scholar] [CrossRef]
- Kim, T.-S.; He, Q.; Kim, K.-W.; Yoon, M.-Y.; Ra, W.-H.; Li, F.P.; Tong, W.; Yu, J.; Oo, W.H.; Choi, B.; et al. Genome-wide resequencing of KRICE_CORE reveals their potential for future breeding, as well as functional and evolutionary studies in the post-genomic era. BMC Genom. 2016, 17, 408. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Woo, D.-U.; Lee, Y.; Jeon, H.-H.; Park, H.; Park, J.-H.; Choi, S.-H.; Lee, C.-M.; Mo, Y.; Kang, Y.-J. RicePedigree: Rice Pedigree Database for Documentation and Assistance in Rice Breeding. Agronomy 2023, 13, 69. https://doi.org/10.3390/agronomy13010069
Woo D-U, Lee Y, Jeon H-H, Park H, Park J-H, Choi S-H, Lee C-M, Mo Y, Kang Y-J. RicePedigree: Rice Pedigree Database for Documentation and Assistance in Rice Breeding. Agronomy. 2023; 13(1):69. https://doi.org/10.3390/agronomy13010069
Chicago/Turabian StyleWoo, Dong-U, Yejin Lee, Ho-Hwi Jeon, Halim Park, Jin-Hwa Park, Sung-Hoon Choi, Chang-Min Lee, Youngjun Mo, and Yang-Jae Kang. 2023. "RicePedigree: Rice Pedigree Database for Documentation and Assistance in Rice Breeding" Agronomy 13, no. 1: 69. https://doi.org/10.3390/agronomy13010069
APA StyleWoo, D.-U., Lee, Y., Jeon, H.-H., Park, H., Park, J.-H., Choi, S.-H., Lee, C.-M., Mo, Y., & Kang, Y.-J. (2023). RicePedigree: Rice Pedigree Database for Documentation and Assistance in Rice Breeding. Agronomy, 13(1), 69. https://doi.org/10.3390/agronomy13010069





