The Evolution of Molecular Genotyping in Plant Breeding
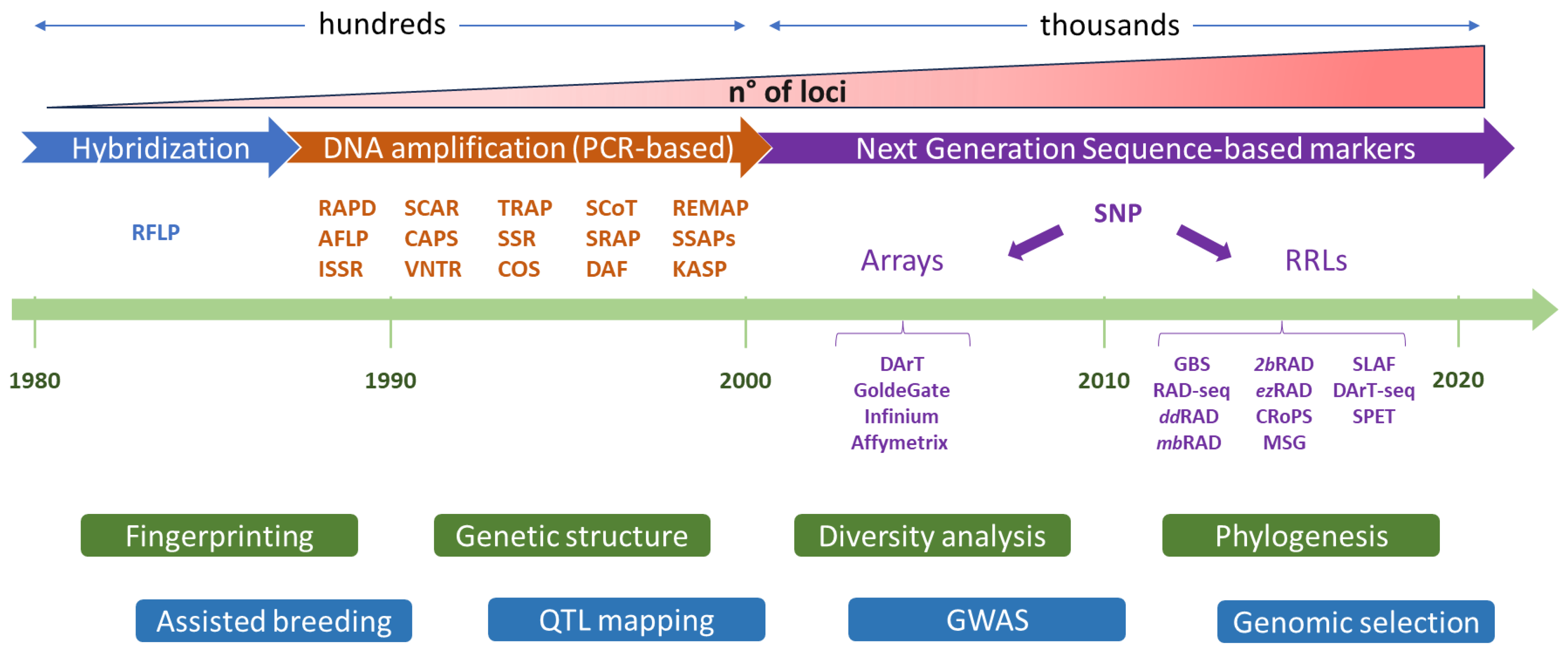
1. The Advent Molecular Markers in Crop Research
2. Platforms for High Throughput Plant Genotyping
3. What’s The Next
Funding
Conflicts of Interest
References
- Andersen, J.R.; Lubberstedt, T. Functional markers in plants. Trends Plant Sci. 2003, 8, 554–560. [Google Scholar] [CrossRef]
- Mondini, L.; Noorani, A.; Pagnotta, M.A. Assessing Plant Genetic Diversity by Molecular Tools. Diversity 2009, 1, 19–35. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphism. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar] [PubMed]
- Kochert, G. RFLP technology. In DNA-Based Markers in Plants; Springer: Dordrecht, The Netherlands, 1994; pp. 8–38. [Google Scholar]
- Gebhardt, C.; Ritter, E.; Debener, T.; Schachtschabel, U.; Walkemeier, B.; Uhrig, H.; Salamini, F. RFLP analysis and linkage mapping in Solanum tuberosum. Theor. Appl. Genet. 1989, 78, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.-W.; Magill, C.; Schertz, K.; Hart, G. A RFLP linkage map of Sorghum bicolor (L.) Moench. Theor. Appl. Genet. 1994, 89, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Brummer, E.C.; Bouton, J.H.; Kochert, G. Development of an RFLP map in diploid alfalfa. Theor. Appl. Genet. 1993, 86, 329–332. [Google Scholar] [CrossRef]
- Tanksley, S.D.; Young, N.D.; Paterson, A.H.; Bonierbale, M.W. RFLP mapping in plant breeding: New tools for an old science. Nat. Biotechnol. 1989, 7, 257–264. [Google Scholar] [CrossRef]
- Domingues, L. PCR: Methods and Protocols, 1st ed.; Springer: New York, NY, USA, 2017; pp. 1–282. [Google Scholar]
- Nadeem, M.A.; Nawaz, M.A.; Shahid, M.Q.; Doğan, Y.; Comertpay, G.; Yıldız, M.; Hatipoğlu, R.; Ahmad, F.; Alsaleh, A.; Labhane, N. DNA molecular markers in plant breeding: Current status and recent advancements in genomic selection and genome editing. Biotechnol. Biotechnol. Equip. 2017, 32, 261–285. [Google Scholar] [CrossRef]
- Garrido-Cardenas, J.A.; Mesa-Valle, C.; Manzano-Agugliaro, F. Trends in plant research using molecular markers. Planta 2018, 247, 543–557. [Google Scholar] [CrossRef]
- Bernardo, R. Molecular Markers and Selection for Complex Traits in Plants: Learning from the Last 20 Years. Crop Sci. 2008, 48, 1649. [Google Scholar] [CrossRef]
- Kao, C.H. Mapping quantitative trait loci using the experimental designs of recombinant inbred populations. Genetics 2006, 174, 1373–1386. [Google Scholar] [CrossRef] [PubMed]
- Shi, A.L.; Chen, P.Y.; Li, D.X.; Zheng, C.M.; Zhang, B.; Hou, A.F. Pyramiding multiple genes for resistance to soybean mosaic virus in soybean using molecular markers. Mol. Breed. 2009, 23, 113–124. [Google Scholar] [CrossRef]
- Kantartzi, S.K. (Ed.) Microsatellites Methods and Protocols. In Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2013; Volume 1006. [Google Scholar]
- Ganal, M.W.; Altmann, T.; Röder, M.S. SNP identification in crop plants. Curr. Opin. Plant Biol. 2009, 12, 211–217. [Google Scholar] [CrossRef]
- Mammadov, J.; Aggarwal, R.; Buyyarapu, R.; Kumpatla, S. SNP Markers and Their Impact on Plant Breeding. Int. J. Plant Genom. 2012, 2012, 728398. [Google Scholar] [CrossRef] [PubMed]
- Tripodi, P. Next generation sequencing technologies to explore the diversity of germplasm resources: Achievements and trends in tomato. Comput. Struct. Biotechnol. J. 2022, 20, 6250–6258. [Google Scholar] [CrossRef]
- Thomson, M.J. High-throughput SNP genotyping to accelerate crop improvement. Plant Breed. Biotechnol. 2014, 2, 195–212. [Google Scholar] [CrossRef]
- Gawroński, P.; Pawełkowicz, M.; Tofil, K.; Uszyński, G.; Sharifova, S.; Ahluwalia, S.; Tyrka, M.; Wędzony, M.; Kilian, A.; Bolibok-Brągoszewska, H. DArT Markers effectively target gene space in the rye genome. Front. Plant Sci. 2016, 7, 1600. [Google Scholar] [CrossRef]
- Mauche, S.; Poirier, O.; Godefroy, T.; Olaso, R.; Gut, I.; Collet, J.P.; Montalescot, G.; Cambien, F. Performance comparison of two microarray platforms to assess differential gene expression in human monocyte and macrophage cells. BMC Genom. 2008, 9, 302. [Google Scholar] [CrossRef]
- Song, Q.; Yan, L.; Quigley, C.; Fickus, E.; Wei, H.; Chen, L.; Dong, F.; Araya, S.; Liu, J.; Hyten, D.; et al. Soybean BARCSoySNP6K: An assay for soybean genetics and breeding research. Plant J. 2020, 104, 800–811. [Google Scholar] [CrossRef]
- Dadshani, S.; Mathew, B.; Ballvora, A.; Mason, A.S.; Léon, J. Detection of breeding signatures in wheat using a linkage disequilibrium-corrected mapping approach. Sci. Rep. 2021, 11, 5527. [Google Scholar] [CrossRef]
- McCouch, S.R.; Wright, M.H.; Tung, C.W.; Maron, L.G.; McNally, K.L.; Fitzgerald, M.; Singh, N.; DeClerck, G.; Agosto-Perez, F.; Korniliev, P.; et al. Open access resources for genome-wide association mapping in rice. Nat. Commun. 2016, 7, 10–532. [Google Scholar] [CrossRef] [PubMed]
- Unterseer, S.; Bauer, E.; Haberer, G.; Seidel, M.; Knaak, C.; Ouzunova, M.; Meitinger, T.; Strom, T.M.; Fries, R.; Pausch, H.; et al. A powerful tool for genome analysis in maize: Development and evaluation of the high density 600 k SNP genotyping array. BMC Genom. 2014, 15, 823. [Google Scholar] [CrossRef]
- Verlouw, J.A.M.; Clemens, E.; de Vries, J.H.; Zolk, O.; Verkerk, A.; Am Zehnhoff-Dinnesen, A.; Medina-Gomez, C.; Lanvers-Kaminsky, C.; Rivadeneira, F.; Langer, T.; et al. A comparison of genotyping arrays. Eur. J. Hum. Genet. 2021, 29, 1611–1624. [Google Scholar] [CrossRef]
- Rasheed, A.; Hao, Y.; Xia, X.; Khan, A.; Xu, Y.; Varshney, R.K.; He, Z. Crop breeding chips and genotyping platforms: Progress, challenges, and perspectives. Mol. Plant 2017, 10, 1047–1064. [Google Scholar] [CrossRef]
- Wang, N.; Yuan, Y.; Wang, H.; Yu, D.; Liu, Y.; Zhang, A.; Gowda, M.; Nair, S.K.; Hao, Z.; Lu, Y.; et al. Applications of genotyping-by-sequencing (GBS) in maize genetics and breeding. Sci. Rep. 2020, 10, 16308. [Google Scholar] [CrossRef]
- Melo, A.T.O.; Bartaula, R.; Hale, I. GBS-SNP-CROP: A reference-optional pipeline for SNP discovery and plant germplasm characterization using variable length, paired-end genotyping-by-sequencing data. BMC Bioinform. 2016, 17, 29. [Google Scholar] [CrossRef]
- Amorese, D.; Armour, C.; Kurn, N. Compositions and Methods for Targeted Nucleic Acid Sequence Enrichment and High Efficiency Library Regeneration. U.S. Patent US9650628B2, 16 May 2017. [Google Scholar]
- Tripodi, P.; Beretta, M.; Peltier, D.; Kalfas, I.; Vasilikiotis, C.; Laidet, A.; Briand, G.; Aichholz, C.; Zollinger, T.; van Treuren, R.; et al. Development and application of Single Primer Enrichment Technology (SPET) SNP assay for population genomics analysis and candidate gene discovery in lettuce. Front. Plant Sci. 2023, 14, 1252777. [Google Scholar] [CrossRef] [PubMed]
- Scheben, A.; Batley, J.; Edwards, D. Genotyping-by-sequencing approaches to characterize crop genomes: Choosing the right tool for the right application. Plant Biotechnol. J. 2017, 15, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Van Treuren, R.; Liu, X.; Zhang, Z.; Chen, J.; Liu, Y.; Dong, S.; Sun, P.; Yang, T.; Lan, T.; et al. Whole-Genome Resequencing of 445 Lactuca Accessions Reveals the Domestication History of Cultivated Lettuce. Nat. Genet. 2021, 53, 752–760. [Google Scholar] [CrossRef]
- Pang, Y.; Liu, C.; Wang, D.; Amand, P.S.; Bernardo, A.; Li, W.; He, F.; Li, L.; Wang, L.; Yuan, X.; et al. High-Resolution Genome-Wide Association Study Identifies Genomic Regions and Candidate Genes for Important Agronomic Traits in Wheat. Mol. Plant 2020, 13, 1311–1327. [Google Scholar] [CrossRef]
- Malmberg, M.M.; Barbulescu, D.M.; Drayton, M.C.; Shinozuka, M.; Thakur, P.; Ogaji, Y.O.; Spangenberg, G.C.; Daetwyler, H.D.; Cogan, N.O.I. Evaluation and Recommendations for Routine Genotyping Using Skim Whole Genome Re-sequencing in Canola. Front. Plant Sci. 2018, 9, 1809. [Google Scholar] [CrossRef] [PubMed]
- Amarasinghe, S.L.; Su, S.; Dong, X.; Zappia, L.; Ritchie, M.E.; Gouil, Q. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020, 21, 30. [Google Scholar] [CrossRef]
- Gomes, B.; Ashley, E.A. Artificial intelligence in molecular medicine. New Engl. J. Med. 2023, 388, 2456–2465. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Sedlazeck, F.J.; Lam, T.W.; Schatz, M.C. A multi-task convolutional deep neural network for variant calling in single molecule sequencing. Nat. Commun. 2019, 10, 998. [Google Scholar] [CrossRef] [PubMed]
- Kawash, J.K.; Smith, S.D.; Karaiskos, S.; Grigoriev, A. ARIADNA: Machine learning method for ancient DNA variant discovery. DNA Res. 2018, 25, 619–627. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tripodi, P. The Evolution of Molecular Genotyping in Plant Breeding. Agronomy 2023, 13, 2569. https://doi.org/10.3390/agronomy13102569
Tripodi P. The Evolution of Molecular Genotyping in Plant Breeding. Agronomy. 2023; 13(10):2569. https://doi.org/10.3390/agronomy13102569
Chicago/Turabian StyleTripodi, Pasquale. 2023. "The Evolution of Molecular Genotyping in Plant Breeding" Agronomy 13, no. 10: 2569. https://doi.org/10.3390/agronomy13102569
APA StyleTripodi, P. (2023). The Evolution of Molecular Genotyping in Plant Breeding. Agronomy, 13(10), 2569. https://doi.org/10.3390/agronomy13102569




