Feasibility of Detecting Sweet Potato (Ipomoea batatas) Virus Disease from High-Resolution Imagery in the Field Using a Deep Learning Framework
Abstract
:1. Introduction
- We propose a novel, fast, one-stage, anchor-free object detector, SPVDet, and its scaled lightweight variant, SPVDet-Nano, which utilizes a single-level feature for simplicity and effectiveness.
- We introduce a bundle of slicing aided hyper inference (SAHI) technologies to bridge the performance gap when inferring on high-resolution images.
- We conduct extensive ablation studies and comparison experiments on benchmark datasets and our self-made SPVD dataset, demonstrating the advancements of SPVDet and SPVDet-Nano, and the feasibility of SPVD detection from ground and aerial RGB images, respectively.
2. Materials and Methods
2.1. Datasets
2.1.1. PASCAL Visual Object Classes (VOC) Challenge 2007 and 2012
2.1.2. Microsoft Common Objects in Context (MS COCO) 2017
2.1.3. SPVD Dataset
2.2. Proposed Method
2.2.1. Systematic Designs of SPVDet
2.2.2. Dealing with High-Resolution Imagery
2.2.3. Implementation Details
3. Results
3.1. Ablation Studies on SPVDet
3.1.1. Principal Components: Backbone, Feature Aggregation Module, and Detection Head
3.1.2. Hyperparameter Fine-Tuning: Dilation Rates, Number of Dynamic Blocks, and Loss Balancing Coefficients
3.2. Performance Comparison with Previous Works: Quantitative Assessment of Generic Object Detection on the MS COCO
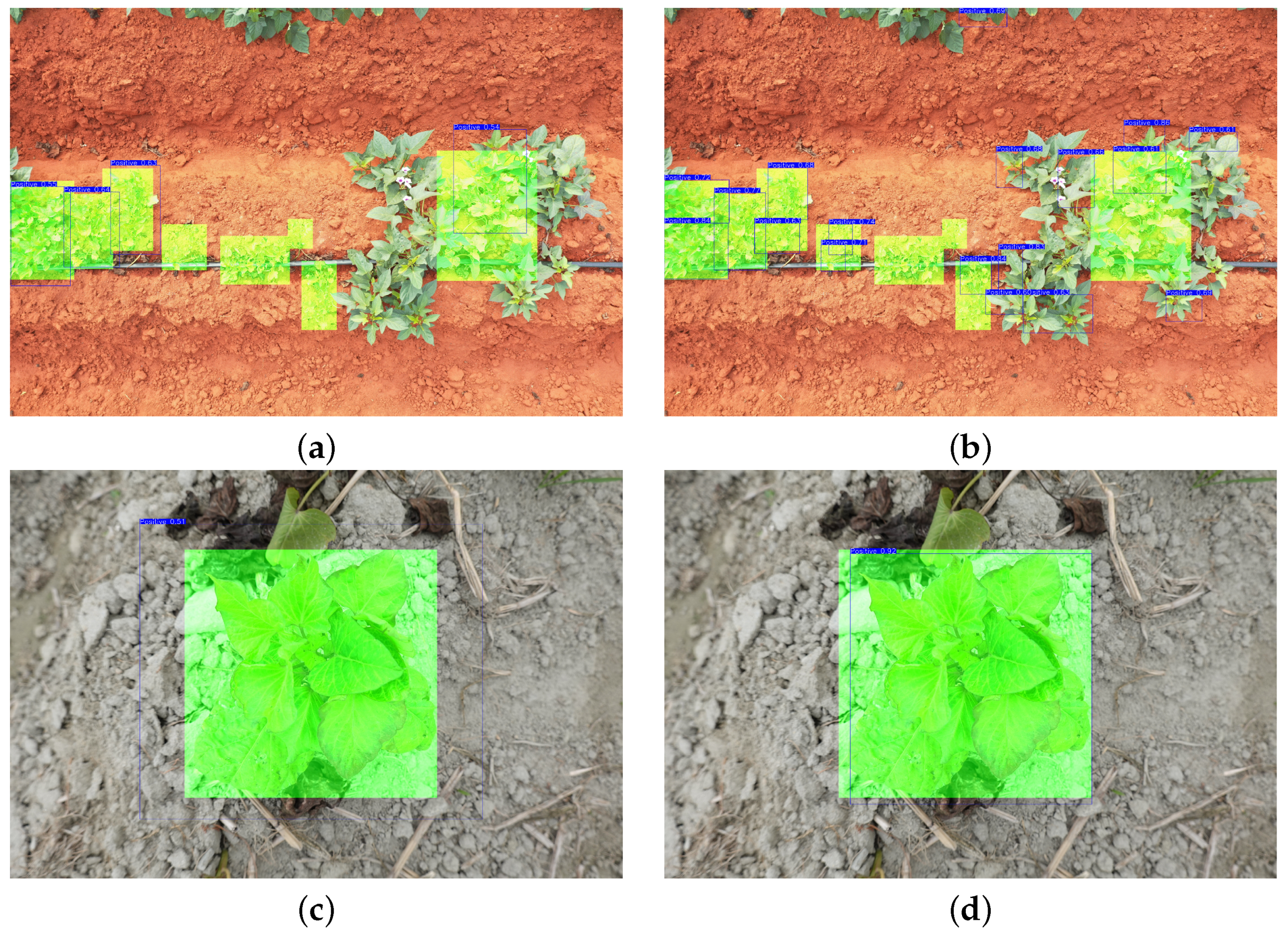
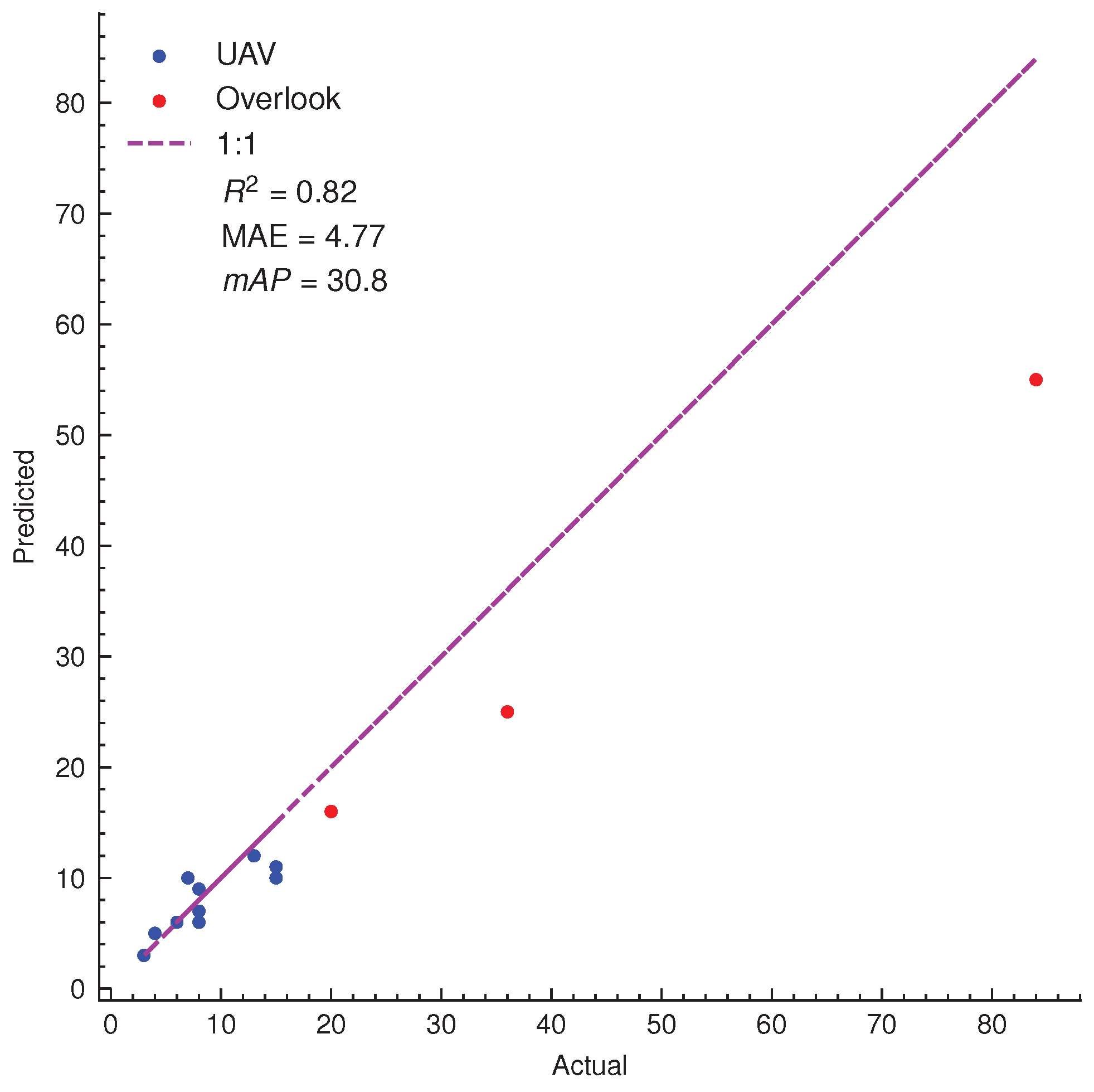
3.3. Assessments of SPVD Detection Performance on Plant Scale from High-Resolution Images in the Field
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. FAOSTAT. 2021. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 25 October 2023).
- Gai, F.; Gong, Y.; Zhang, P. Production and Deployment of Virus-Free Sweetpotato in China. Crop Prot. 2000, 19, 105–111. [Google Scholar] [CrossRef]
- Clark, C.A.; Hoy, M.W. Effects of Common Viruses on Yield and Quality of Beauregard Sweetpotato in Louisiana. Plant Dis. 2006, 90, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.A.C. Global Plant Virus Disease Pandemics and Epidemics. Plants 2021, 10, 233. [Google Scholar] [CrossRef] [PubMed]
- Clark, C.A.; Davis, J.A.; Abad, J.A.; Cuellar, W.J.; Fuentes, S.; Kreuze, J.F.; Gibson, R.W.; Mukasa, S.B.; Tugume, A.K.; Tairo, F.D.; et al. Sweetpotato Viruses: 15 Years of Progress on Understanding and Managing Complex Diseases. Plant Dis. 2012, 96, 168–185. [Google Scholar] [CrossRef]
- David, M.; Kante, M.; Fuentes, S.; Eyzaguirre, R.; Diaz, F.; De Boeck, B.; Mwanga, R.O.M.; Kreuze, J.; Grüneberg, W.J. Early-Stage Phenotyping of Sweet Potato Virus Disease Caused by Sweet Potato Chlorotic Stunt Virus and Sweet Potato Virus C to Support Breeding. Plant Dis. 2023, 107, 2061–2069. [Google Scholar] [CrossRef]
- Tatineni, S.; Hein, G.L. Plant Viruses of Agricultural Importance: Current and Future Perspectives of Virus Disease Management Strategies. Phytopathology 2023, 113, 117–141. [Google Scholar] [CrossRef]
- Wang, Y.M.; Ostendorf, B.; Gautam, D.; Habili, N.; Pagay, V. Plant Viral Disease Detection: From Molecular Diagnosis to Optical Sensing Technology—A Multidisciplinary Review. Remote Sens. 2022, 14, 1542. [Google Scholar] [CrossRef]
- Galieni, A.; D’Ascenzo, N.; Stagnari, F.; Pagnani, G.; Xie, Q.; Pisante, M. Past and Future of Plant Stress Detection: An Overview From Remote Sensing to Positron Emission Tomography. Front. Plant Sci. 2021, 11, 609155. [Google Scholar] [CrossRef]
- Singh, A.; Jones, S.; Ganapathysubramanian, B.; Sarkar, S.; Mueller, D.; Sandhu, K.; Nagasubramanian, K. Challenges and Opportunities in Machine-Augmented Plant Stress Phenotyping. Trends Plant Sci. 2021, 26, 53–69. [Google Scholar] [CrossRef]
- Ghosh, D.; Chakraborty, S.; Kodamana, H.; Chakraborty, S. Application of Machine Learning in Understanding Plant Virus Pathogenesis: Trends and Perspectives on Emergence, Diagnosis, Host-Virus Interplay and Management. Virol. J. 2022, 19, 42. [Google Scholar] [CrossRef]
- Sozzi, M.; Cantalamessa, S.; Cogato, A.; Kayad, A.; Marinello, F. Automatic Bunch Detection in White Grape Varieties Using YOLOv3, YOLOv4, and YOLOv5 Deep Learning Algorithms. Agronomy 2022, 12, 319. [Google Scholar] [CrossRef]
- Bouguettaya, A.; Zarzour, H.; Kechida, A.; Taberkit, A.M. A Survey on Deep Learning-Based Identification of Plant and Crop Diseases from UAV-based Aerial Images. Clust. Comput. 2023, 26, 1297–1317. [Google Scholar] [CrossRef]
- Kaur, S.; Pandey, S.; Goel, S. Plants Disease Identification and Classification through Leaf Images: A Survey. Arch. Comput. Methods Eng. 2019, 26, 507–530. [Google Scholar] [CrossRef]
- Sambasivam, G.; Opiyo, G.D. A Predictive Machine Learning Application in Agriculture: Cassava Disease Detection and Classification with Imbalanced Dataset Using Convolutional Neural Networks. Egypt. Inform. J. 2021, 22, 27–34. [Google Scholar] [CrossRef]
- Oishi, Y.; Habaragamuwa, H.; Zhang, Y.; Sugiura, R.; Asano, K.; Akai, K.; Shibata, H.; Fujimoto, T. Automated Abnormal Potato Plant Detection System Using Deep Learning Models and Portable Video Cameras. Int. J. Appl. Earth Obs. Geoinf. 2021, 104, 102509. [Google Scholar] [CrossRef]
- Wang, X.; Liu, J.; Zhu, X. Early Real-Time Detection Algorithm of Tomato Diseases and Pests in the Natural Environment. Plant Methods 2021, 17, 43. [Google Scholar] [CrossRef]
- Li, K.; Zhang, L.; Li, B.; Li, S.; Ma, J. Attention-Optimized DeepLab V3 + for Automatic Estimation of Cucumber Disease Severity. Plant Methods 2022, 18, 109. [Google Scholar] [CrossRef]
- Liu, L.; Ouyang, W.; Wang, X.; Fieguth, P.; Chen, J.; Liu, X.; Pietikäinen, M. Deep Learning for Generic Object Detection: A Survey. Int. J. Comput. Vis. 2020, 128, 261–318. [Google Scholar] [CrossRef]
- Sharma, V.; Mir, R.N. A Comprehensive and Systematic Look up into Deep Learning Based Object Detection Techniques: A Review. Comput. Sci. Rev. 2020, 38, 100301. [Google Scholar] [CrossRef]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar] [CrossRef]
- Chen, Z.; Wu, R.; Lin, Y.; Li, C.; Chen, S.; Yuan, Z.; Chen, S.; Zou, X. Plant Disease Recognition Model Based on Improved YOLOv5. Agronomy 2022, 12, 365. [Google Scholar] [CrossRef]
- Ma, L.; Yu, Q.; Yu, H.; Zhang, J. Maize Leaf Disease Identification Based on YOLOv5n Algorithm Incorporating Attention Mechanism. Agronomy 2023, 13, 521. [Google Scholar] [CrossRef]
- Mao, R.; Wang, Z.; Li, F.; Zhou, J.; Chen, Y.; Hu, X. GSEYOLOX-s: An Improved Lightweight Network for Identifying the Severity of Wheat Fusarium Head Blight. Agronomy 2023, 13, 242. [Google Scholar] [CrossRef]
- Ge, Z.; Liu, S.; Wang, F.; Li, Z.; Sun, J. YOLOX: Exceeding YOLO Series in 2021. arXiv 2021, arXiv:2107.08430. [Google Scholar]
- Liu, W.; Zhai, Y.; Xia, Y. Tomato Leaf Disease Identification Method Based on Improved YOLOX. Agronomy 2023, 13, 1455. [Google Scholar] [CrossRef]
- Kouadio, L.; El Jarroudi, M.; Belabess, Z.; Laasli, S.E.; Roni, M.Z.K.; Amine, I.D.I.; Mokhtari, N.; Mokrini, F.; Junk, J.; Lahlali, R. A Review on UAV-Based Applications for Plant Disease Detection and Monitoring. Remote Sens. 2023, 15, 4273. [Google Scholar] [CrossRef]
- Shahi, T.B.; Xu, C.Y.; Neupane, A.; Guo, W. Recent Advances in Crop Disease Detection Using UAV and Deep Learning Techniques. Remote Sens. 2023, 15, 2450. [Google Scholar] [CrossRef]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal Loss for Dense Object Detection. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2999–3007. [Google Scholar] [CrossRef]
- Tian, Z.; Shen, C.; Chen, H.; He, T. FCOS: Fully Convolutional One-Stage Object Detection. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Republic of Korea, 27 October–2 November 2019; pp. 9626–9635. [Google Scholar] [CrossRef]
- Redmon, J.; Farhadi, A. YOLOv3: An Incremental Improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar]
- Bochkovskiy, A.; Wang, C.Y.; Liao, H.Y.M. YOLOv4: Optimal Speed and Accuracy of Object Detection. arXiv 2020, arXiv:2004.10934. [Google Scholar]
- Chen, Q.; Wang, Y.; Yang, T.; Zhang, X.; Cheng, J.; Sun, J. You Only Look One-level Feature. In Proceedings of the 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, TN, USA, 20–25 June 2021; pp. 13034–13043. [Google Scholar] [CrossRef]
- Yang, J.; Wang, K.; Li, R.; Qin, Z.; Perner, P. A Novel Fast Combine-and-Conquer Object Detector Based on Only One-Level Feature Map. Comput. Vis. Image Underst. 2022, 224, 103561. [Google Scholar] [CrossRef]
- Dai, X.; Chen, Y.; Xiao, B.; Chen, D.; Liu, M.; Yuan, L.; Zhang, L. Dynamic Head: Unifying Object Detection Heads with Attentions. In Proceedings of the 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, TN, USA, 20–25 June 2021; pp. 7369–7378. [Google Scholar] [CrossRef]
- Everingham, M.; Van Gool, L.; Williams, C.K.I.; Winn, J.; Zisserman, A. The Pascal Visual Object Classes (VOC) Challenge. Int. J. Comput. Vis. 2010, 88, 303–338. [Google Scholar] [CrossRef]
- Lin, T.Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft COCO: Common Objects in Context. In Proceedings of the Computer Vision—ECCV, Zurich, Switzerland, 6–12 September 2014; Lecture Notes in Computer Science. Fleet, D., Pajdla, T., Schiele, B., Tuytelaars, T., Eds.; Springer: Cham, Switzerland, 2014; pp. 740–755. [Google Scholar] [CrossRef]
- Akyon, F.C.; Onur Altinuc, S.; Temizel, A. Slicing Aided Hyper Inference and Fine-Tuning for Small Object Detection. In Proceedings of the 2022 IEEE International Conference on Image Processing (ICIP), Bordeaux, France, 16–19 October 2022; pp. 966–970. [Google Scholar] [CrossRef]
- Zhang, L.; Gao, X. Transfer Adaptation Learning: A Decade Survey. IEEE Trans. Neural Netw. Learn. Syst. 2022, 1–22. [Google Scholar] [CrossRef]
- Gulzar, Y. Fruit Image Classification Model Based on MobileNetV2 with Deep Transfer Learning Technique. Sustainability 2023, 15, 1906. [Google Scholar] [CrossRef]
- Gulzar, Y.; Ünal, Z.; Aktaş, H.; Mir, M.S. Harnessing the Power of Transfer Learning in Sunflower Disease Detection: A Comparative Study. Agriculture 2023, 13, 1479. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Wang, C.Y.; Bochkovskiy, A.; Liao, H.Y.M. YOLOv7: Trainable Bag-of-Freebies Sets New State-of-the-Art for Real-Time Object Detectors. arXiv 2022, arXiv:2207.02696. [Google Scholar]
- Feng, C.; Zhong, Y.; Gao, Y.; Scott, M.R.; Huang, W. TOOD: Task-aligned One-stage Object Detection. In Proceedings of the 2021 IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, BC, Canada, 11–17 October 2021; pp. 3490–3499. [Google Scholar] [CrossRef]
- Li, X.; Lv, C.; Wang, W.; Li, G.; Yang, L.; Yang, J. Generalized Focal Loss: Towards Efficient Representation Learning for Dense Object Detection. IEEE Trans. Pattern Anal. Mach. Intell. 2023, 45, 3139–3153. [Google Scholar] [CrossRef]
- Rezatofighi, H.; Tsoi, N.; Gwak, J.; Sadeghian, A.; Reid, I.; Savarese, S. Generalized Intersection Over Union: A Metric and a Loss for Bounding Box Regression. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 658–666. [Google Scholar] [CrossRef]
- Duan, K.; Bai, S.; Xie, L.; Qi, H.; Huang, Q.; Tian, Q. CenterNet: Keypoint Triplets for Object Detection. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Republic of Korea, 27 October–2 November 2019; pp. 6568–6577. [Google Scholar] [CrossRef]
- Huang, H.; Han, H.; Lei, Y.; Qiao, H.; Tang, D.; Han, Y.; Deng, Z.; Mao, L.; Wu, X.; Zhang, K.; et al. Application of Grafting Method in Resistance Identification of Sweet Potato Virus Disease and Resistance Evaluation of Elite Sweet Potato [Ipomoea batatas (L.) Lam] Varieties. Plants 2023, 12, 957. [Google Scholar] [CrossRef]
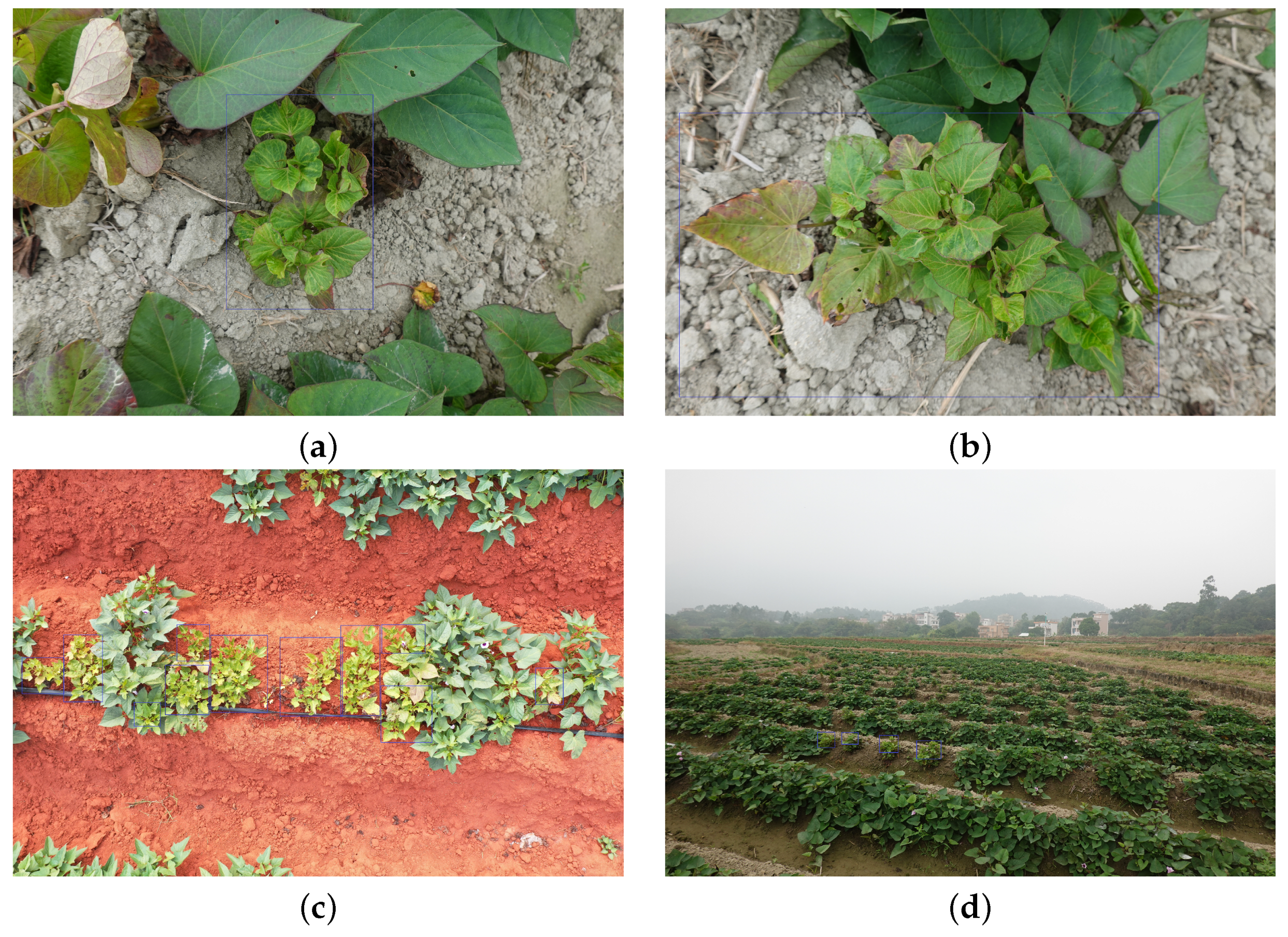
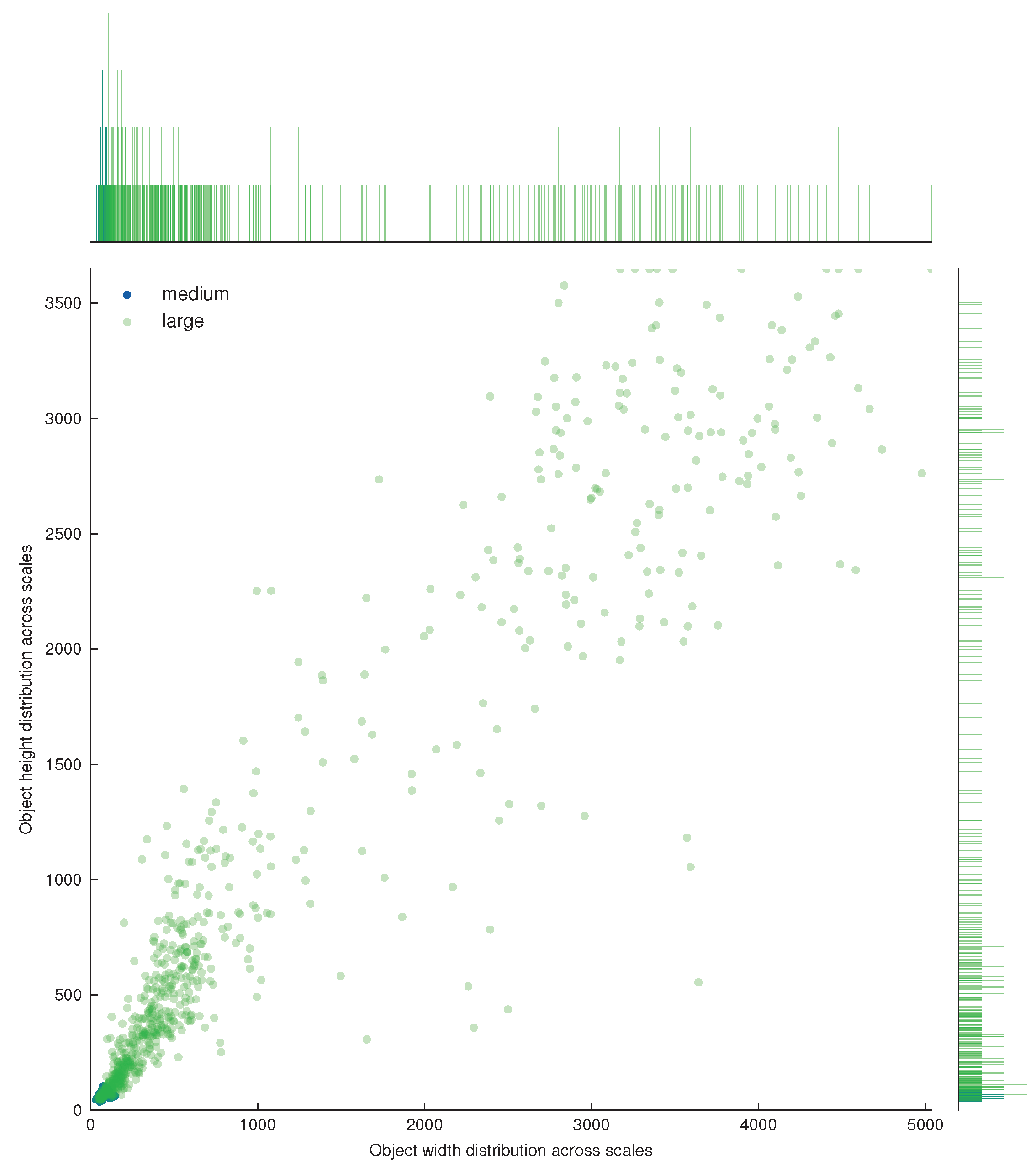

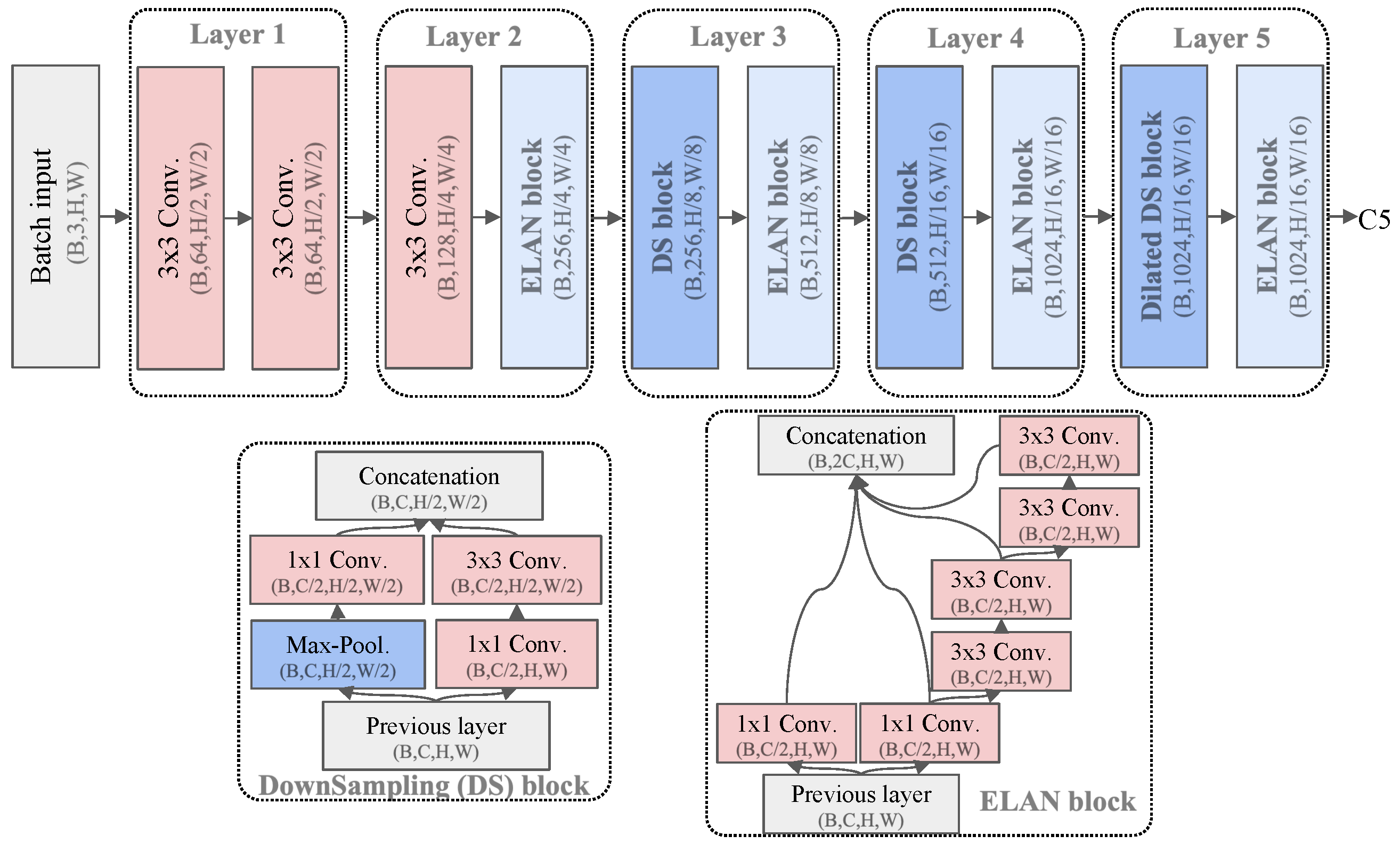
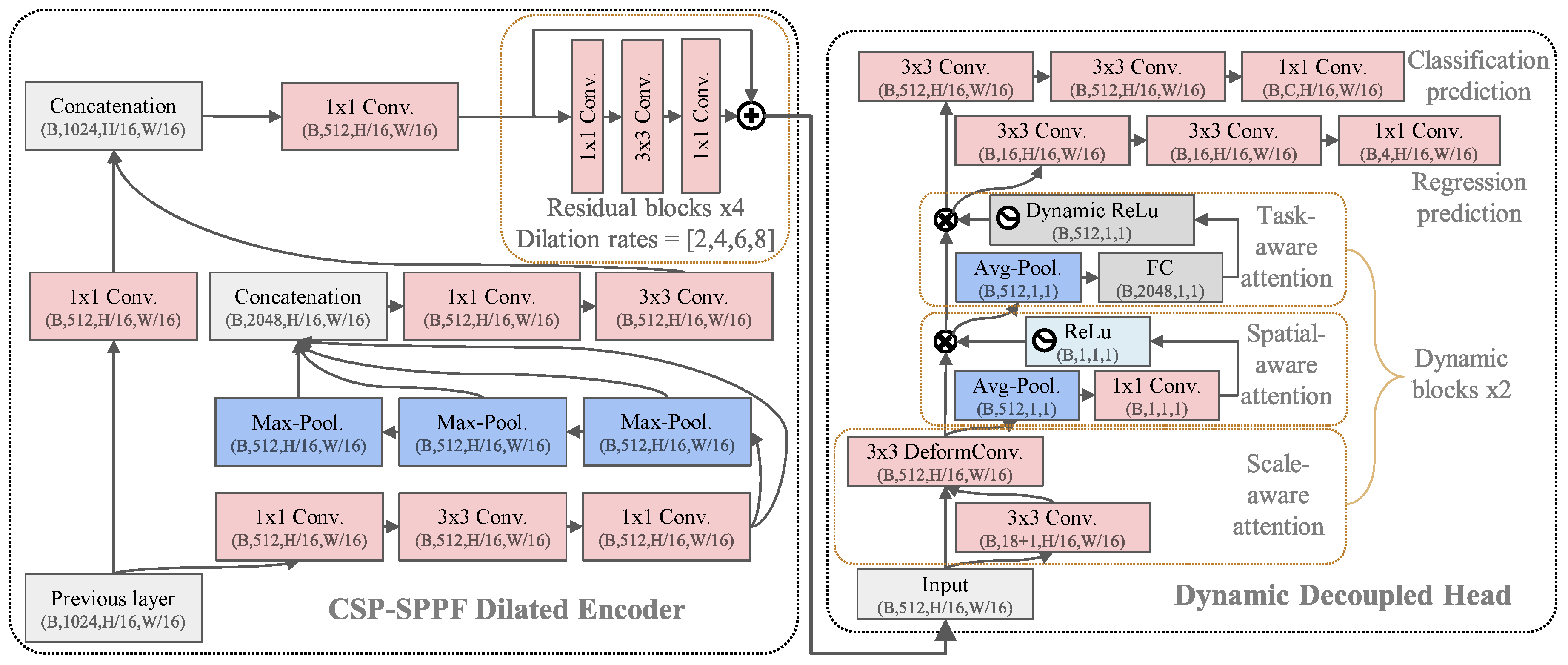
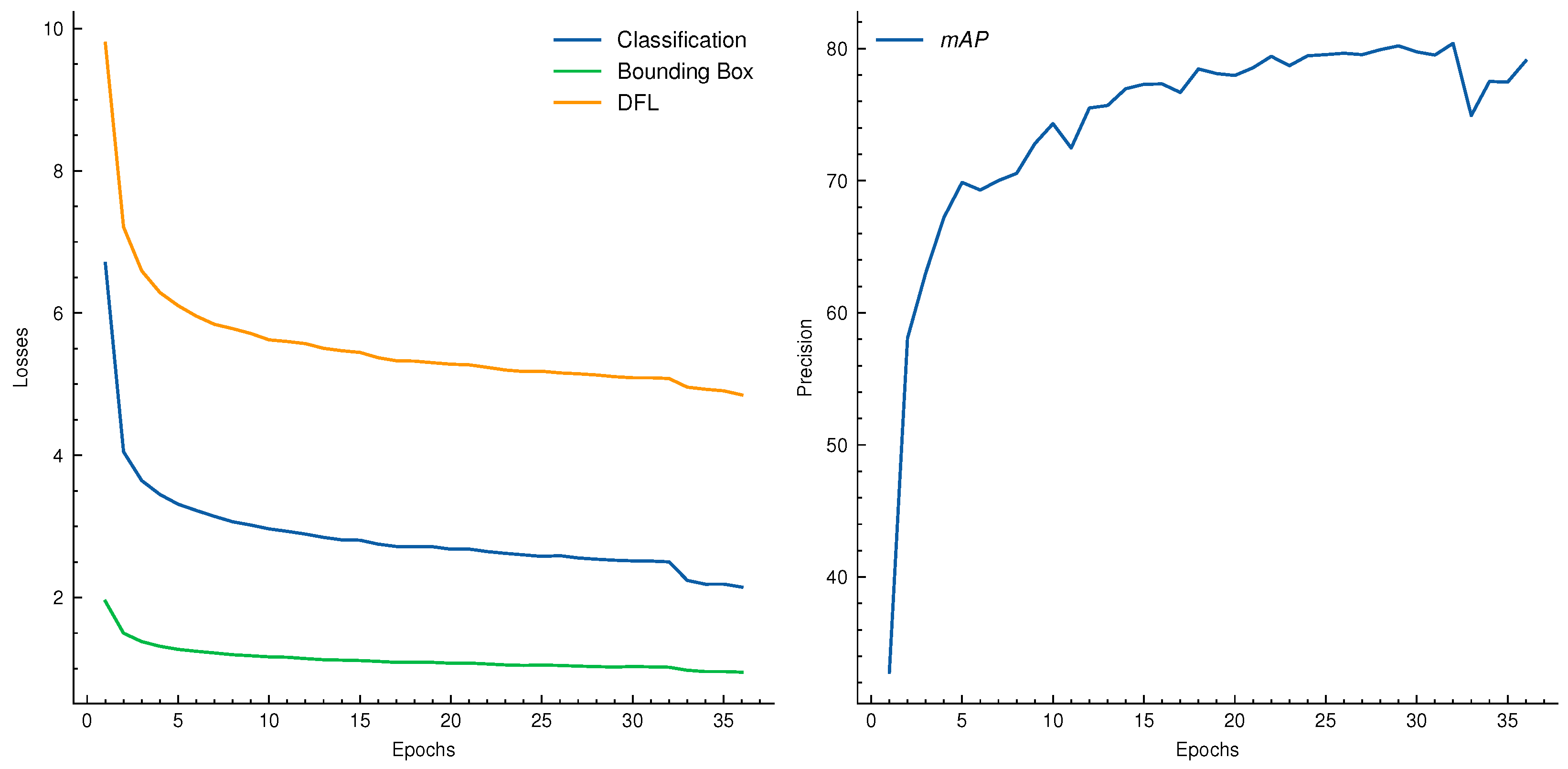
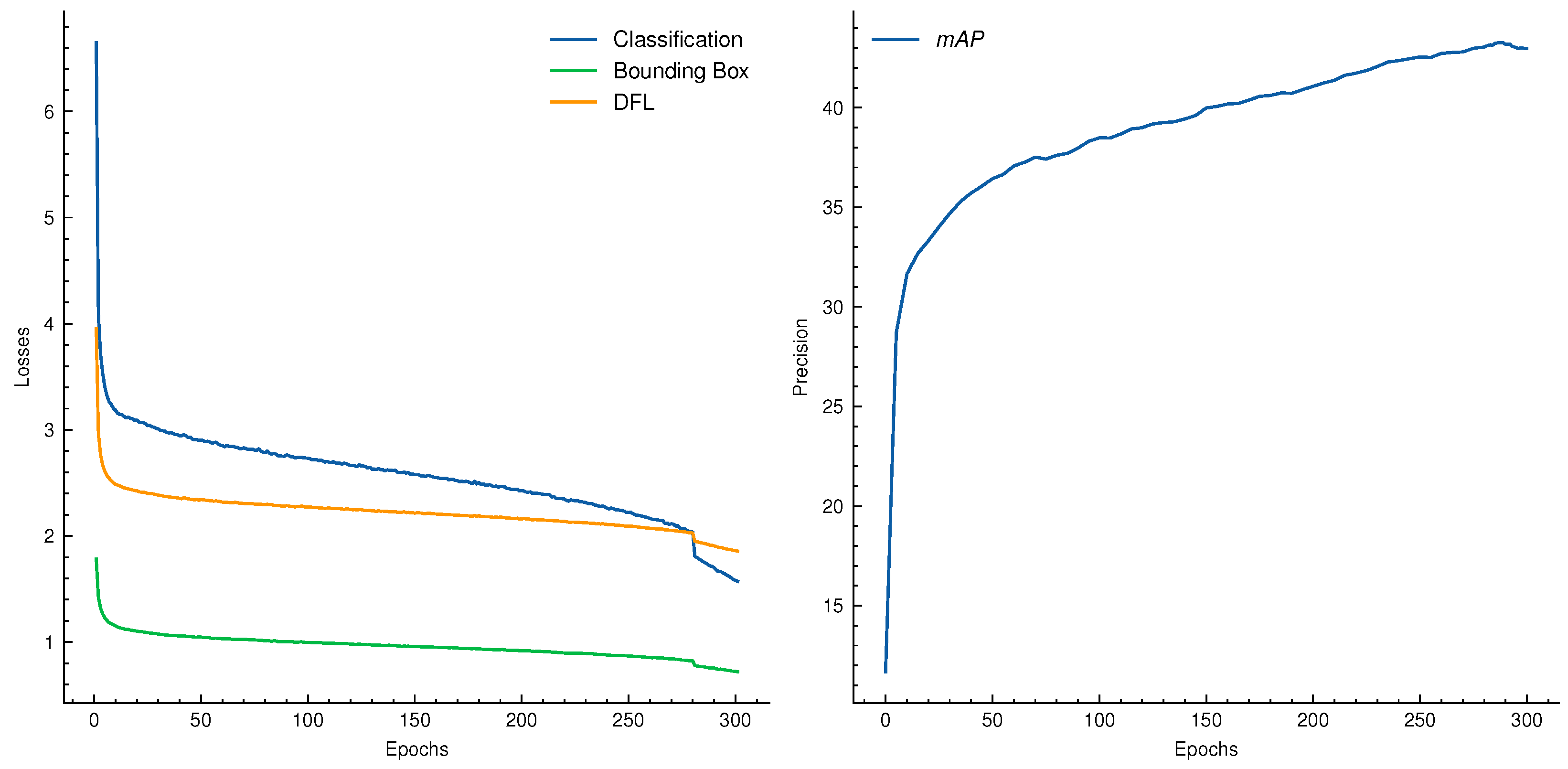
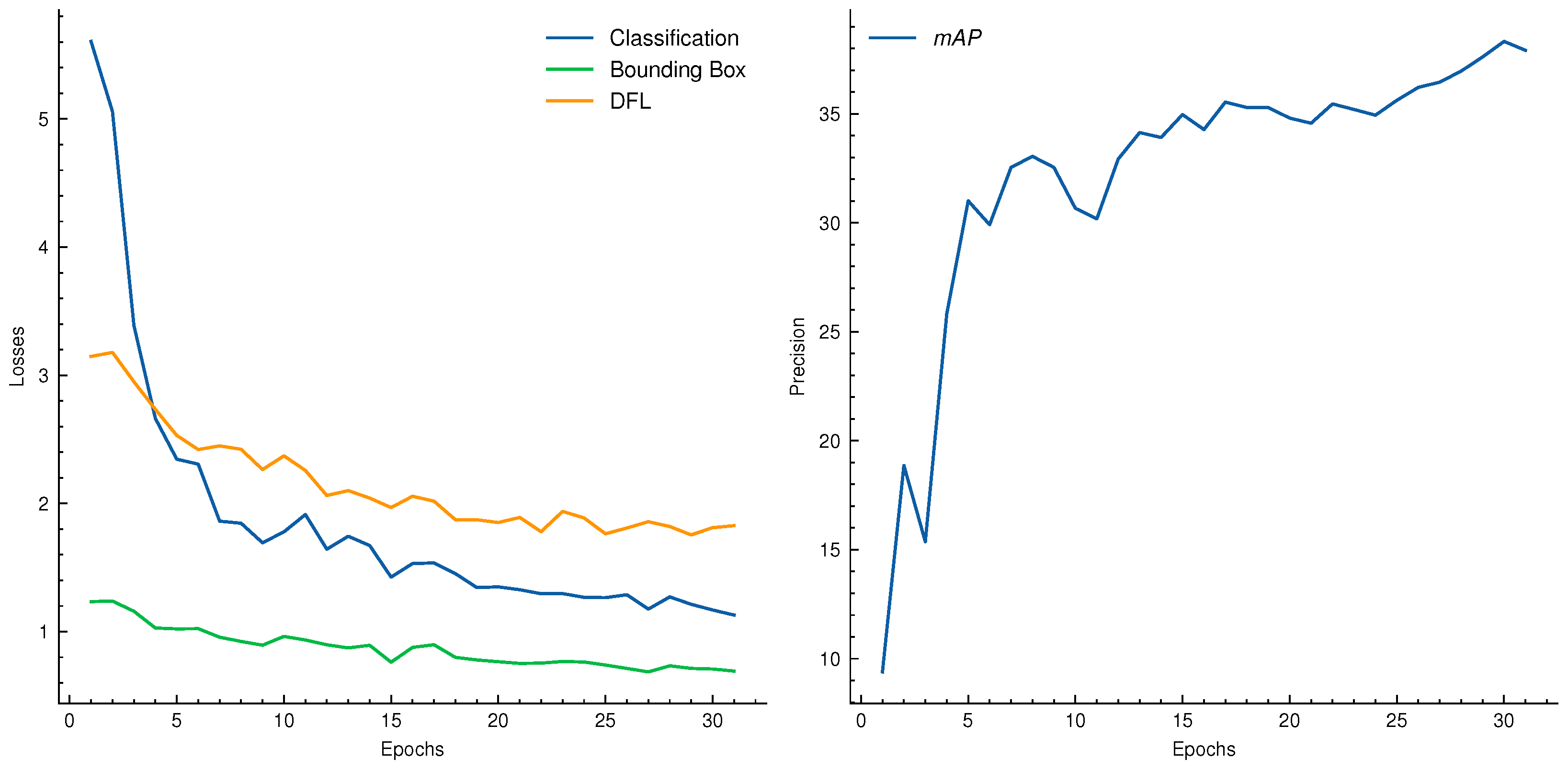
| Dataset | Subset | Resolution | No. Images | No. BBoxes | Small | Medium | Large |
|---|---|---|---|---|---|---|---|
| SPVD | training | 5472 × 3648 (0.24 cm/pixel) | 211 | 748 | 0 | 69 | 679 |
| validation | 54 | 161 | 0 | 15 | 146 | ||
| test | 68 | 295 | 1 | 45 | 249 |
| Backbone | Feature Aggregation | Detection Head | Backbone Performance | Detector Performance | ||||
|---|---|---|---|---|---|---|---|---|
| Top1-Acc | FLOPs | Params | mAP | FLOPs | Params | |||
| ResNet-50 | Vanilla DE | Vanilla DH | 76.1% | 90 G | 20 M | 76.04 | 118 G | 29 M |
| Dynamic DH | 76.46 | 119 G | 30 M | |||||
| CSP-SPPF DE | Vanilla DH | 77.54 | 133 G | 34 M | ||||
| Dynamic DH | 77.70 | 134 G | 34 M | |||||
| CSPDarkNet-53 | Vanilla DE | Vanilla DH | 75.0% | 125 G | 27 M | 78.18 | 153 G | 36 M |
| Dynamic DH | 78.54 | 153 G | 37 M | |||||
| CSP-SPPF DE | Vanilla DH | 79.12 | 168 G | 41 M | ||||
| Dynamic DH | 79.33 | 168 G | 42 M | |||||
| CSPDarkNet-L | Vanilla DE | Vanilla DH | 75.1% | 118 G | 27 M | 79.31 | 146 G | 37 M |
| Dynamic DH | 79.41 | 146 G | 37 M | |||||
| CSP-SPPF DE | Vanilla DH | 79.79 | 161 G | 41 M | ||||
| Dynamic DH | 80.08 | 161 G | 42 M | |||||
| CSPELANNet | Vanilla DE | Vanilla DH | 75.8% | 102 G | 19 M | 79.64 | 129 G | 28 M |
| Dynamic DH | 80.03 | 129 G | 29 M | |||||
| CSP-SPPF DE | Vanilla DH | 80.34 | 144 G | 32 M | ||||
| Dynamic DH | 80.57 | 144 G | 33 M | |||||
| Dilation Rates | Number of Dynamic Head Blocks | Loss Balancing Coefficients | FLOPs | Params | mAP | ||
|---|---|---|---|---|---|---|---|
| Classification | IoU | DFL | |||||
| [1, 2, 3, 4] | 2 | 1.0 | 1.0 | 1.0 | 138 G | 31 M | 79.97 |
| 1.0 | 5.0 | 1.5 | 81.17 | ||||
| 1.0 | 7.5 | 2.0 | 80.71 | ||||
| 1.0 | 10.0 | 2.5 | 80.02 | ||||
| 4 | 1.0 | 1.0 | 1.0 | 139 G | 32 M | 80.12 | |
| 1.0 | 5.0 | 1.5 | 80.98 | ||||
| 1.0 | 7.5 | 2.0 | 80.89 | ||||
| 1.0 | 10.0 | 2.5 | 80.68 | ||||
| [2, 4, 6, 8] | 2 | 1.0 | 1.0 | 1.0 | 138 G | 31 M | 80.12 |
| 1.0 | 5.0 | 1.5 | 80.66 | ||||
| 1.0 | 7.5 | 2.0 | 80.85 | ||||
| 1.0 | 10.0 | 2.5 | 80.07 | ||||
| 4 | 1.0 | 1.0 | 1.0 | 139 G | 32 M | 80.43 | |
| 1.0 | 5.0 | 1.5 | 80.86 | ||||
| 1.0 | 7.5 | 2.0 | 80.76 | ||||
| 1.0 | 10.0 | 2.5 | 80.52 | ||||
| [4, 6, 8, 10] | 2 | 1.0 | 1.0 | 1.0 | 138 G | 31 M | 80.05 |
| 1.0 | 5.0 | 1.5 | 80.92 | ||||
| 1.0 | 7.5 | 2.0 | 80.94 | ||||
| 1.0 | 10.0 | 2.5 | 80.50 | ||||
| 4 | 1.0 | 1.0 | 1.0 | 139 G | 32 M | 80.39 | |
| 1.0 | 5.0 | 1.5 | 80.75 | ||||
| 1.0 | 7.5 | 2.0 | 80.98 | ||||
| 1.0 | 10.0 | 2.5 | 80.64 | ||||
| Category | Model | Backbone | Size | AP | FPS | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| One Level | CenterNet | ResNet101 | 512 × 512 | 34.6 | 53.0 | 36.9 | - | - | - | 45 |
| YOLO | ResNet101 | 8,001,333 | 39.8 | 59.4 | 42.9 | 20.5 | 45.5 | 54.9 | 21 | |
| CC-Det | ResNet101 | 512 × 512 | 40.6 | 59.4 | 44.2 | 22.6 | 45.7 | 55.1 | 50 | |
| Feature Pyramid | RetinaNet | ResNet-101-FPN | 8,001,333 | 39.1 | 59.1 | 42.3 | 21.8 | 42.7 | 50.2 | 15 |
| FCOS | ResNet-101-FPN | 8,001,333 | 41.5 | 60.7 | 45.0 | 24.4 | 44.8 | 51.6 | 17 | |
| YOLOv3 | DarkNet-53 | 608 × 608 | 33.0 | 57.9 | 34.4 | 18.3 | 35.4 | 41.9 | 76 | |
| YOLOv4 | CSPDarkNet-53 | 608 × 608 | 43.5 | 65.7 | 47.3 | 26.7 | 46.7 | 53.3 | 57 | |
| One Level | SPVDet (ours) | CSPELANNet | 512 × 512 | 41.8 | 59.1 | 44.9 | 18.6 | 46.7 | 64.7 | 180 |
| 608 × 608 | 43.8 | 62.3 | 47.5 | 22.3 | 50.3 | 66.6 | 157 | |||
| SPVDet-Nano (ours) | CSPELANNet-Nano | 512 × 512 | 31.1 | 47.7 | 32.7 | 9.4 | 33.2 | 53.5 | 245 | |
| 608 × 608 | 33.8 | 51.4 | 35.8 | 12.8 | 37.5 | 54.4 | 232 |
| Model Setup | Metric Threshold | Patch Size = 640 | Patch Size = 480 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AP | AP | ||||||||||||
| SPVDet + Fl | - | 16.8 | 33.0 | 15.5 | 0.0 | 14.3 | 17.8 | 16.8 | 33.0 | 15.5 | 0.0 | 14.3 | 17.8 |
| SPVDet + SAHI + Fl | IoS = 0.5 | 30.8 | 47.8 | 32.0 | 60.0 | 7.8 | 34.8 | 26.0 | 38.3 | 26.5 | 60.0 | 4.5 | 29.8 |
| IoU = 0.5 | 28.2 | 42.3 | 29.5 | 60.0 | 7.8 | 31.9 | 25.8 | 39.0 | 25.9 | 60.0 | 4.5 | 29.6 | |
| SPVDet-Nano + Fl | - | 12.2 | 34.1 | 4.8 | 0.0 | 1.7 | 14.1 | 12.2 | 34.1 | 4.8 | 0.0 | 1.7 | 14.1 |
| SPVDet-Nano + SAHI + Fl | IoS = 0.5 | 15.5 | 25.4 | 16.9 | 0.0 | 2.7 | 17.9 | 13.3 | 21.1 | 14.2 | 0.0 | 1.6 | 15.6 |
| IoU = 0.5 | 15.1 | 24.3 | 16.4 | 0.0 | 2.2 | 17.5 | 11.1 | 17.6 | 11.8 | 0.0 | 1.8 | 12.8 | |
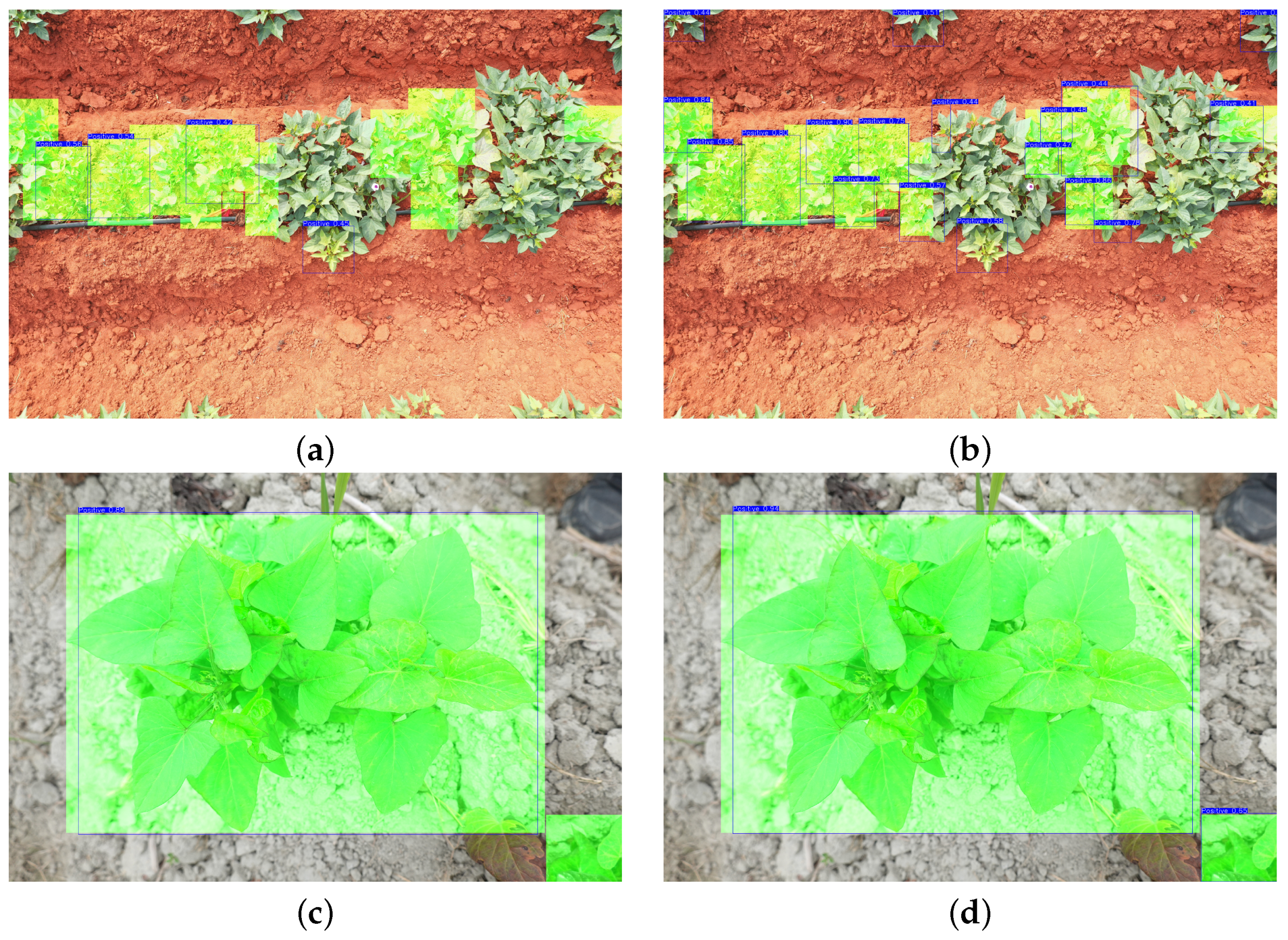
| Predicted | Close-Up View | UAV View | Overlook View | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Actual | Accuracy | Actual | Accuracy | Actual | Accuracy | ||||
| Foreground | Background | Foreground | Background | Foreground | Background | ||||
| Foreground | 57 | 6 | 78.1% | 72 | 7 | 76.6% | 84 | 12 | 55.3% |
| Background | 11 | Null | 15 | Null | 56 | Null | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, F.; Ding, Z.; Song, Q.; Xiao, J.; Zheng, J.; Li, H.; Luo, Z.; Wang, Z.; Yue, X.; Huang, L. Feasibility of Detecting Sweet Potato (Ipomoea batatas) Virus Disease from High-Resolution Imagery in the Field Using a Deep Learning Framework. Agronomy 2023, 13, 2801. https://doi.org/10.3390/agronomy13112801
Zeng F, Ding Z, Song Q, Xiao J, Zheng J, Li H, Luo Z, Wang Z, Yue X, Huang L. Feasibility of Detecting Sweet Potato (Ipomoea batatas) Virus Disease from High-Resolution Imagery in the Field Using a Deep Learning Framework. Agronomy. 2023; 13(11):2801. https://doi.org/10.3390/agronomy13112801
Chicago/Turabian StyleZeng, Fanguo, Ziyu Ding, Qingkui Song, Jiayi Xiao, Jianyu Zheng, Haifeng Li, Zhongxia Luo, Zhangying Wang, Xuejun Yue, and Lifei Huang. 2023. "Feasibility of Detecting Sweet Potato (Ipomoea batatas) Virus Disease from High-Resolution Imagery in the Field Using a Deep Learning Framework" Agronomy 13, no. 11: 2801. https://doi.org/10.3390/agronomy13112801
APA StyleZeng, F., Ding, Z., Song, Q., Xiao, J., Zheng, J., Li, H., Luo, Z., Wang, Z., Yue, X., & Huang, L. (2023). Feasibility of Detecting Sweet Potato (Ipomoea batatas) Virus Disease from High-Resolution Imagery in the Field Using a Deep Learning Framework. Agronomy, 13(11), 2801. https://doi.org/10.3390/agronomy13112801






