Pyramiding Rice Blast Resistance Gene Pi2 and Fragrance Gene badh2
Abstract
:1. Introduction
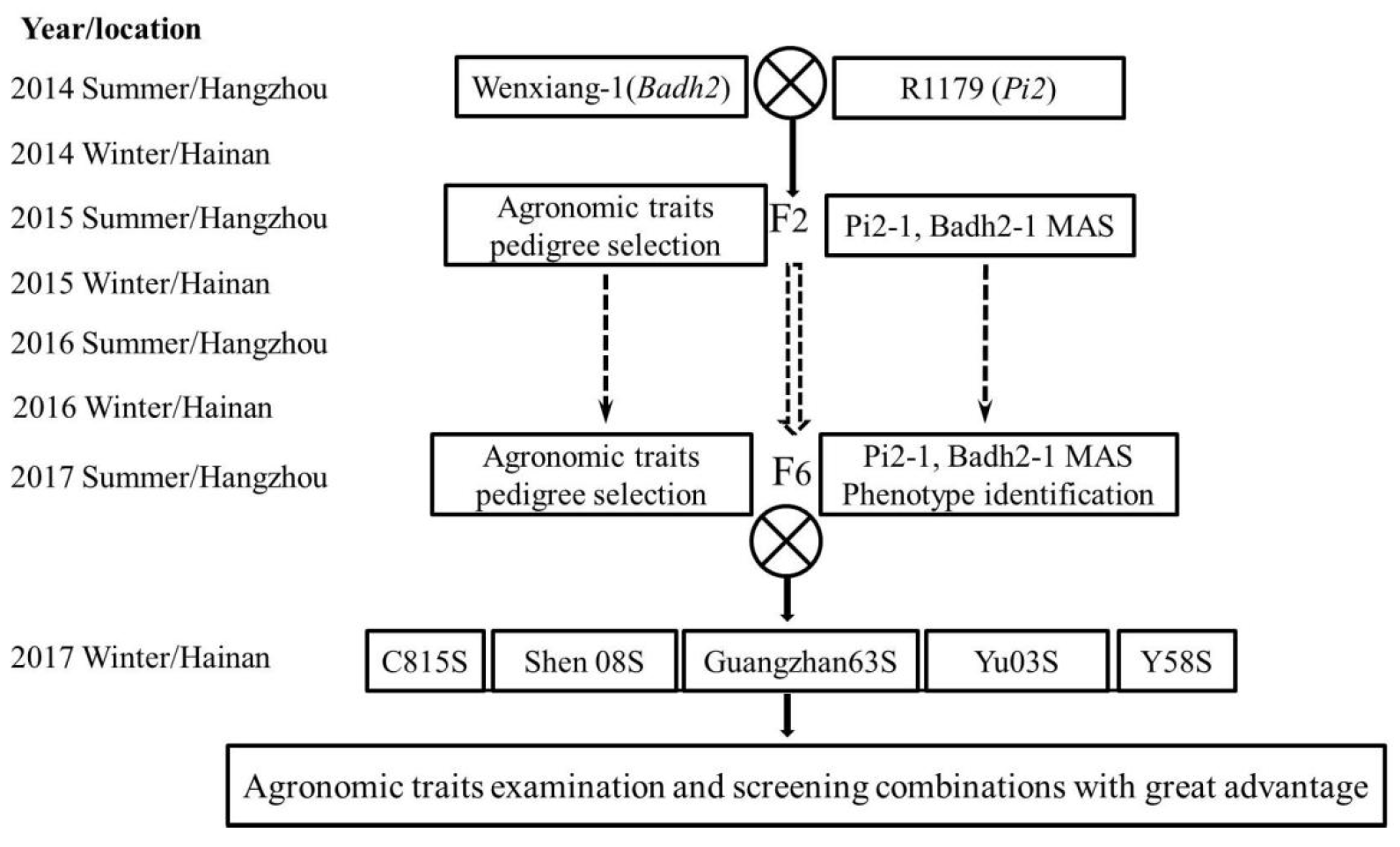
2. Materials and Methods
2.1. Experimental Materials
2.2. Phenotype Identification of Fragrance and the Amylose Content of Parents and Selected New Rice Varieties
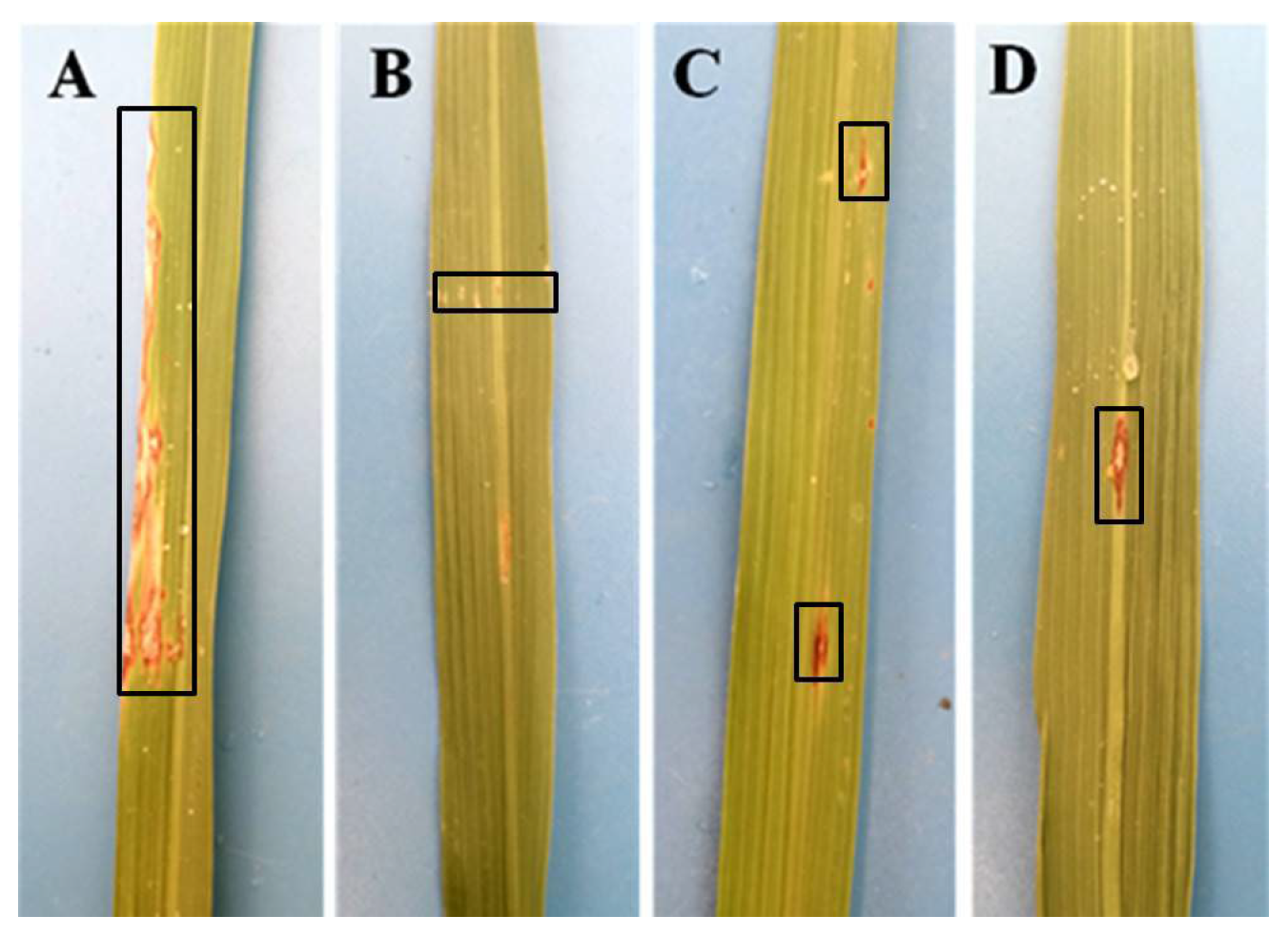
2.3. Phenotype Identification of Rice Blast Resistance
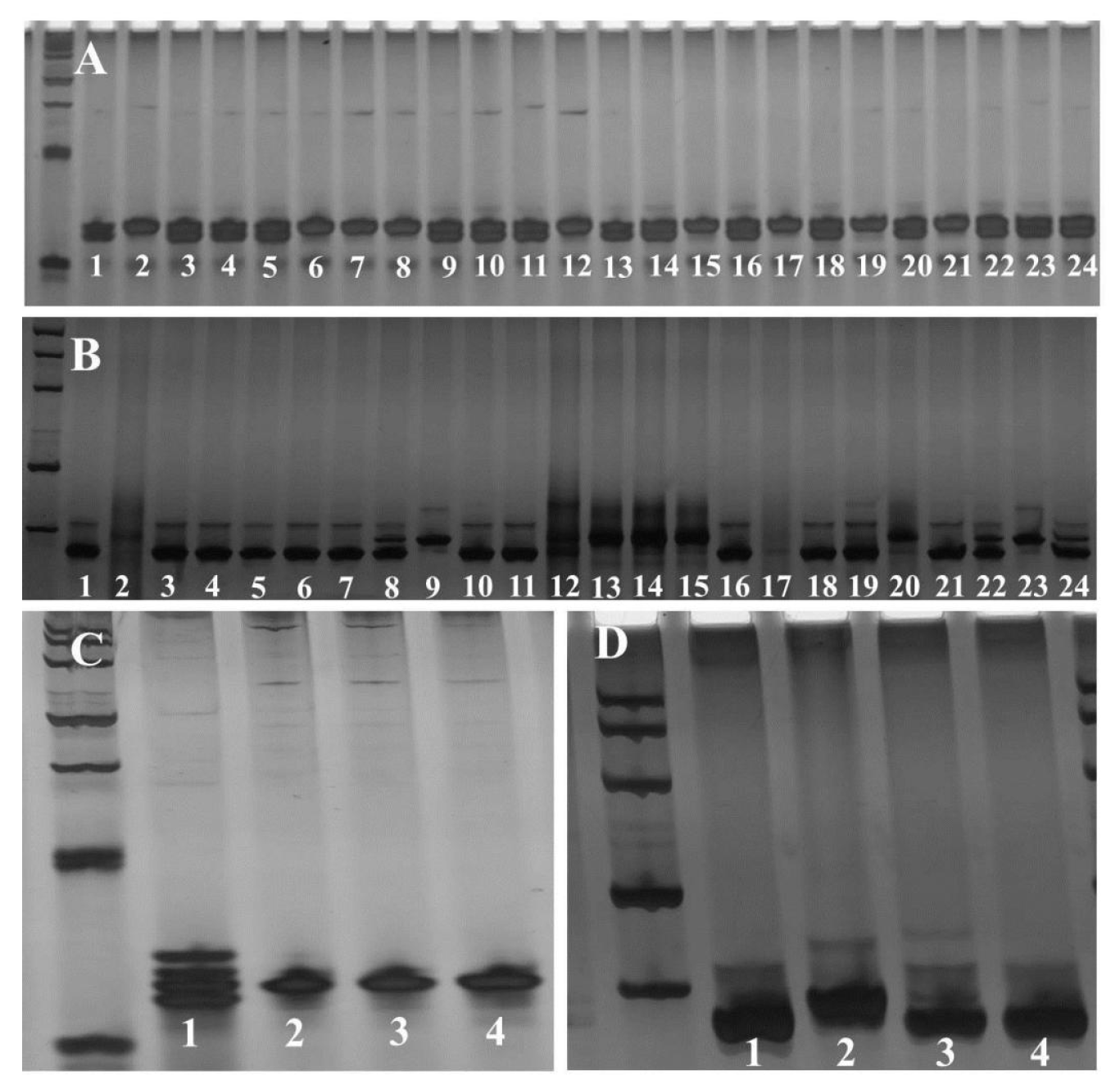
2.4. Genotype Classification and Genetic Composition Analysis among Segregation Population Individuals and Selected Elite Rice Varieties
2.5. MAS of Rice Varieties with Both Fragrance and High-Level Rice Blast Resistance
2.6. Evaluation of the New Hybrid Rice Lines
3. Results
3.1. Agronomic Traits of the Two Parents
3.2. MAS of Newly Bred Rice Restore Lines Using Functional Markers Pi2-1 and Badh2-1
3.3. Rice Blast Resistance Identification and Fragrance Examination
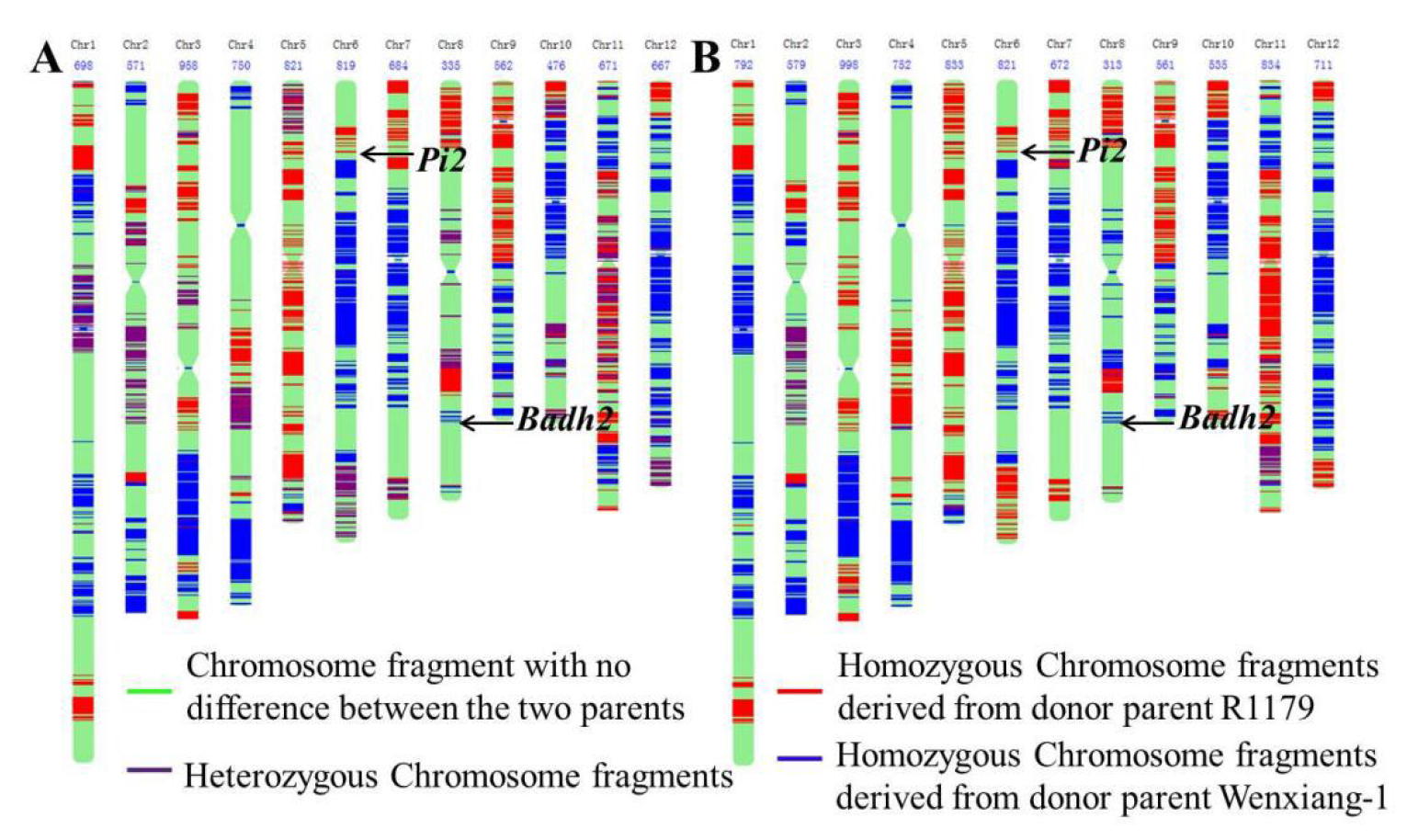
3.4. Genotype Identification and Genetic Composition Analysis
3.5. The Agronomic Performance of Hybrid Rice Using R365 and R403 as the Male Parents
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sheng, Z.; Li, Q.; Li, W.; Chen, X.; Wei, L.; Xie, G.; Jiao, G.; Shao, J.; Wang, S. Identification of a three-base deletion in the Pi2 locus, and development of functional marker for marker-assisted resistance selection. Euphytica 2017, 213, 202. [Google Scholar] [CrossRef]
- Khush, G.; Jena, K. Current status and future prospects for research on blast resistance in rice (Oryza sativa L.). In Advances in Genetics, Genomics and Control of Rice Blast Disease; Wang, G.L., Valent, B., Eds.; Springer: New York, NY, USA, 2009; pp. 1–10. [Google Scholar]
- Talbot, N.J. On the Trail of a Cereal Killer: Exploring the Biology of Magnaporthe grisea. Annu. Rev. Microbiol. 2003, 57, 177–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skamnioti, P.; Gurr, S. Against the Grain: Safe Guarding Rice from Rice Blast Disease. Trends Biotechnol. 2009, 27, 141–150. [Google Scholar] [CrossRef] [Green Version]
- Feng, X.; Wang, C.; Nan, J.; Zhang, X.; Wang, R.; Jiang, G.; Yuan, Q.; Lin, S. Updating the elite rice variety Kongyu 131 by improving the Gn1a Locus. Rice 2017, 10, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angeles-Shim, R.B.; Reyes, V.P.; del Valle, M.M.; Lapis, R.S.; Shim, J.; Sunohara, H.; Jena, K.K.; Ashikari, M.; Doi, K. Marker-Assisted Introgression of Quantitative Resistance Gene Pi21 Confers Broad Spectrum Resistance to Rice Blast. Rice Sci. 2020, 27, 113–123. [Google Scholar] [CrossRef]
- Thanasilungura, K.; Kranto, S.; Monkham, T.; Chankaew, S.; Sanitchon, J. Improvement of a RD6 Rice Variety for Blast Resistance and Salt Tolerance through Marker-Assisted Backcrossing. Agronomy 2020, 10, 1118. [Google Scholar] [CrossRef]
- Hayashi, K.; Yoshida, H. Refunctionalization of the Ancient Rice Blast Disease Resistance Gene Pit by the Recruitment of a Retrotransposon as a Promoter. Plant J. 2009, 57, 413–425. [Google Scholar] [CrossRef]
- Lin, F.; Chen, S.; Que, Z.Q.; Wang, L.; Liu, X.Q.; Pan, Q.H. The Blast Resistance Gene Pi37 Encodes a Nucleotide Binding Site-Leucine-Rich Repeat Protein and Is a Member of a Resistance Gene Cluster on Rice Chromosome 1. Genetics 2007, 177, 1871–1880. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, A.; Hayashi, N.; Miyao, A.; Hirochika, H. Unique Features of the Rice Blast Resistance Pish Locus Revealed by Large Scale Retrotransposon-Tagging. BMC Plant Biol. 2010, 10, 175. [Google Scholar] [CrossRef] [Green Version]
- Fukuoka, S.; Yamamoto, S.-I.; Mizobuchi, R.; Yamanouchi, U.; Ono, K.; Kitazawa, N.; Yasuda, N.; Fujita, Y.; Nguyen, T.T.T.; Koizumi, S.; et al. Multiple functional polymorphisms in a single disease resistance gene in rice enhance durable resistance to blast. Sci. Rep. 2014, 4, 4550. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Lei, C.L.; Xu, X.T.; Hao, K.; Wang, J.L.; Cheng, Z.J.; Ma, X.D.; Zhou, K.N.; Zhang, X.; Guo, X.P.; et al. Pi64, Encoding a Novel CC-NBS-LRR Protein, Confers Resistance to Leaf and Neck Blast in Rice. Mol. Plant Microbe Interact. 2015, 28, 558–568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.-X.; Yano, M.; Yamanouchi, U.; Iwamoto, M.; Monna, L.; Hayasaka, H.; Katayose, Y.; Sasaki, T. The Pib Gene for Rice Blast Resistance Belongs to the Nucleotide Binding and Leucine-Rich Repeat Class of Plant Disease Resistance Genes. Plant J. 1999, 19, 55–64. [Google Scholar] [CrossRef]
- Li, W.T.; Zhu, Z.W.; Chern, M.S.; Yin, J.J.; Yang, C.; Ran, L.; Cheng, M.P.; He, M.; Wang, K.; Wang, J.; et al. A Natural Allele of a Transcription Factor in Rice Confers Broad-Spectrum Blast Resistance. Cell 2017, 170, 114–126.e15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukuoka, S.; Saka, N.; Koga, H.; Ono, K.; Shimizu, T.; Ebana, K.; Hayashi, N.; Takahashi, A.; Hirochika, H.; Okuno, K.; et al. Loss of Function of a Proline-Containing Protein Confers Durable Disease Resistance in Rice. Science 2009, 325, 998–1001. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Hayashi, N.; Wang, C.-T.; Fukuoka, S.; Kawasaki, S.; Takatsuji, H.; Jiang, C.-J. Rice Blast Resistance Gene Pikahei-1(t), a Member of a Resistance Gene Cluster on Chromosome 4, Encodes a Nucleotide-Binding Site and Leucine-Rich Repeat Protein. Mol. Breed. 2014, 34, 691–700. [Google Scholar] [CrossRef]
- Zhou, B.; Qu, S.H.; Liu, G.F.; Dolan, M.; Sakai, H.; Lu, G.D.; Bellizzi, M.; Wang, G.-L. The Eight Amino-Acid Differences within Three Leucine-Rich Repeats between Pi2 and Piz-t Resistance Proteins Determine the Resistance Specificity to Magnaporthe grisea. Mol. Plant Microbe Interact. 2006, 19, 1216–1228. [Google Scholar] [CrossRef] [Green Version]
- Qu, S.H.; Liu, G.F.; Zhou, B.; Bellizzi, M.; Zeng, L.R.; Dai, L.Y.; Han, B.; Wang, G.-L. The Broad-Spectrum Blast Resistance Gene Pi9 Encodes a Nucleotide-Binding Site—Leucine-Rich Repeat Protein and Is a Member of a Multigene Family in Rice. Genetics 2006, 172, 1901–1914. [Google Scholar] [CrossRef] [Green Version]
- Deng, Y.W.; Zhai, K.R.; Xie, Z.; Yang, D.Y.; Zhu, X.D.; Liu, J.Z.; Wang, X.; Qin, P.; Yang, Y.Z.; Zhang, G.M.; et al. Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 2017, 355, 962–965. [Google Scholar] [CrossRef]
- Chen, X.W.; Shang, J.J.; Chen, D.X.; Lei, C.L.; Zou, Y.; Zhai, W.X.; Liu, G.Z.; Xu, J.C.; Ling, Z.Z.; Cao, G.; et al. A B-Lectin Receptor Kinase Gene Conferring Rice Blast Resistance. Plant J. 2006, 46, 794–804. [Google Scholar] [CrossRef]
- Shang, J.J.; Tao, Y.; Chen, X.W.; Zou, Y.; Lei, C.L.; Wang, J.; Li, X.B.; Zhao, X.F.; Zhang, M.J.; Lu, Z.K.; et al. Identification of a New Rice Blast Resistance Gene; Pid3, by Genomewide Comparison of Paired Nucleo-Tide-Binding Site-Leucine-Rich Repeat Genes and Their Pseudogene Alleles between the Two Sequenced Rice Genomes. Genetics 2009, 182, 1303–1311. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Shi, Y.F.; Liu, W.Z.; Chai, R.Y.; Fu, Y.P.; Zhuang, J.Y.; Wu, J.L. A Pid3 Allele from Rice Cultivar Gumei2 Confers Resistance to Magnaporthe oryzae. J. Genet. Genom. 2011, 38, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Lü, Q.M.; Xu, X.; Shang, J.J.; Jiang, G.H.; Pang, Z.Q.; Zhou, Z.Z.; Wang, J.; Liu, Y.; Li, T.; Li, X.B.; et al. Functional Analysis of Pid3-A4, an Ortholog of Rice Blast Resistance Gene Pid3 Revealed by Allele Mining in Common Wild Rice. Phytopathology 2013, 103, 594–599. [Google Scholar]
- Zhu, X.Y.; Chen, S.; Yang, J.Y.; Zhou, S.C.; Zeng, L.X.; Han, J.L.; Su, J.; Wang, L.; Pan, Q.H. The Identification of Pi50(t), a New Member of the Rice Blast Resistance Pi2/Pi9 Multigene Family. Theor. Appl. Genet. 2012, 124, 1295–1304. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Q.; Lin, F.; Wang, L.; Pan, Q.H. The in silico mapbased cloning of Pi36, a rice coiled-coil-nucleotide-binding site-leucine-rich repeat gene that confers race-specific resistance to the blast fungus. Genetics 2007, 176, 2541–2549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berruyer, R.; Adreit, H.; Milazzo, J.; Gaillard, S.; Berger, A.; Dioh, W.; Lebrun, M.-H.; Tharreau, D. Identification and fine mapping of Pi33, the rice resistance gene corresponding to the Magnaporthe grisea avirulence gene ACE1. Theor. Appl. Genet. 2003, 107, 1139–1147. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Song, M.Y.; Seo, Y.S.; Kim, H.K.; Ko, S.; Cao, P.J.; Suh, J.P.; Yi, G.; Roh, J.H.; Lee, S.; et al. Rice Pi5-mediated resistance to Magnaporthe oryzae requires the presence of two coiled-coil-nucleotide-binding-leucine-rich repeat genes. Genetics 2009, 181, 1627–1638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takagi, H.; Uemura, A.; Yaegashi, H.; Tamiru, M.; Abe, A.; Mitsuo, C.; Utsushi, H.; Natsume, S.; Kanzaki, H.; Matsumura, H.; et al. MutMap-Gap: Whole-genome resequencing of mutant F2 progeny bulk combined with de novo assembly of gap regions identifies the rice blast resistance gene Pii. New Phytol. 2013, 200, 276–283. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, B.; Zhu, X.Y.; Yang, J.Y.; Bordeos, A.; Wang, G.; Leach, J.E.; Leung, H. Fine-mapping and molecular marker development for Pi56(t), a NBS-LRR gene conferring broad-spectrum resistance to Magnaporthe oryzae in rice. Theor. Appl. Genet. 2013, 126, 985–998. [Google Scholar] [CrossRef]
- Zhou, X.G.; Liao, H.C.; Chern, M.; Yin, J.J.; Chen, Y.F.; Wang, J.P.; Zhu, X.B.; Chen, Z.X.; Yuan, C.; Zhao, W.; et al. Loss of function of a rice TPR-domain RNA-binding protein confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. USA 2018, 115, 3174–3179. [Google Scholar] [CrossRef] [Green Version]
- Okuyama, Y.; Kanzaki, H.; Abe, A.; Yoshida, K.; Tamiru, M.; Saitoh, H.; Fujibe, T.; Matsumura, H.; Shenton, M.; Galam, D.C.; et al. A multifaceted genomics approach allows the isolation of the rice Piablast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant J. 2011, 66, 467–479. [Google Scholar] [CrossRef]
- Chauhan, R.S.; Farman, M.L.; Zhang, H.B.; Leong, S.A. Genetic and physical mapping of a rice blast resistance locus; Pi-CO39(t), that corresponds to the avirulence gene AVR1-CO39 of Magnaporthe grisea. Mol. Genet. Genom. 2002, 267, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Sharma, T.R.; Madhav, M.S.; Singh, B.K.; Shanker, P.; Jana, T.K.; Dalal, V.; Pandit, A.; Singh, A.; Gaikwad, K.; Upreti, H.C.; et al. High-resolution mapping; cloning and molecular characterization of the Pi-kh gene of rice; which confers resistance to Magnaporthe grisea. Mol. Genet. Genom. 2005, 274, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Soubam, D.; Singh, P.K.; Thakur, S.; Singh, N.K.; Sharma, T.R. A novel blast resistance gene, Pi54rh cloned from wild species of rice, Oryza rhizomatis confers broad spectrum resistance to Magnaporthe oryzae. Funct. Integr. Genom. 2012, 12, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Devanna, N.B.; Vijayan, J.; Sharma, T.R. The Blast Resistance Gene Pi54of Cloned from Oryza officinalis Interacts with Avr-Pi54 through Its Novel Non-LRR Domains. PLoS ONE 2014, 9, e104840. [Google Scholar] [CrossRef] [Green Version]
- Zhai, C.; Lin, F.; Dong, Z.; He, X.; Yuan, B.; Zeng, X.; Wang, L.; Pan, Q. The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication. New Phytol. 2011, 189, 321–334. [Google Scholar] [CrossRef]
- Hua, L.X.; Wu, J.Z.; Chen, C.X.; Wu, W.H.; He, X.Y.; Lin, F.; Wang, L.; Ashikawa, I.; Matsumoto, T.; Wang, L.; et al. The isolation of Pi1, an allele at the Pik locus which confers broad spectrum resistance to rice blast. Theor. Appl. Genet. 2012, 125, 1047–1055. [Google Scholar] [CrossRef]
- Zhai, C.; Zhang, Y.; Yao, N.; Lin, F.; Liu, Z.; Dong, Z.Q.; Wang, L.; Pan, Q.H. Function and Interaction of the Coupled Genes Responsible for Pik-h Encoded Rice Blast Resistance. PLoS ONE 2014, 9, e98067. [Google Scholar] [CrossRef]
- Ashikawa, I.; Hayashi, N.; Yamane, H.; Kanamori, H.; Wu, J.Z.; Matsumoto, T.; Ono, K.; Yano, M. Two Adjacent Nucleo-Tide-Binding Site-Leucine-Rich Repeat Class Genes Are Required to Confer Pikm-Specific Rice Blast Resistance. Genetics 2008, 180, 2267–2276. [Google Scholar] [CrossRef] [Green Version]
- Yuan, B.; Zhai, C.; Wang, W.J.; Zeng, X.S.; Xu, X.K.; Hu, H.Q.; Lin, F.; Wang, L.; Pan, Q.H. The Pik-p Resistance to Magnaporthe oryzae in Rice Is Mediated by a Pair of Closely Linked CC-NBS-LRR Genes. Theor. Appl. Genet. 2011, 122, 1017–1028. [Google Scholar] [CrossRef]
- Chen, J.; Peng, P.; Tian, J.S.; He, Y.G.; Zhang, L.P.; Liu, Z.X.; Yin, D.D.; Zhang, Z.H. Pike, a rice blast resistance allele consisting of two adjacent NBS-LRR genes, was identified as a novel allele at the Pik locus. Mol. Breed. 2015, 35, 117. [Google Scholar] [CrossRef]
- Fjellstrom, R.G.; Conaway-Bormans, C.A.; McClung, A.M.; Marchetti, M.A.; Shank, A.R.; Park, W.D. Development of DNA Markers Suitable for Marker Assisted Selection of Three Pi Genes Conferring Resistance to Multiple Pyricularia grisea Pathotypes. Crop Sci. 2004, 44, 1790–1798. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, N.; Inoue, H.; Kato, T.; Funao, T.; Shirota, M.; Shimizu, T.; Kanamori, H.; Yamane, H.; Hayano-Saito, Y.; Matsumoto, T.; et al. Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J. 2010, 64, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Bryan, G.T.; Wu, K.-S.; Farrall, L.; Jia, Y.; Hershey, H.P.; McAdams, S.A.; Faulk, K.N.; Donaldson, G.K.; Tarchini, R.; Valent, B. A Single Amino Acid Difference Distinguishes Resistant and Susceptible Alleles of the Rice Blast Resistance Gene Pi-ta. Plant Cell 2000, 12, 2033–2046. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.J.; Wang, X.Y.; Jia, Y.L.; Minkenberg, B.; Wheatley, M.; Fan, J.B.; Jia, M.H.; Famoso, A.; Edwards, J.D.; Wamishe, Y.; et al. The rice blast resistance gene Ptr encodes an atypical protein required for broad-spectrum disease resistance. Nat. Commun. 2018, 9, 2039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, Z.; Zhang, Y. Why fragrance rice produced in Thailand can be sold worldwide? World Agric China 2003, 2, 33–36, (Chinese, In English abstract). [Google Scholar]
- Widjaja, R.; Craske, J.D.; Wootton, M. Comparative Studies on Volatile Components of Non-Fragrant and Fragrant Rices. J. Sci. Food Agric. 1996, 70, 151–161. [Google Scholar] [CrossRef]
- Jezussek, M.; Juliano, B.O.; Schieberle, P. Comparison of Key Aroma Compounds in Cooked Brown Rice Varieties Based on Aroma Extract Dilution Analyses. J. Agric. Food Chem. 2002, 50, 1101–1105. [Google Scholar] [CrossRef]
- Bradbury, L.M.T.; Fitzgerald, T.L.; Henry, R.J.; Jin, Q.; Waters, D.L.E. The gene for fragrance in rice. Plant Biotechnol. J. 2005, 3, 363–370. [Google Scholar] [CrossRef]
- Fitzgerald, M.A.; Hamilton, N.R.S.; Calingacion, M.N.; Verhoeven, H.A.; Butardo, V.M. Is there a second fragrance gene in rice? Plant Biotechnol. J. 2008, 6, 416–423. [Google Scholar] [CrossRef]
- Shi, W.; Yang, Y.; Chen, S.; Xu, M. Discovery of a new fragrance allele and the development of functional markers for the breeding of fragrant rice varieties. Mol. Breed. 2008, 22, 185–192. [Google Scholar] [CrossRef]
- Kovach, M.J.; Calingacion, M.N.; Fitzgerald, M.A.; McCouch, S.R. The origin and evolution of fragrance in rice (Oryza sativa L.). Proc. Natl. Acad. Sci. USA 2009, 106, 14444–14449. [Google Scholar] [CrossRef] [Green Version]
- Shao, G.N.; Tang, A.; Tang, S.Q.; Luo, J.; Jiao, G.A.; Wu, J.L.; Hu, P.S. A new deletion mutation of fragrant gene and the development of three molecular markers for fragrance in rice. Plant Breed. 2011, 130, 172–176. [Google Scholar] [CrossRef]
- Shao, G.; Tang, S.; Chen, M.; Wei, X.; He, J.; Luo, J.; Jiao, G.; Hu, Y.; Xie, L.; Hu, P. Haplotype variation at Badh2, the gene determining fragrance in rice. Genomics 2013, 101, 157–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Xiao, W.; Wang, W.; Feng, A.; Zhu, X.; Chen, S.; Chen, Z. Analysis of a major rice blast resistance gene in the rice restorer line Hanghui 1179. Euphytica 2017, 213, 143. [Google Scholar] [CrossRef]
- Ying, X.; Xu, X.; Chen, M.; Ouyang, Y.; Zhu, Z.; Min, J. Determination of 2-acetyl-1-pyrroline in aroma rice using gas chromatography-mass spectrometry. Chin. J. Chromatogr. 2010, 28, 782–785. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Lu, Y.; Shao, Y.; Zhang, G.; Xiao, P.; Shen, S.; Corke, H.; Bao, J. Molecular marker assisted selection for improvement of the eating, cooking and sensory quality of rice (Oryza sativa L.). J. Cereal Sci. 2010, 51, 159–164. [Google Scholar] [CrossRef]
- Ramkumar, G.; Srinivasarao, K.; Madhan, M.; Sudarshan, I.; Sivaranjani, A.; Gopalakrishna, K.; Neeraja, C.; Balachandran, S.; Sundaram, R.; Prasad, M.; et al. Development and validation of functional marker targeting an InDel in the major rice blast disease resistance gene Pi54 (Pikh). Mol Breed. 2011, 27, 129–135. [Google Scholar] [CrossRef]
- Jiang, J.; Yang, D.; Ali, J.; Mou, T. Molecular marker-assisted pyramiding of broad-spectrum disease resistance genes, Pi2 and Xa23, into GZ63-4S, an elite thermo-sensitive genic male-sterile line in rice. Mol. Breed. 2015, 35, 83–94. [Google Scholar] [CrossRef]
- Xu, J.; Wang, B.; Wu, Y.; Du, P.; Wang, J.; Wang, M.; Yi, C.; Gu, M.; Liang, G. Fine mapping and candidate gene analysis of ptgms2-1, the photoperiod-thermo-sensitive genic male sterile gene in rice (Oryza sativa L.). Theor. Appl. Genet. 2010, 122, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Ma, T.; Zhang, A.; Ong, K.H.; Luo, Z.; Li, Z.; Yang, J.; Yin, Z. Marker-assisted breeding of Chinese elite rice cultivar 9311 for disease resistance to rice blast and bacterial blight and tolerance to submergence. Mol. Breed. 2017, 37, 106. [Google Scholar] [CrossRef]
- Luo, Y.; Yin, Z. Marker-assisted breeding of Thai fragrance rice for semi-dwarf phenotype, submergence tolerance and disease resistance to rice blast and bacterial blight. Mol. Breed. 2013, 32, 709–721. [Google Scholar] [CrossRef]
- Zhang, T.; Zheng, J.; Xu, J.; Jiang, K.; Wu, X.; Wang, X. Genetic diversity of aromatic rice varieties based on markers of functional genes and SSR. Sci. Agric. Sin. 2008, 41, 625–635, (Chinese, In English abstract). [Google Scholar]
- Chen, C.; Li, H.; Liu, L.; Chen, Y.; Luo, Q. Breeding status and development strategy of fragrant rice in Guangxi province. China Rice 2017, 23, 117–120, (Chinese, In English abstract). [Google Scholar]
- Mi, J.; Yang, D.; Chen, Y.; Jiang, J.; Mou, H.; Huang, J.; Ouyang, Y.; Mou, T. Accelerated molecular breeding of a novel P/TGMS line with broad-spectrum resistance to rice blast and bacterial blight in two-line hybrid rice. Rice 2018, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; He, H.; Zhou, F.; Yu, H.; Deng, X.W. Development of genomics-based genotyping platforms and their applications in rice breeding. Curr. Opin. Plant Biol. 2013, 16, 247–254. [Google Scholar] [CrossRef]
- Yu, H.; Xie, W.; Li, J. A whole-genome SNP array (RICE6K) for genomic breeding in rice. Plant Biotechnol. J. 2014, 12, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Collard, B.; Mackill, D.J. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. B Biol. Sci. 2008, 363, 557–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Marker | Gene of Interest | Chr. | Primer (5′-3′) | Type of Marker | Reference |
|---|---|---|---|---|---|
| Pi2-1 | Pi2 | 6 | F: 5′ TTGGATTGGAGCATTATTCG 3′ R: 5′ GCATGTGCTAGACTCTTGGGT 3′ | Indel | Sheng et al., 2017 [1] |
| Badh2-1 | badh2 | 8 | F: 5′ GTTGCATTTACTGGGAGT 3′ F: 5′ GAAAAGGACAACATTGAGAA 3′ | Indel | Shao et al., 2013 [54] |
| Variety | Pi2 Genotype | badh2 Genotype | Rice Blast ResistancePhenotype | 2-AP Content mg/kg |
|---|---|---|---|---|
| R1179 | + | - | 3 (R) | / |
| Wenxiang-1 | - | + | 7 (S) | 0.867 |
| R365 | + | + | 3 (R) | 0.563 |
| R403 | + | + | 4 (R) | 0.618 |
| Variety | Plant Height/cm | Number of Effective Panicles | Panicle Length/cm | Grain Number Per Panicle | Seed-Setting Rate/% | 1000-Grain Weight/g | Grain Length/Width | Yield Per Plant/g | Amylose Content (%) | 2-AP Content (mg/kg) |
|---|---|---|---|---|---|---|---|---|---|---|
| R1179 | 114.00 ± 2.15 | 12.00 ± 1.12 | 27.30 ± 1.21 | 247.33 ± 16.41 | 88.48 ± 3.51 | 25.31 ± 0.51 | 3.57 | 41.22 ± 3.51 | 16.54 ± 1.3 | / |
| Wenxiang-1 | 122.30 ± 1.86 | 11.00 ± 1.33 | 25.48 ± 2.13 | 156.67 ± 12.11 | 75.07 ± 5.62 | 29.11 ± 0.45 | 3.77 | 30.94 ± 4.12 | 24.1 ± 2.1 | 0.867 |
| R365 | 125.80 ± 1.73 | 17.00 ± 0.95 | 26.36 ± 1.82 | 240.33 ± 20.32 | 90.08 ± 6.13 | 25.04 ± 0.53 | 3.80 | 42.13 ± 3.89 | 13.65 ± 1.5 | 0.563 |
| R403 | 129.50 ± 2.43 | 13.00 ± 1.76 | 29.94 ± 1.43 | 259.31 ± 21.42 | 87.67 ± 4.22 | 25.53 ± 0.47 | 3.54 | 45.08 ± 4.21 | 15.5 ± 1.6 | 0.618 |
| Variety | The Number of Homozygous Loci Derived from Donor Parent Wenxiang-1 | The Percentage of Homozygous Loci Derived from Donor Parent Wenxiang-1 | The Number of Homozygous Loci Derived from Donor Parent R1179 | The Percentage of Homozygous Loci Derived from Donor Parent R1179 | The Number of Heterozygous Loci |
|---|---|---|---|---|---|
| R365 | 2493 | 40.67% | 3988 | 59.33% | 1531 |
| R403 | 3714 | 46.26% | 4342 | 53.74% | 345 |
| Growth Duration | Number of Effective Panicles | Number of Grains Per Panicle | Seed-Setting Rate | Yield Per Hectare ton/ha | 1000-Grain Weight (g) | Grain Length/Width | Amylose Content (%) | 2-AP Content (mg/kg) | Rice Blast Resistance | |
|---|---|---|---|---|---|---|---|---|---|---|
| C815S/R365 | 135.33 ± 2.52 | 10.67 ± 0.58 | 179.67 ± 3.21 | 83.4 ± 1.86 ** | 9.93 ± 0.15 * | 24.93 ± 0.15 ** | 3.43 ± 0.15 | 17.33 ± 0.45 | 0.219 ± 0.004 | 4 (MR) |
| Y58S/R365 | 131 ± 2 | 8.33 ± 1.15 | 171.67 ± 3.51 | 83.56 ± 1.62 | 9.5 ± 0.26 | 26.4 ± 0.36 | 3.6 ± 0.1 | 24.67 ± 0.86 | 0.234 ± 0.012 | 4 (MR) |
| Shen08S/R365 | 130 ± 1 | 6.67 ± 0.58 | 173 ± 7 | 82.2 ± 0.95 | 9.37 ± 0.35 | 24.13 ± 0.31 | 3.43 ± 0.06 | 24.23 ± 0.4 | 0.365 ± 0.014 | 4 (MR) |
| Guangzhan63S/R403 | 135.33 ± 0.58 | 9.67 ± 1.15 | 195 ± 2.65 | 79 ± 2 | 9.17 ± 0.12 | 26.23 ± 0.15 | 3.5 ± 0.2 | 18.3 ± 0.36 | 0.414 ± 0.038 | 3 (R) |
| Yu03S/R403 | 138.33 ± 3.05 | 12 ± 1 | 182.67 ± 3.51 | 79.37 ± 0.74 * | 9.6 ± 0.17 * | 25.1 ± 0.12 ** | 3.57 ± 0.25 | 17.67 ± 0.21 | 0.237 ± 0.068 | 3 (R) |
| Fengliangyou-4 | 133.67 ± 1.15 | 11.67 ± 0.58 | 184.33 ± 7.09 | 78.37 ± 0.93 | 9.17 ± 0.21 | 27.57 ± 0.21 | 3.3 ± 0.1 | 16.77 ± 0.35 | / | 5 (MS) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Tang, S.; Guo, N.; An, R.; Ren, Z.; Hu, S.; Wei, X.; Jiao, G.; Xie, L.; Wang, L.; et al. Pyramiding Rice Blast Resistance Gene Pi2 and Fragrance Gene badh2. Agronomy 2023, 13, 589. https://doi.org/10.3390/agronomy13020589
Wang Y, Tang S, Guo N, An R, Ren Z, Hu S, Wei X, Jiao G, Xie L, Wang L, et al. Pyramiding Rice Blast Resistance Gene Pi2 and Fragrance Gene badh2. Agronomy. 2023; 13(2):589. https://doi.org/10.3390/agronomy13020589
Chicago/Turabian StyleWang, Yakun, Shengjia Tang, Naihui Guo, Ruihu An, Zongliang Ren, Shikai Hu, Xiangjin Wei, Guiai Jiao, Lihong Xie, Ling Wang, and et al. 2023. "Pyramiding Rice Blast Resistance Gene Pi2 and Fragrance Gene badh2" Agronomy 13, no. 2: 589. https://doi.org/10.3390/agronomy13020589
APA StyleWang, Y., Tang, S., Guo, N., An, R., Ren, Z., Hu, S., Wei, X., Jiao, G., Xie, L., Wang, L., Chen, Y., Zhao, F., Tang, S., Hu, P., & Sheng, Z. (2023). Pyramiding Rice Blast Resistance Gene Pi2 and Fragrance Gene badh2. Agronomy, 13(2), 589. https://doi.org/10.3390/agronomy13020589







