A Pleiotropic Drug Resistance Transporter TaABCG36 Contributes to Defense against Puccinia triticina in Triticum aestivum
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials, Pt Pathotype, and Primers
2.2. Pt and Phytohormone Treatments
2.3. Genomic DNA, RNA Extraction, and cDNA Synthesis
2.4. Isolation and Characterization of TaABCG36
2.5. Quantitative Real-Time PCR
2.6. Subcellular Localization of TaABCG36
2.7. BSMV-Mediated TaABCG36 Gene Silencing
2.8. Histopathological Observations of Fungal Growth and Host Response
3. Results
3.1. Isolation and Characterization of TaABCG36
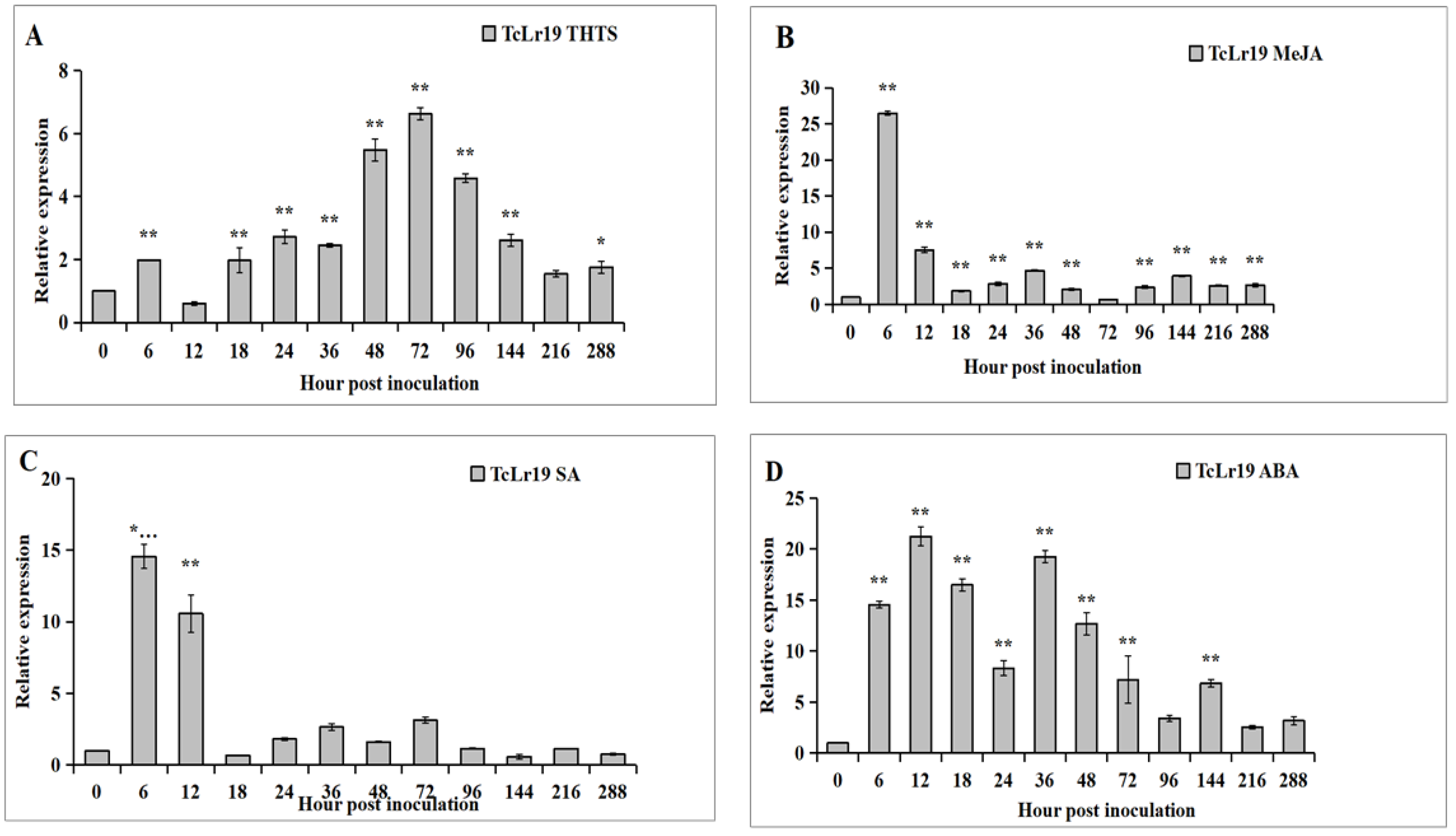
3.2. Expression Patterns of TaABCG36 in TcLr19
3.3. TaABCG36 Is a Plasma-Membrane-Localized Protein
3.4. TaABCG36-Silenced Plants Showed Decreased Leaf Rust Resistance
3.5. Histopathological Analysis of TaABCG36 Knockdown Leaves
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rea, P.A. Plant ATP-binding cassette transporters. Annu. Rev. Plant Biol. 2007, 58, 347–375. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Park, J.; Choi, H.; Burla, B.; Kretzschmar, T.; Lee, Y.; Martinoia, E. Plant ABC transporters. Arab. Book/Am. Soc. Plant Biol. 2011, 9, e0153. [Google Scholar] [CrossRef] [PubMed]
- Hollenstein, K.; Frei, D.C.; Locher, K.P. Structure of an ABC transporter in complex with its binding protein. Nature 2007, 446, 213–216. [Google Scholar] [CrossRef] [PubMed]
- Locher, K.P. Structure and mechanism of ABC transporters. Curr. Opin. Struct. Biol. 2004, 14, 426–431. [Google Scholar] [CrossRef]
- Dudler, R.; Hertig, C. Structure of an MDR-like gene from Arabidopsis thaliana: Evolutionary implications. J. Biol. Chem. 1992, 267, 5882–5888. [Google Scholar] [CrossRef]
- Garcia, O.; Bouige, P.; Forestier, C.; Dassa, E. Inventory and comparative analysis of rice and Arabidopsis ATP-binding cassette (ABC) systems. J. Mol. Biol. 2004, 343, 249–265. [Google Scholar] [CrossRef]
- Ofori, P.A.; Mizuno, A.; Suzuki, M.; Martinoia, E.; Reuscher, S.; Aoki, K.; Shibata, D.; Otagaki, S.; Matsumoto, S.; Shiratake, K. Genome-wide analysis of ATP binding cassette (ABC) transporters in tomato. PLoS ONE 2018, 13, e0200854. [Google Scholar] [CrossRef]
- Cakir, B.; Kilickaya, O. Whole-genome survey of the putative ATP-binding cassette transporter family genes in Vitis vinifera. PLoS ONE 2013, 8, e78860. [Google Scholar] [CrossRef]
- Shi, M.Y.; Wang, S.S.; Zhang, Y.; Wang, S.; Zhao, J.; Feng, H.; Sun, P.P.; Fang, C.B.; Xie, X.B. Genome-wide characterization and expression analysis of ATP-binding cassette (ABC) transporters in strawberry reveal the role of FvABCC11 in cadmium tolerance. Sci. Hortic. 2020, 271, 109464. [Google Scholar] [CrossRef]
- Chen, P.J.; Li, Y.; Zhao, L.H.; Hou, Z.M.; Yan, M.K.; Hu, B.Y.; Liu, Y.H.; Azam, S.M.; Zhang, Z.Y.; Rahman, Z.; et al. Genome-wide identification and expression profiling of ATP-binding cassette (ABC) transporter gene family in pineapple (Ananas comosus (L.) Merr.) reveal the role of AcABCG38 in pollen development. Front. Plant Sci. 2017, 8, 2150. [Google Scholar] [CrossRef]
- Nogia, P.; Pati, P.K. Plant secondary metabolite transporters: Diversity, functionality, and their modulation. Front. Plant Sci. 2021, 12, 758202. [Google Scholar] [CrossRef]
- Dhara, A.; Raichaudhuri, A. ABCG transporter proteins with beneficial activity on plants. Phytochemis 2021, 184, 112663. [Google Scholar] [CrossRef]
- Francisco, R.M.; Regalado, A.; Ageorges, A.; Burla, B.J.; Bassin, B.; Eisenach, C.; Zarrouk, O.; Vialet, S.; Marlin, T.; Chaves, M.M.; et al. ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-Glucosides. Plant Cell 2013, 25, 1840–1854. [Google Scholar] [CrossRef]
- Demurtas, O.C.; Francisco, R.; Diretto, G.; Ferrante, P.; Frusciante, S.; Pietrella, M.; Aprea, G.; Borghi, L.; Feeney, M.; Frigerio, L.; et al. ABCC transporters mediate the vacuolar accumulation of crocins in Saffron stigmas. Plant Cell 2019, 31, 2789–2804. [Google Scholar] [CrossRef]
- Fu, X.; Liu, H.; Hassani, D.; Peng, B.; Yan, X.; Wang, Y.; Wang, C.; Li, L.; Liu, P.; Pan, Q.; et al. AaABCG40 enhances artemisinin content and modulates drought tolerance in Artemisia annua. Front. Plant Sci. 2020, 11, 950. [Google Scholar] [CrossRef]
- Dahuja, A.; Kumar, R.R.; Sakhare, A.; Watts, A.; Singh, B.; Goswami, S.; Sachdev, A.; Praveen, S. Role of ATP-binding cassette transporters in maintaining plant homeostasis under abiotic and biotic stresses. Physiol. Plant. 2021, 171, 785–801. [Google Scholar] [CrossRef]
- Khare, D.; Choi, H.; Huh, S.U.; Bassin, B.; Kim, J.; Martinoia, E.; Sohn, K.H.; Paek, K.H.; Lee, Y. Arabidopsis ABCG34 contributes to defense against necrotrophic pathogens by mediating the secretion of camalexin. Proc. Natl. Acad. Sci. USA 2017, 114, 5712–5720. [Google Scholar] [CrossRef]
- Cho, C.H.; Jang, S.; Choi, B.Y.; Hong, D.; Choi, D.S.; Choi, S.; Kim, H.; Han, S.K.; Kim, S.; Kim, M.S.; et al. Phylogenetic analysis of ABCG subfamily proteins in plants: Functional clustering and coevolution with ABCGs of pathogens. Physiol. Plant. 2021, 172, 1422–1438. [Google Scholar] [CrossRef]
- Bultreys, A.; Trombik, T.; Drozak, A.; Boutry, M. Nicotiana plumbaginifolia plants silenced for the ATP-binding cassette transporter gene NpPDR1 show increased susceptibility to a group of fungal and oomycete pathogens. Mol. Plant Pathol. 2009, 10, 651–663. [Google Scholar] [CrossRef]
- Stukkens, Y.; Bultreys, A.; Grec, S.; Trombik, T.; Vanham, D.; Boutry, M. NpPDR1, a pleiotropic drug resistance-type ATP-binding cassette transporter from Nicotiana plumbaginifolia, plays a major role in plant pathogen defense. Plant Physiol. 2005, 139, 341–352. [Google Scholar] [CrossRef]
- Pierman, B.; Toussaint, F.; Bertin, A.; Lévy, D.; Smargiasso, N.; De Pauw, E.; Boutry, M. Activity of the purified plant ABC transporter NtPDR1 is stimulated by diterpenes and sesquiterpenes involved in constitutive and induced defenses. J. Biol. Chem. 2017, 292, 19491–19502. [Google Scholar] [CrossRef] [PubMed]
- Shibata, Y.; Ojika, M.; Sugiyama, A.; Yazaki, K.; Jones, D.A.; Kawakita, K.; Takemptp, D. The full-size ABCG transporters Nb-ABCG1 and Nb-ABCG2 function in pre- and postinvasion defense against Phytophthora infestans in Nicotiana benthamiana. The Plant Cell 2016, 28, 1163–1181. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.; Gomi, K.; Kaku, H.; Abe, H.; Seto, H.; Nakatsu, S.; Neya, M.; Kobayashi, M.; Nakaho, K.; Ichinose, Y. Identification of natural diterpenes that inhibit bacterial wilt disease in tobacco, tomato and Arabidopsis. Plant Cell Physiol. 2012, 53, e1432–e1444. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Song, N.; Ma, L.; Fang, D.H.; Wu, J.S. NaPDR1 and NaPDR1-like are essential for the resistance of Nicotiana attenuata against fungal pathogen Alternaria alternata. Plant Divers. 2018, 40, 68–73. [Google Scholar] [CrossRef]
- Stein, M.; Dittgen, J.; Sanchez-Rodruguez, C.; Hou, B.H.; Molina, A.; Schulze-Lefert, P.; Lipka, V.; Somerville, S. Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to non-host resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 2006, 18, 731–746. [Google Scholar] [CrossRef]
- Shang, Y.; Xiao, J.; Ma, L.L.; Wang, H.Y.; Qi, Z.J.; Chen, P.D.; Liu, D.J.; Wang, X.E. Characterization of a PDR type ABC transporter gene from wheat (Triticum aestivum L.). Chin. Sci. Bull. 2009, 18, 3249–3257. [Google Scholar] [CrossRef]
- Muhovski, Y.; Jacquemin, J.M.; Batoko, H. Identification and differential induction of ABCG transporter genes in wheat cultivars challenged by a deoxynivalenol-producing Fusarium graminearum strain. Mol. Biol. Rep. 2014, 41, 6181–6194. [Google Scholar] [CrossRef]
- Wang, G.P.; Hou, W.Q.; Zhang, L.; Wu, H.Y.; Zhao, L.F.; Du, X.Y.; Ma, X.; Li, A.F.; Wang, H.W.; Kong, L.R. Functional analysis of a wheat pleiotropic drug resistance gene involved in Fusarium head blight resistance. J. Integr. Agric. 2016, 15, 2215–2227. [Google Scholar] [CrossRef]
- Walter, S.; Kahla, A.; Arunachalam, C.; Perochon, A.; Khan, M.R.; Scofield, S.R.; Doohan, F.M. A wheat ABC transporter contributes to both grain formation and mycotoxin tolerance. J. Exp. Bot. 2015, 66, 2583–2593. [Google Scholar] [CrossRef]
- Theodoulou, F.L.; Clark, I.M.; He, X.L.; Pallett, K.E.; Cole, D.J.; Hallahan, D.L. Co-induction of glutathione-S-transferases and multidrug resistance associated protein by xenobiotics in wheat. Pest Manag. Sci. 2003, 59, 202–214. [Google Scholar] [CrossRef]
- Bhati, K.K.; Sharma, S.; Aggarwal, S.; Kaur, M.; Shukla, V.; Kaur, J.; Mantri, S.; Pandey, A.K. Genome-wide identification and expression characterization of ABCC-MRP transporters in hexaploid wheat. Front. Plant Sci. 2015, 6, 488. [Google Scholar] [CrossRef]
- Wang, X.; Wang, C.; Sheng, H.; Wang, Y.; Zeng, J.; Kang, H.; Fan, X.; Sha, L.; Zhang, H.; Zhou, Y. Transcriptome-wide identification and expression analyses of ABC transporters in dwarf polish wheat under metal stresses. Biol. Plant. 2017, 61, 293–304. [Google Scholar] [CrossRef]
- Rajagopalan, N.; Lu, Y.; Burton, I.W.; Monteil-Rivera, F.; Halasz, A.; Reimer, E.; Tweidt, R.; Brûlé-Babel, A.; Kutcher, H.R.; You, F.M.; et al. A phenylpropanoid diglyceride associates with the leaf rust resistance Lr34res gene in wheat. Phytochemistry 2020, 178, 112456. [Google Scholar] [CrossRef]
- Sarma, D.; Knott, D.R. The transfer of leaf-rust resistance from Agropyron to Triticum by irradiation. Can. J. Genet. Cytol. 1966, 8, 137–143. [Google Scholar] [CrossRef]
- Gupta, S.K.; Charpe, A.; Prabhu, K.V.; Haque, Q.M. Identification and validation of molecular markers linked to the leaf rust resistance gene Lr19 in wheat. Theor. Appl. Genet. 2006, 113, 1027–1036. [Google Scholar] [CrossRef]
- Saini, R.G.; Kaur, L. Adult plant leaf rust (Puccinia recondita tririci) resistance of known Lr genes against three virulence variants of race 77 from Indian sub-continent. Ind. J. Agric. Res. 1998, 68, 776–779. [Google Scholar]
- Prins, R.; Groenewald, J.Z.; Marais, G.F.; Snape, J.W.; Koebner, R.M.D. AFLP and STS tagging of Lr19, a gene conferring resistance to leaf rust in wheat. Theor. Appl. Genet. 2001, 103, 618–624. [Google Scholar] [CrossRef]
- Rai, A.; Singh, A.M.; Ahlawat, A.K.; Kumar, R.R.; Raghunandan, K.; Saini, S.; Ganjewala, D.; Shukla, R.B. Quality evaluation of near isogenic lines of the wheat variety carrying Sr26, Lr19 and Yr10 genes. J. Cereal Sci. 2019, 88, 110–117. [Google Scholar] [CrossRef]
- Rosewarne, G.; Bonnett, D.; Rebetzke, G.; Lonergan, P.; Larkin, P. The potential of Lr19 and Bdv2 translocations to improve yield and disease resistance in the high rainfall wheat zones of Australia. Agronomy 2015, 5, 55–70. [Google Scholar] [CrossRef]
- Wang, F.; Yuan, S.T.; Wu, W.Y.; Yang, Y.Q.; Cui, Z.C.; Wang, H.Y.; Liu, D.Q. TaTLP1 interacts with TaPR1 to contribute to wheat defense responses to leaf rust fungus. PLoS Genet. 2020, 16, e1008713. [Google Scholar] [CrossRef]
- Li, J.Y.; Wang, X.D.; Zhang, L.R.; Meng, Q.F.; Zhang, N.; Yang, W.X.; Liu, D.Q. A wheat NBS-LRR gene TaRGA19 participates in Lr19-mediated resistance to Puccinia triticina. Plant Physiol. Bioch. 2017, 119, 1–8. [Google Scholar]
- Pourkhorshid, Z.; Dadkhodaie, A.; Shamloo-Dashtpagerdi, R. Molecular analyses in wheat and Aegilops tauschii reveal a new orthologue of the leaf rust resistance gene Lr19 on chromosome 7DL of Ae. tauschii. J. Phytopathol. 2022, 170, 255–263. [Google Scholar] [CrossRef]
- Gennaro, A.; Koebner, R.M.D.; Ceoloni, C. A candidate for Lr19, an exotic gene conditioning leaf rust resistance in wheat. Funct. Integr. Genom. 2009, 9, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Long, D.L.; Kolmer, J.A. A north American system of nomenclature for Puccinia recondite f. sp. tritici. Phytopathology 1989, 79, 525–529. [Google Scholar] [CrossRef]
- Lück, S.; Kreszies, T.; Strickert, M.; Schweizer, P.; Kuhlmann, M.; Douchkov, D. siRNA-Finder (si-Fi) software for RNAi-target design and off-target prediction. Front. Plant Sci. 2019, 10, 1023. [Google Scholar] [CrossRef]
- Zhang, L.R.; Yang, W.X.; Liu, D.Q. TaRAR1 is required for Lr24-mediated wheat leaf rust resistance. Agric. Sci. China 2011, 10, 1732–1738. [Google Scholar] [CrossRef]
- Wang, X.D.; Wang, X.J.; Deng, L.; Chang, H.T.; Dubcovsky, J.; Feng, H.; Han, Q.M.; Huang, L.L.; Kang, Z.S. Wheat TaNPSN SNARE homologues are involved in vesicle-mediated resistance to stripe rust (Puccinia striiformis f. sp. tritici). J. Exp. Bot. 2014, 65, 4807. [Google Scholar] [CrossRef]
- Yan, X.C.; Li, M.M.; Zhang, P.P.; Yin, G.H.; Zhang, H.Z.; Gebrewhid, T.W.; Zhang, J.P.; Dong, L.L.; Liu, D.Q.; Liu, Z.Y.; et al. High-temperature wheat leaf rust resistance gene Lr13 exhibits pleiotropic effects on hybrid necrosis. Mol. Plant 2021, 14, 1029–1032. [Google Scholar] [CrossRef]
- Zhao, J.J.; Bi, W.S.; Zhao, S.Q.; Su, J.; Li, M.Y.; Ma, L.S.; Yu, X.M.; Wang, X.D. Wheat apoplast-localized lipid transfer protein TaLTP3 enhances defense responses against Puccinia triticina. Front. Plant Sci. 2021, 12, 771806. [Google Scholar] [CrossRef]
- Van den Brüle, S.; Muller, A.; Fleming, A.J.; Smart, C.C. The ABC transporter SpTUR2 confers resistance to the antifungal diterpene sclareol. Plant J. 2002, 30, 649–662. [Google Scholar] [CrossRef]
- Decottignies, A.; Grant, A.M.; Nichols, J.W.; de Wet, H.; McIntosh, D.B.; Goffeau, A. ATPase and multidrug transport activities of the overexpressed yeast ABC protein Yor1p. J. Biol. Chem. 1998, 273, 12612–12622. [Google Scholar] [CrossRef]
- Campos-Salinas, J.; León-Guerrero, D.; González-Rey, E.; Delgado, M.; Castanys, S.; Pérez-Victoria, J.M.; Gamarro, F. LABCG2, a new ABC transporter implicated in phosphatidylserine exposure, is involved in the infectivity and pathogenicity of Leishmania. PLoS Negl. Trop. Dis. 2013, 7, e2179. [Google Scholar] [CrossRef]
- Deppe, J.P.; Rabbat, R.; Hörtensteiner, S.; Keller, B.; Martinoia, E.; Lopéz-Marqués, R.L. The wheat ABC transporter Lr34 modifies the lipid environment at the plasma membrane. J. Biol. Chem. 2018, 293, 18667–18679. [Google Scholar] [CrossRef]
- Bai, Z.Y.; Wang, D.M.; Hou, C.Y.; Liu, N.; Han, S.F.; Ma, L.H. Microstructure and ultrastructure infected by wheat rust fungus. Chin. J. Cell Biol. 2003, 25, 393–397. [Google Scholar]




| Primers | Primer Sequence (5′-3′) | Primer Application |
|---|---|---|
| 3′-GSP1 | CAAGAGCAAGAGCCATCCAGCAGC | RACE amplification |
| 5′-GSP2 | AGCTCCTTTCTTATAGCTCTCCCGGTGT | |
| ABCG-F | ATGGACGCGACGGCGGAAATCCAC | Full length amplification |
| ABCG-R | CATTCGGGTGGTGCAAAATGT | |
| GAPDH-F | AACTGCCTTGCTCCTCTTG | Real-time quantitative PCR |
| GAPDH-R | CATCAAACCCTCAACAATGC | |
| qABCG-F | TTCGATGACATCATCCTCCT | |
| qABCG-R | GCACCCAGTATTGCTTTTGA | |
| V-ABCG-F1 | ATATTAATTAATTCGATGACATCATCCTCCT | Virus-induced gene silencing |
| V-ABCG-R1 | TATGCGGCCGCGCACCCAGTATTGCTTTTGA | |
| V-ABCG-F2 | ATATTAATTAAGAAAGATGCATTGGTTGGTC | |
| V-ABCG-R2 | TATGCGGCCGCTCGTGCAAACCACAGTTCTA | |
| V-ABCG-F3 | ATATTAATTAAACAACTGGTGAGATGCTGGT | |
| V-ABCG-R3 | TATGCGGCCGCAGGAGGATGATGTCATCGAA | |
| Y-ABCG-F | GCTCTAGAATGCCGACGATCGAGGTGCGGTTCG | Subcellular localization |
| Y-ABCG-R | TCCCCCGGGTACCTCTTCTGGAAGTTGAGCTTCA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, N.; Hu, Y.; Wu, Y.; Mapuranga, J.; Yuan, Y.; Yang, W. A Pleiotropic Drug Resistance Transporter TaABCG36 Contributes to Defense against Puccinia triticina in Triticum aestivum. Agronomy 2023, 13, 607. https://doi.org/10.3390/agronomy13020607
Zhang N, Hu Y, Wu Y, Mapuranga J, Yuan Y, Yang W. A Pleiotropic Drug Resistance Transporter TaABCG36 Contributes to Defense against Puccinia triticina in Triticum aestivum. Agronomy. 2023; 13(2):607. https://doi.org/10.3390/agronomy13020607
Chicago/Turabian StyleZhang, Na, Yaya Hu, Yanhui Wu, Johannes Mapuranga, Ying Yuan, and Wenxiang Yang. 2023. "A Pleiotropic Drug Resistance Transporter TaABCG36 Contributes to Defense against Puccinia triticina in Triticum aestivum" Agronomy 13, no. 2: 607. https://doi.org/10.3390/agronomy13020607
APA StyleZhang, N., Hu, Y., Wu, Y., Mapuranga, J., Yuan, Y., & Yang, W. (2023). A Pleiotropic Drug Resistance Transporter TaABCG36 Contributes to Defense against Puccinia triticina in Triticum aestivum. Agronomy, 13(2), 607. https://doi.org/10.3390/agronomy13020607






