Genomic and Transcriptomic Characterization of Alternaria alternata during Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Culture Conditions
2.2. Plant Preparation and Inoculation Experiment
2.3. RNA Extraction, Library Preparation, and Illumina Sequencing
2.4. Genome Annotation and Functional Analysis
2.5. Transcriptome Analysis
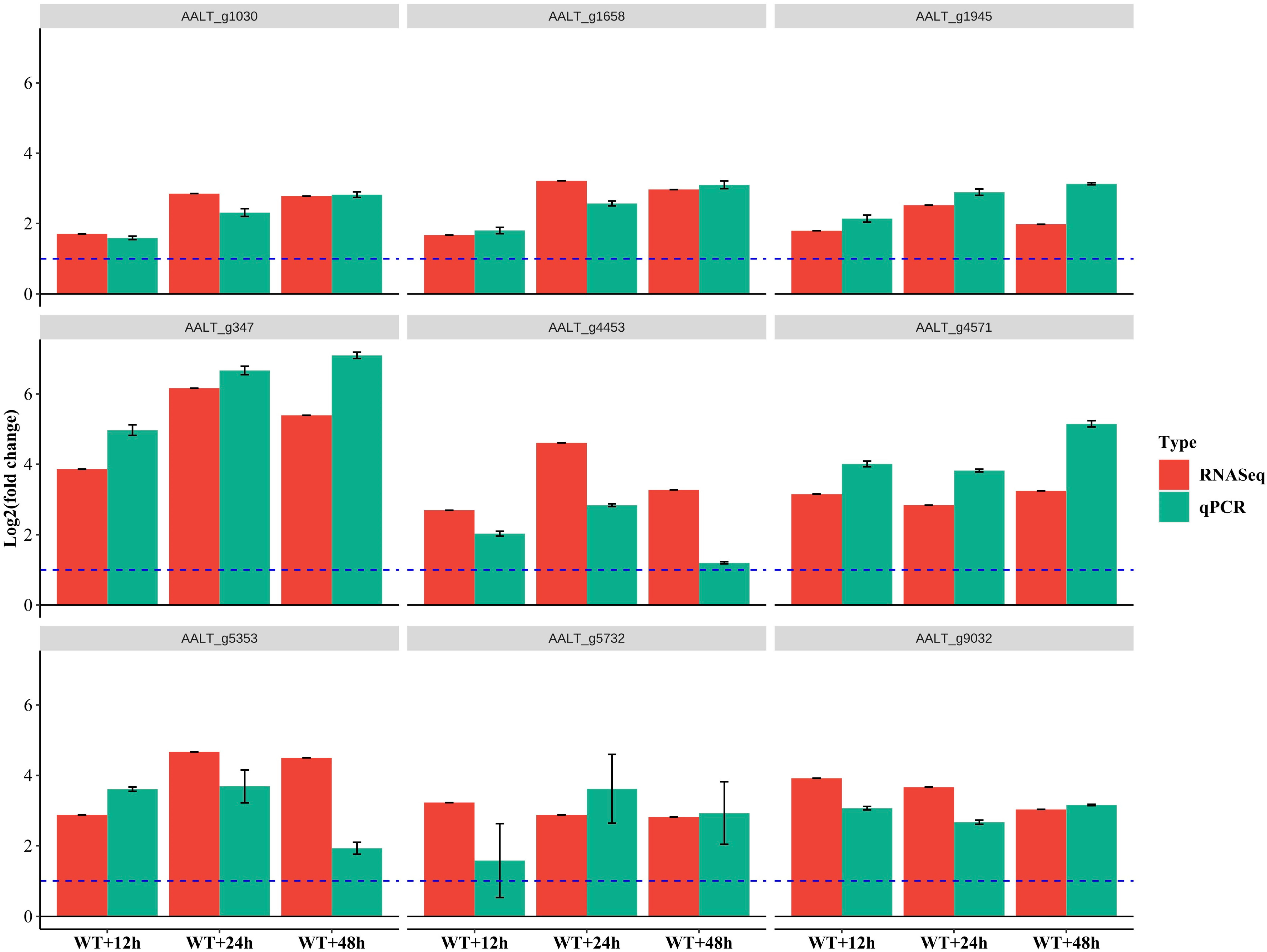
2.6. RT-qPCR Analysis
3. Results
3.1. Genomic Characterization of Alternaria alternata Z7
3.2. Gene Expression Patterns of A. alternata during Infection
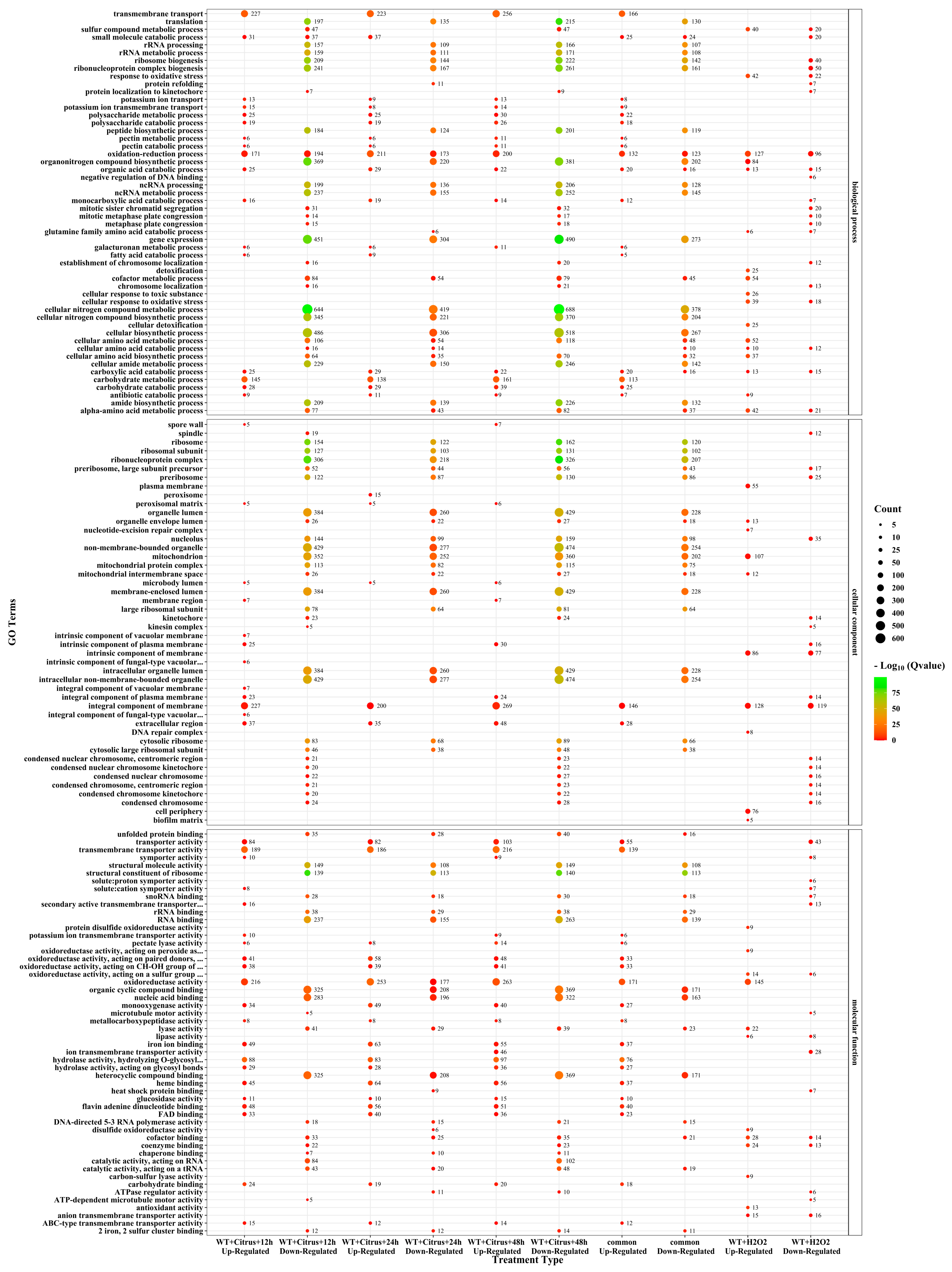
3.3. Multiple Critical Genes in GO Categories Were Affected in A. alternata during Infection
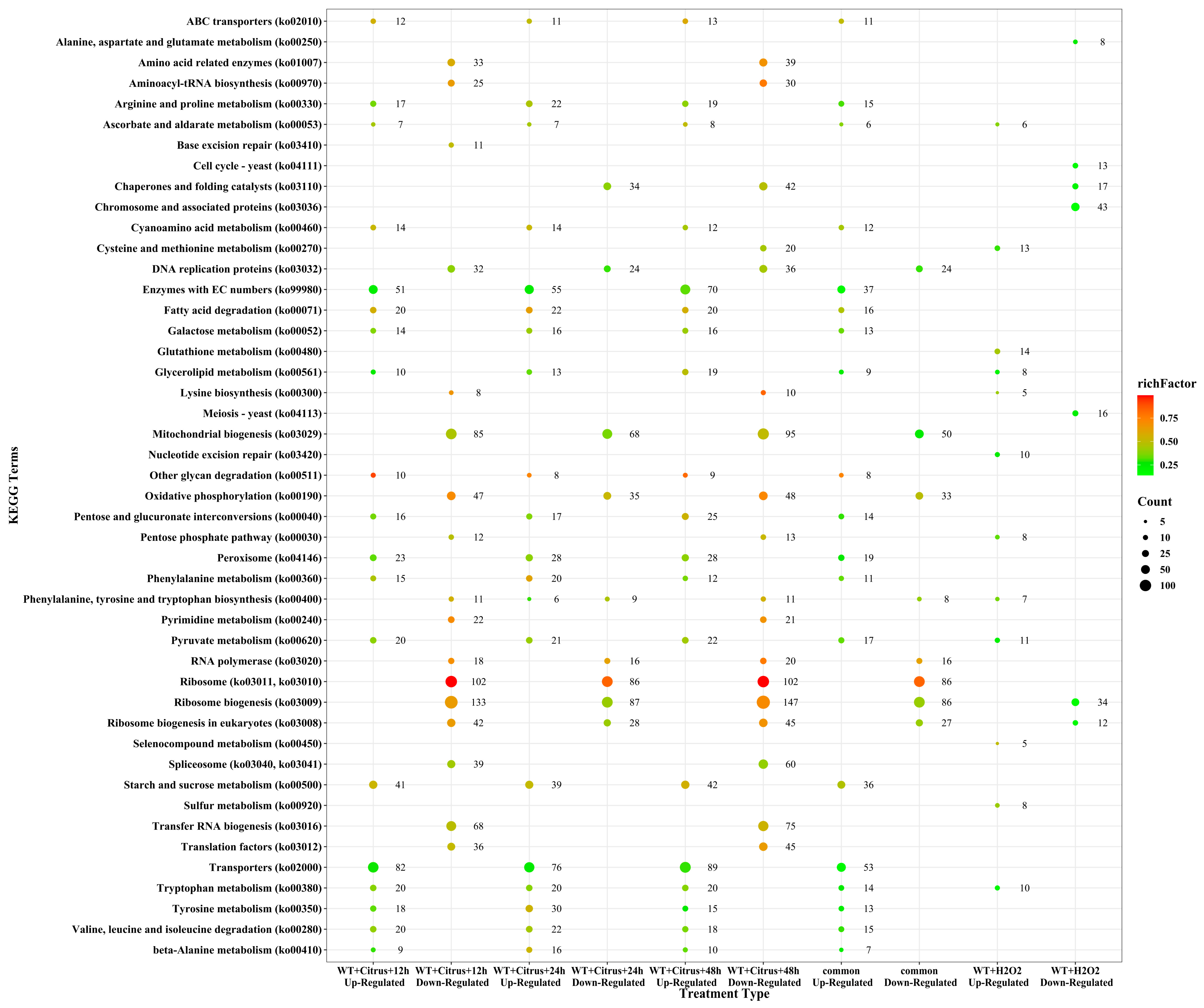
3.4. Multiple Critical Genes Pathways Were Affected in A. alternata during Infection
3.5. Multiple Critical SM Gene Clusters Were Regulated in A. alternata during Infection
3.6. Multiple Critical Genes Related to Plant-Pathogen Interaction Were Affected in Alternaria alternata during Infection
3.7. Multiple Critical Genes Related to Peroxisome Were Activated in Alternaria alternata during Infection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stahl, E.A.; Bishop, J.G. Plant-pathogen arms races at the molecular level. Curr. Opin. Plant Biol. 2000, 3, 299–304. [Google Scholar] [CrossRef]
- Agrawal, A.A.; Heil, M. Synthesizing specificity: Multiple approaches to understanding the attack and defense of plants. Trends Plant Sci. 2012, 17, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Frantzeskakis, L.; Di Pietro, A.; Rep, M.; Schirawski, J.; Wu, C.H.; Panstruga, R. Rapid evolution in plant-microbe interactions—A molecular genomics perspective. New Phytol. 2020, 225, 1134–1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wojtaszek, P. Oxidative burst: An early plant response to pathogen infection. Biochem. J. 1997, 322, 681–692. [Google Scholar] [CrossRef] [Green Version]
- Camejo, D.; Guzmán-Cedeño, Á.; Moreno, A. Reactive oxygen species, essential molecules, during plant-pathogen interactions. Plant Physiol. Biochem. 2016, 103, 10–23. [Google Scholar] [CrossRef]
- Rausher, M.D. Co-evolution and plant resistance to natural enemies. Nature 2001, 411, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Shetty, N.P.; Jørgensen, H.J.L.; Jensen, J.D.; Collinge, D.B.; Shetty, H.S. Roles of reactive oxygen species in interactions between plants and pathogens. Eur. J. Plant Pathol. 2008, 121, 267–280. [Google Scholar] [CrossRef]
- Mittler, R. ROS are good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef] [Green Version]
- Scheel, D. Oxidative burst and the role of reactive oxygen species in plant-pathogen interactions. In Oxidative Stress in Plants; Inze, D., Van Montagu, M., Eds.; Taylor and Francis: New York, NY, USA, 2002; pp. 137–153. [Google Scholar]
- Zurbriggen, M.D.; Carrillo, N.; Hajirezaei, M.-R. ROS signaling in the hypersensitive response: When, where and what for? Plant Signal. Behav. 2010, 5, 393–396. [Google Scholar] [CrossRef] [Green Version]
- Hofius, D.; Tsitsigiannis, D.I.; Jones, J.D.; Mundy, J. Inducible cell death in plant immunity. Semin. Cancer Biol. 2007, 17, 166–187. [Google Scholar] [CrossRef]
- Rodriguez-Moreno, L.; Ebert, M.K.; Bolton, M.D.; Thomma, B.P. Tools of the crook-infection strategies of fungal plant pathogens. Plant J. 2018, 93, 664–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, S.; Shan, L.; He, P. Microbial signature-triggered plant defense responses and early signaling mechanisms. Plant Sci. 2014, 228, 118–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, G.A.; Terol, J.; Ibanez, V.; López-García, A.; Pérez-Román, E.; Borredá, C.; Domingo, C.; Tadeo, F.R.; Carbonell-Caballero, J.; Alonso, R. Genomics of the origin and evolution of Citrus. Nature 2018, 554, 311–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woudenberg, J.; Groenewald, J.; Binder, M.; Crous, P. Alternaria redefined. Stud. Mycol. 2013, 75, 171–212. [Google Scholar] [CrossRef] [Green Version]
- Tsuge, T.; Harimoto, Y.; Akimitsu, K.; Ohtani, K.; Kodama, M.; Akagi, Y.; Egusa, M.; Yamamoto, M.; Otani, H. Host-selective toxins produced by the plant pathogenic fungus Alternaria alternata. FEMS Microbiol. Rev. 2013, 37, 44–66. [Google Scholar] [CrossRef]
- Isshiki, A.; Akimitsu, K.; Nishio, K.; Tsukamoto, M.; Yamamoto, H. Purification and characterization of an endopolygalacturonase from the rough lemon pathotype of Alternaria alternata, the cause of citrus brown spot disease. Physiol. Mol. Plant Pathol. 1997, 51, 155–167. [Google Scholar] [CrossRef]
- Chung, K.-R. Stress response and pathogenicity of the necrotrophic fungal pathogen Alternaria alternata. Scientifica 2012, 2012, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Masunaka, A.; Ohtani, K.; Peever, T.; Timmer, L.; Tsuge, T.; Yamamoto, M.; Yamamoto, H.; Akimitsu, K. An isolate of Alternaria alternata that is pathogenic to both tangerines and rough lemon and produces two host-selective toxins, ACT-and ACR-toxins. Phytopathology 2005, 95, 241–247. [Google Scholar] [CrossRef] [Green Version]
- Izumi, Y.; Ohtani, K.; Miyamoto, Y.; Masunaka, A.; Fukumoto, T.; Gomi, K.; Tada, Y.; Ichimura, K.; Peever, T.; Akimitsu, K. A polyketide synthase gene, ACRTS2, is responsible for biosynthesis of host-selective ACR-toxin in the rough lemon pathotype of Alternaria alternata. Mol. Plant-Microbe Interact. 2012, 25, 1419–1429. [Google Scholar] [CrossRef] [Green Version]
- Huang, F.; Fu, Y.; Nie, D.; Stewart, J.E.; Peever, T.L.; Li, H. Identification of a novel phylogenetic lineage of Alternaria alternata causing citrus brown spot in China. Fungal Biol. 2015, 119, 320–330. [Google Scholar] [CrossRef]
- Huang, S.; Jia, Z.; Li, H.; Zhang, S.; Shen, J.; Gai, Y.; Jiao, C.; Sun, X.; Duan, S.; Wang, M. ACT-Toxin, the Key Effector for the Virulence of Alternaria alternata Tangerine Pathotype to Specific Citrus Species. Agronomy 2022, 12, 3181. [Google Scholar] [CrossRef]
- Timmer, L.; Solel, Z.; Gottwald, T.; Ibanez, A.; Zitko, S. Environmental factors affecting production, release, and field populations of conidia of Alternaria alternata, the cause of brown spot of citrus. Phytopathology 1998, 88, 1218–1223. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.L.; Yu, P.L.; Chung, K.R. The glutathione peroxidase-mediated reactive oxygen species resistance, fungicide sensitivity and cell wall construction in the citrus fungal pathogen Alternaria alternata. Environ. Microbiol. 2016, 18, 923–935. [Google Scholar] [CrossRef]
- Ma, H.; Wang, M.; Gai, Y.; Fu, H.; Zhang, B.; Ruan, R.; Chung, K.-R.; Li, H. Thioredoxin and glutaredoxin systems required for oxidative stress resistance, fungicide sensitivity, and virulence of Alternaria alternata. Appl. Environ. Microbiol. 2018, 84, e00086-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Ma, H.; Zheng, F.; Chen, Y.; Wang, M.; Jiao, C.; Li, H.; Gai, Y. The transcription regulator ACTR controls ACT-toxin biosynthesis and pathogenicity in the tangerine pathotype of Alternaria alternata. Microbiol. Res. 2021, 248, 126747. [Google Scholar] [CrossRef]
- Gai, Y.; Liu, B.; Ma, H.; Li, L.; Chen, X.; Moenga, S.; Riely, B.; Fayyaz, A.; Wang, M.; Li, H. The methionine biosynthesis regulator AaMetR contributes to oxidative stress tolerance and virulence in Alternaria alternata. Microbiol. Res. 2019, 219, 94–109. [Google Scholar] [CrossRef]
- Gai, Y.; Li, L.; Ma, H.; Riely, B.K.; Liu, B.; Li, H. Critical Role of MetR/MetB/MetC/MetX in Cysteine and Methionine Metabolism, Fungal Development, and Virulence of Alternaria alternata. Appl. Environ. Microbiol. 2021, 87, e01911-20. [Google Scholar] [CrossRef]
- Lin, C.-H.; Yang, S.L.; Chung, K.-R. The YAP1 homolog-mediated oxidative stress tolerance is crucial for pathogenicity of the necrotrophic fungus Alternaria alternata in citrus. Mol. Plant-Microbe Interact. 2009, 22, 942–952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gai, Y.; Ma, H.; Chen, Y.; Li, L.; Cao, Y.; Wang, M.; Sun, X.; Jiao, C.; Riely, B.K.; Li, H. Chromosome-scale genome sequence of Alternaria alternata causing Alternaria Brown Spot of Citrus. Mol. Plant Microbe Interact. 2021, 34, 726–732. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Sun, X.; Yu, D.; Xu, J.; Chung, K.; Li, H. Genomic and transcriptomic analyses of the tangerine pathotype of Alternaria alternata in response to oxidative stress. Sci. Rep. 2016, 6, srep32437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gai, Y.; Li, L.; Liu, B.; Ma, H.; Chen, Y.; Zheng, F.; Sun, X.; Wang, M.; Jiao, C.; Li, H. Distinct and essential roles of bZIP transcription factors in the stress response and pathogenesis in Alternaria alternata. Microbiol. Res. 2022, 256, 126915. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [Green Version]
- Guy, L.; Roat Kultima, J.; Andersson, S.G. genoPlotR: Comparative gene and genome visualization in R. Bioinformatics 2010, 26, 2334–2335. [Google Scholar] [CrossRef] [Green Version]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Wang, Z.; Dan, Z.; Zhang, L.; Xu, M.; Yang, G.; Chai, M.; Li, Z.; Xie, H.; Cong, L. Transcriptome Analysis of Fusarium Root-Rot-Resistant and-Susceptible Alfalfa (Medicago sativa L.) Plants during Plant-Pathogen Interactions. Genes 2022, 13, 788. [Google Scholar] [CrossRef] [PubMed]
- Deboever, E.; Deleu, M.; Mongrand, S.; Lins, L.; Fauconnier, M.-L. Plant-pathogen interactions: Underestimated roles of phyto-oxylipins. Trends Plant Sci. 2020, 25, 22–34. [Google Scholar] [CrossRef] [Green Version]
- Mendoza-Martínez, A.E.; Cano-Dominguez, N.; Aguirre, J. Yap1 homologs mediate more than the redox regulation of the antioxidant response in filamentous fungi. Fungal Biol. 2020, 124, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Herrero, E.; Ros, J.; Bellí, G.; Cabiscol, E. Redox control and oxidative stress in yeast cells. Biochim. Et Biophys. Acta (BBA) Gen. Subj. 2008, 1780, 1217–1235. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.L.; Wang, C.L.; Chen, P.Y.; Lee, M.H. YAP1 homologue-mediated redox sensing is crucial for a successful infection by Monilinia fructicola. Mol. Plant Pathol. 2017, 18, 783–797. [Google Scholar] [CrossRef]
- Voolstra, C.R.; Ziegler, M. Adapting with microbial help: Microbiome flexibility facilitates rapid responses to environmental change. BioEssays 2020, 42, e2000004. [Google Scholar] [CrossRef]
- Leal, S.M.; Vareechon, C.; Cowden, S.; Cobb, B.A.; Latgé, J.-P.; Momany, M.; Pearlman, E. Fungal antioxidant pathways promote survival against neutrophils during infection. J. Clin. Investig. 2012, 122, 2482–2498. [Google Scholar] [CrossRef] [Green Version]
- Torres, M.A. ROS in biotic interactions. Physiol. Plant. 2010, 138, 414–429. [Google Scholar] [CrossRef]
- Wang, M.; Yang, X.; Ruan, R.; Fu, H.; Li, H. Csn5 is required for the conidiogenesis and pathogenesis of the Alternaria alternata tangerine pathotype. Front. Microbiol. 2018, 9, 508. [Google Scholar] [CrossRef] [Green Version]
- Fu, H.; Chung, K.R.; Gai, Y.; Mao, L.; Li, H. The basal transcription factor II H subunit Tfb5 is required for stress response and pathogenicity in the tangerine pathotype of Alternaria alternata. Mol. Plant Pathol. 2020, 21, 1337–1352. [Google Scholar] [CrossRef] [PubMed]





| Gene ID | Protein ID | WT + 12 h | WT + 24 h | WT + 48 h | WT + H2O2 | Annotation |
|---|---|---|---|---|---|---|
| AALT_g8702 | OWY49743.1 | −3.59 | −4.57 | −6.59 | −1.16 | CBM18, carbohydrate binding module family 18 |
| AALT_g9592 | OWY42369.1 | −4.42 | −2.14 | −4.00 | −1.84 | FTR1, iron permease FTR1 family |
| AALT_g6701 | OWY55098.1 | −3.51 | −2.61 | −3.97 | 2.05 | ERG3, sterol desaturase family |
| AALT_g260 | OWY42744.1 | −3.48 | −2.28 | −4.23 | −0.61 | ARG1, arginosuccinate synthase |
| AALT_g3024 | OWY52659.1 | −3.34 | −3.98 | −2.27 | −0.73 | Glycoside hydrolase family 28 protein |
| AALT_g3675 | OWY44810.1 | −3.51 | −2.48 | −3.18 | −0.01 | Jacalin like lectin domain |
| AALT_g5000 | OWY57849.1 | −2.27 | −2.82 | −3.32 | −0.67 | bZIP transcription factor |
| AALT_g7369 | OWY48574.1 | −3.45 | −1.70 | −2.93 | −0.01 | GPP1, haloacid dehalogenase like hydrolase |
| AALT_g7743 | OWY50139.1 | −2.77 | −1.99 | −3.17 | 0.13 | ERG10, belongs to the thiolase family |
| AALT_g7699 | OWY48045.1 | −2.59 | −2.33 | −2.94 | 1.28 | ERG11, belongs to the cytochrome P450 family |
| AALT_g2814 | OWY52449.1 | −2.12 | −2.37 | −3.15 | 0.09 | CCP1, belongs to the peroxidase family |
| AALT_g4788 | OWY57637.1 | −2.57 | −1.79 | −3.04 | −0.94 | URA5, phosphoribosyl transferase domain |
| AALT_g3969 | OWY45104.1 | −3.08 | −1.11 | −3.03 | −0.57 | ARG4, argininosuccinate lyase C terminal |
| AALT_g1579 | OWY41826.1 | −2.39 | −2.42 | −2.19 | 0.65 | Major facilitator superfamily |
| AALT_g5087 | OWY57936.1 | −1.88 | −2.12 | −2.96 | −0.21 | ARO4, stereospecific condensation of PEP |
| AALT_g9214 | OWY57068.1 | −2.36 | −1.77 | −2.69 | −0.31 | cyp1, PPIases accelerate the folding of proteins. |
| AALT_g9127 | OWY51854.1 | −2.14 | −1.43 | −2.80 | 0.26 | CDC7, protein tyrosine kinase |
| AALT_g3183 | OWY52818.1 | −2.08 | −1.48 | −2.47 | 0.14 | Mitochondrial PGP phosphatase |
| AALT_g6591 | OWY47556.1 | 2.20 | 2.79 | 1.29 | −0.24 | Belongs to the thiolase family |
| AALT_g8160 | OWY48788.1 | 2.41 | 2.18 | 2.03 | 1.04 | cytochrome P450 |
| AALT_g1296 | OWY41543.1 | 1.87 | 2.25 | 2.57 | 0.03 | Belongs to the glycosyl hydrolase 31 family |
| AALT_g6581 | OWY47546.1 | 2.25 | 2.11 | 2.34 | −0.06 | Homeodomain |
| AALT_g5268 | OWY58117.1 | 2.21 | 2.39 | 2.11 | −1.10 | CHS3, chitin synthase |
| AALT_g374 | OWY42858.1 | 2.53 | 1.58 | 2.95 | 0.55 | NCR1, Niemann Pick C1 N terminus |
| AALT_g4774 | OWY57623.1 | 2.55 | 2.39 | 2.27 | 0.03 | MaoC like domain |
| AALT_g4259 | OWY50973.1 | 1.92 | 2.22 | 3.20 | −0.44 | cyp3, PPIases accelerate the folding of proteins. |
| AALT_g8336 | OWY45374.1 | 2.39 | 1.42 | 3.68 | −0.20 | Alcohol dehydrogenase GroES like domain |
| AALT_g6866 | OWY55263.1 | 1.73 | 3.09 | 2.74 | −0.59 | Enoyl (acyl carrier protein) reductase |
| AALT_g65 | OWY42549.1 | 2.49 | 2.42 | 2.92 | −0.35 | Zinc finger transcription factor |
| AALT_g12014 | OWY45860.1 | 2.92 | 1.32 | 3.68 | −2.07 | Fungal specific transcription factor domain |
| AALT_g3883 | OWY45018.1 | 3.15 | 2.49 | 2.30 | −0.12 | Fungal specific transcription factor domain |
| AALT_g11132 | OWY55775.1 | 2.04 | 3.38 | 2.73 | 0.62 | Chitin binding domain |
| AALT_g6936 | OWY55333.1 | 2.50 | 3.71 | 2.34 | −0.13 | the iron ascorbate dependent oxidoreductase family |
| AALT_g10705 | OWY51536.1 | 3.70 | 2.70 | 2.63 | −2.75 | plyE, polysaccharide lyase family 3 protein |
| AALT_g5513 | OWY43764.1 | 3.71 | 2.74 | 3.90 | −0.41 | C2H2 type zinc finger |
| AALT_g6798 | OWY55195.1 | 3.27 | 3.49 | 4.22 | 0.17 | Basic region leucine zipper |
| AALT_g2197 | OWY46214.1 | 3.90 | 3.36 | 3.78 | −0.42 | Glycosyl hydrolase family 61 |
| AALT_g9490 | OWY49079.1 | 2.97 | 4.35 | 4.33 | 0.92 | CDR1, belongs to the ABC transporter superfamily |
| AALT_g1240 | OWY41487.1 | 4.00 | 4.29 | 3.56 | −0.31 | ENA2, belongs to the cation transport ATPase |
| AALT_g1011 | OWY41258.1 | 4.03 | 3.97 | 3.95 | −0.05 | C2H2 type zinc finger |
| AALT_g5353 | OWY43604.1 | 2.88 | 4.67 | 4.50 | −0.44 | Belongs to the thiolase family |
| AALT_g5423 | OWY43674.1 | 3.63 | 3.60 | 4.86 | 0.75 | scytalone dehydratase |
| AALT_g2730 | OWY46747.1 | 4.64 | 4.64 | 3.16 | 1.81 | polyketide synthase |
| AALT_g9423 | OWY49012.1 | 3.16 | 3.20 | 6.11 | 0.54 | Polysaccharide lyase family 1 protein |
| AALT_g6502 | OWY47467.1 | 3.17 | 4.67 | 4.71 | −0.63 | Polysaccharide lyase family 3 protein |
| AALT_g9572 | OWY42349.1 | 6.23 | 2.92 | 4.05 | −1.50 | brlA, zinc finger |
| AALT_g4000 | OWY45135.1 | 4.02 | 4.97 | 4.72 | −0.20 | ICL1, the isocitrate lyase PEP mutase superfamily. |
| AALT_g8883 | OWY54340.1 | 5.14 | 3.98 | 4.75 | −0.66 | the glycosyl hydrolase 11 (cellulase G) family |
| AALT_g1567 | OWY41814.1 | 3.53 | 6.08 | 4.49 | 0.36 | FAD binding domain |
| AALT_g3819 | OWY44954.1 | 3.84 | 4.78 | 6.69 | −0.37 | Polysaccharide lyase family 3 protein |
| AALT_g6230 | OWY47195.1 | 4.38 | 4.75 | 6.50 | 0.06 | the ABC transporter superfamily. ABCG family |
| AALT_g600 | OWY43084.1 | 4.57 | 5.71 | 5.45 | −0.14 | CDR1, the ABC transporter superfamily |
| AALT_g9863 | OWY52019.1 | 7.15 | 5.42 | 4.93 | −0.98 | EXG2, the glycosyl hydrolase 5 (cellulase A) family |
| AALT_g11942 | OWY51608.1 | 4.65 | 6.50 | 6.64 | 0.00 | KR domain |
| AALT_g2054 | OWY46071.1 | 5.91 | 6.53 | 6.60 | −1.02 | ENA2, the cation transport ATPase family |
| AALT_g9836 | OWY51992.1 | 5.81 | 5.87 | 8.22 | 0.39 | Pectate lyase |
| AALT_g9484 | OWY49073.1 | 5.23 | 6.91 | 8.24 | −0.41 | Polysaccharide lyase family 3 protein |
| AALT_g11182 | OWY53848.1 | 6.33 | 8.90 | 6.87 | −2.50 | SnoaL like domain |
| AALT_g11588 | OWY44559.1 | 7.27 | 8.13 | 7.22 | 0.96 | GMC oxidoreductase |
| AALT_g8391 | OWY45429.1 | 7.29 | 8.33 | 7.63 | 0.67 | Ricin b lectin |
| AALT_g3371 | OWY53006.1 | 9.60 | 8.07 | 7.13 | −0.04 | Fungal trichothecene efflux pump (TRI12) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gai, Y.; Niu, Q.; Kong, J.; Li, L.; Liang, X.; Cao, Y.; Zhou, X.; Sun, X.; Ma, H.; Wang, M.; et al. Genomic and Transcriptomic Characterization of Alternaria alternata during Infection. Agronomy 2023, 13, 809. https://doi.org/10.3390/agronomy13030809
Gai Y, Niu Q, Kong J, Li L, Liang X, Cao Y, Zhou X, Sun X, Ma H, Wang M, et al. Genomic and Transcriptomic Characterization of Alternaria alternata during Infection. Agronomy. 2023; 13(3):809. https://doi.org/10.3390/agronomy13030809
Chicago/Turabian StyleGai, Yunpeng, Qichen Niu, Jinchao Kong, Lei Li, Xingxing Liang, Yuwei Cao, Xianqi Zhou, Xuepeng Sun, Haijie Ma, Mingshuang Wang, and et al. 2023. "Genomic and Transcriptomic Characterization of Alternaria alternata during Infection" Agronomy 13, no. 3: 809. https://doi.org/10.3390/agronomy13030809
APA StyleGai, Y., Niu, Q., Kong, J., Li, L., Liang, X., Cao, Y., Zhou, X., Sun, X., Ma, H., Wang, M., Shrivastava, N., Li, H., & Jiao, C. (2023). Genomic and Transcriptomic Characterization of Alternaria alternata during Infection. Agronomy, 13(3), 809. https://doi.org/10.3390/agronomy13030809








