Improving Deep Learning Classifiers Performance via Preprocessing and Class Imbalance Approaches in a Plant Disease Detection Pipeline
Abstract
:1. Introduction
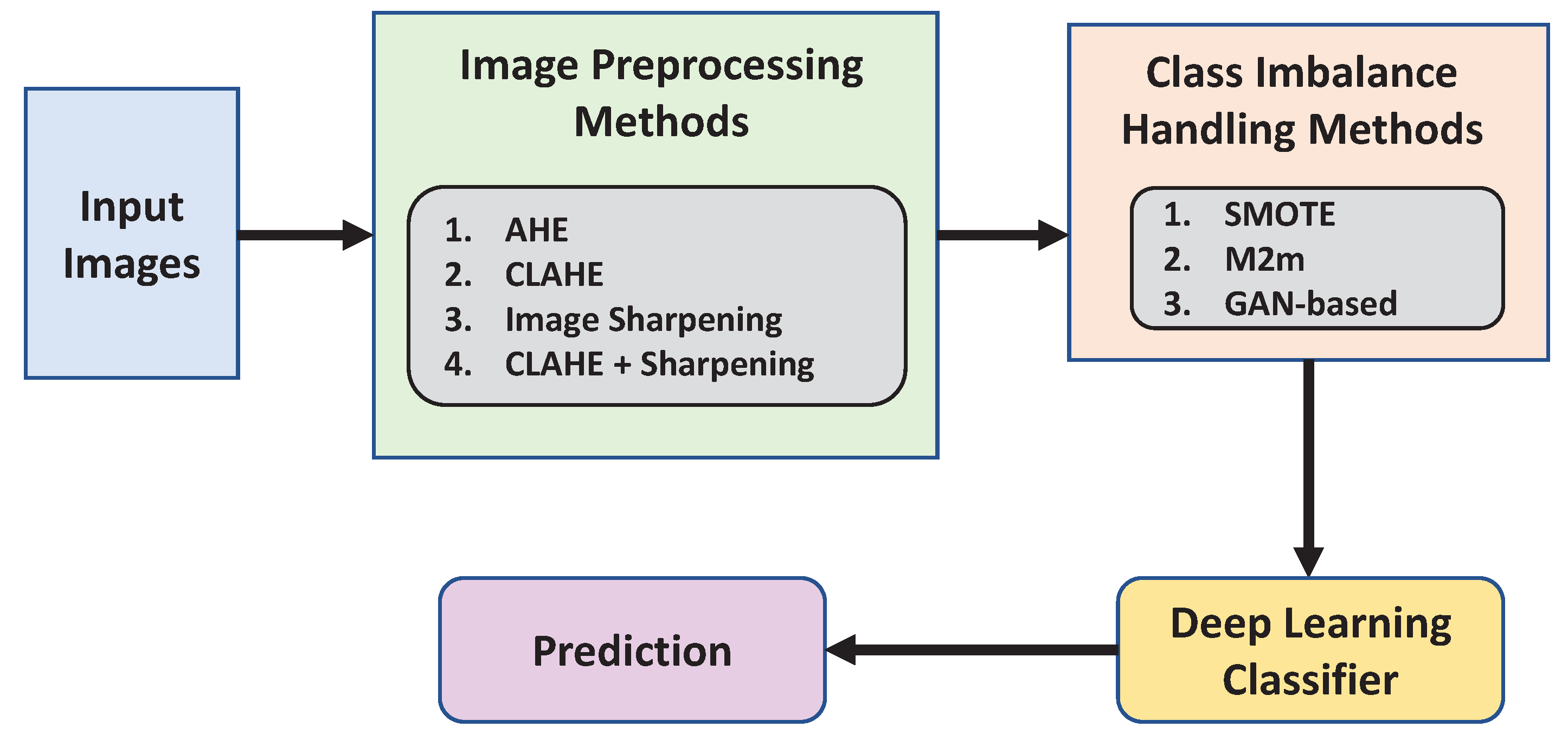
- To increase the pertinent features in the plant leaf, we conduct a qualitative and quantitative evaluation of a variety of image pre-processing approaches (described in Section 2.1. We discovered that contrast limited adaptive histogram equalization (CLAHE), in conjunction with image sharpening, can give a deep classifier a notable significant improvement.
- We examine the effectiveness of data balancing approaches across many categories, focusing on how adding new synthetic data affects the classifier’s ability to learn. With the use of three data augmentation techniques—synthetic minority oversampling technique (SMOTE), generative adversarial networks (GAN), and major-to-minor translation (M2m)—we conducted extensive tests on the data set.
- We investigate the detection of bacterial wilt disease on tomato plants, which is unique due to its symptoms.
2. Materials and Methods
2.1. Image Preprocessing Techniques
2.1.1. Adaptive Histogram Equalization (AHE)
2.1.2. Contrast Limited Adaptive Histogram Equalization (CLAHE)
2.1.3. Image Sharpening (SH)
2.1.4. CLAHE + Sharpening (CL + SH)
2.2. Addressing Class Imbalance
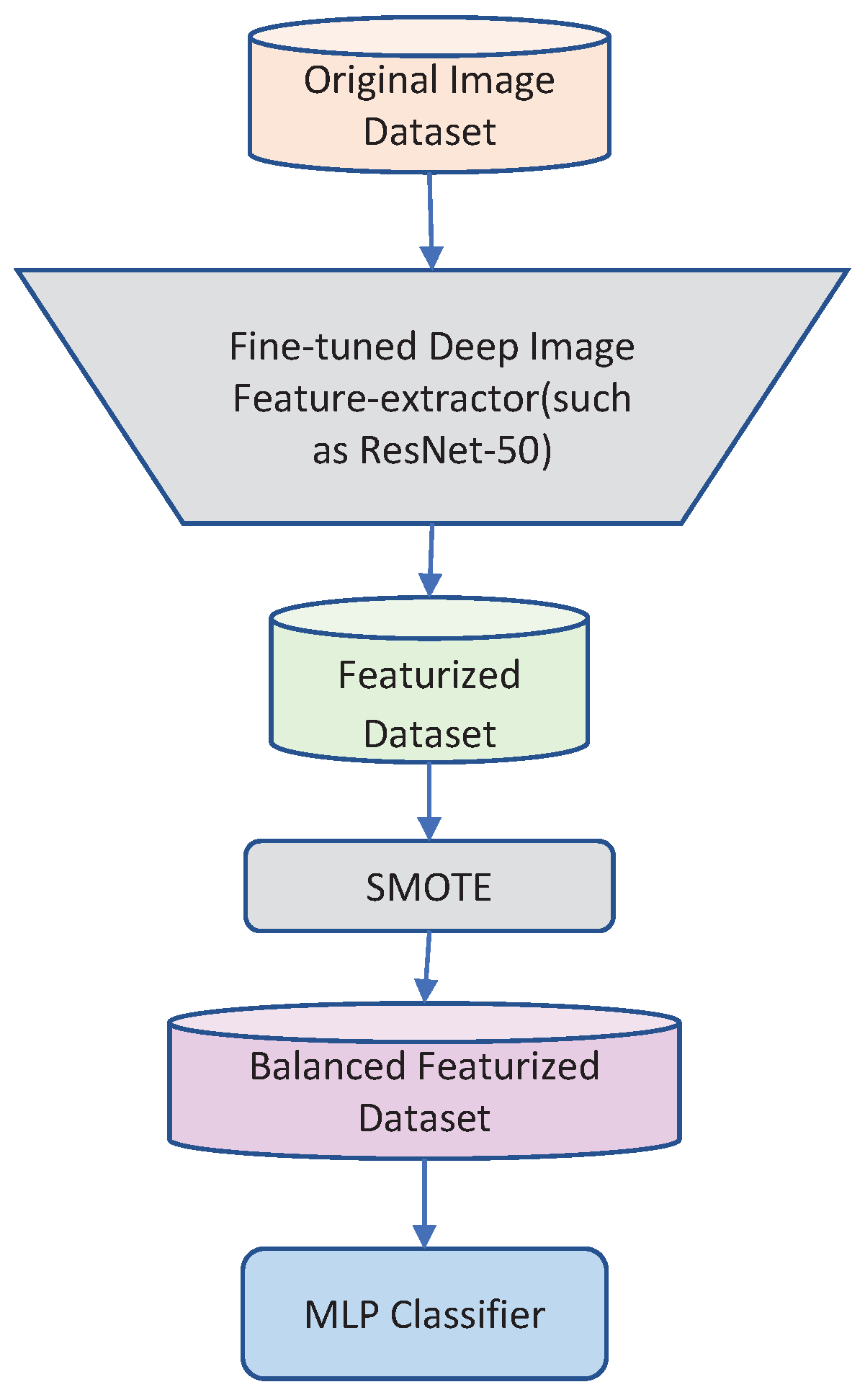
2.2.1. Synthetic Minority Oversampling Technique (SMOTE)
2.2.2. Major-to-Minor Translation (M2m)
2.2.3. Generative Adversarial Networks
2.3. Deep Classifier Selection
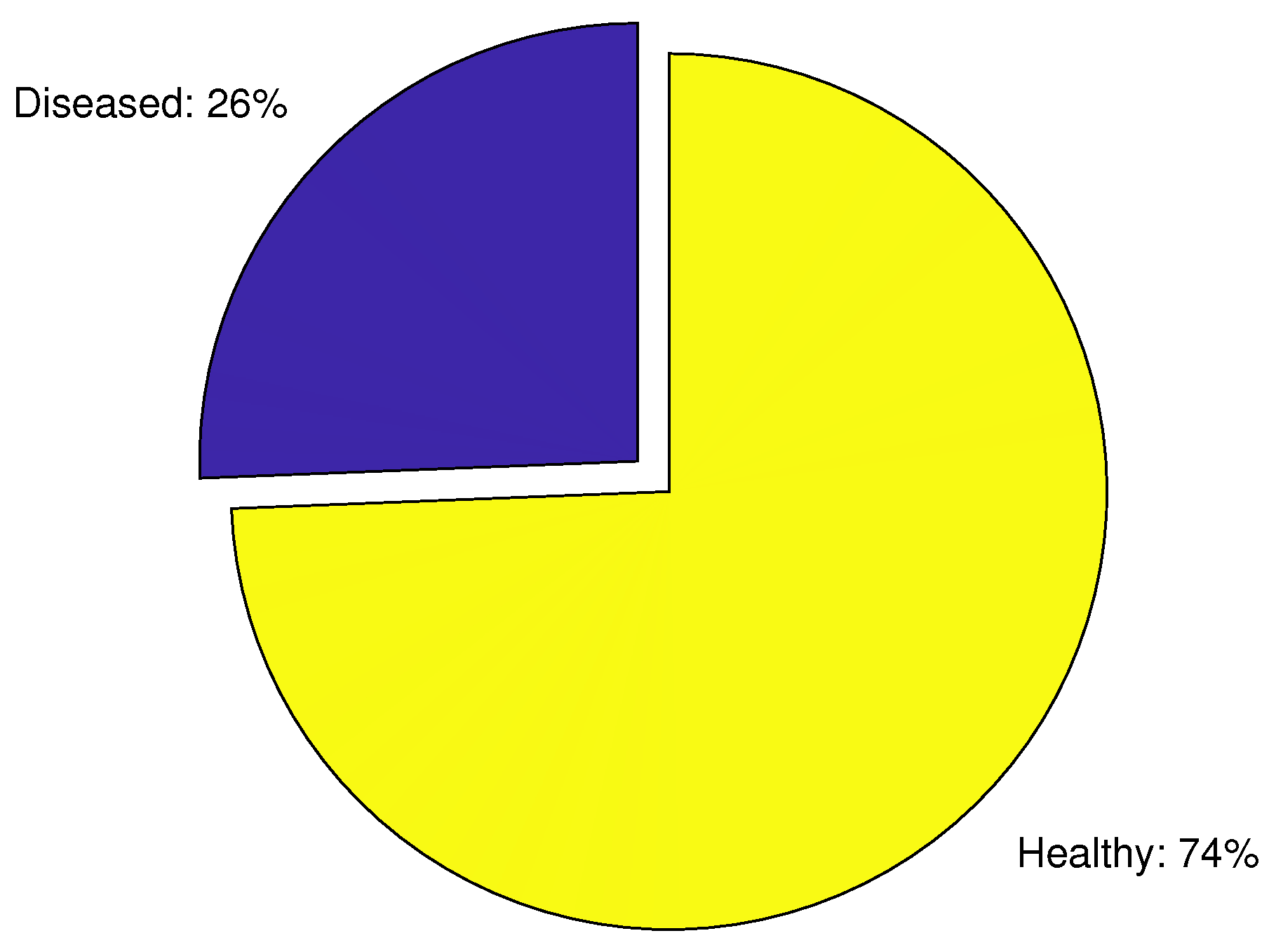
2.4. Data Preparation Phase
2.5. Disease Inoculation Procedure
2.6. Image Acquisition
3. Results and Discussion
3.1. Experimental Protocol
3.1.1. Average Classification Accuracy (ACA)
3.1.2. Precision
3.1.3. F1-Score
3.1.4. Area under Curve (AUC)
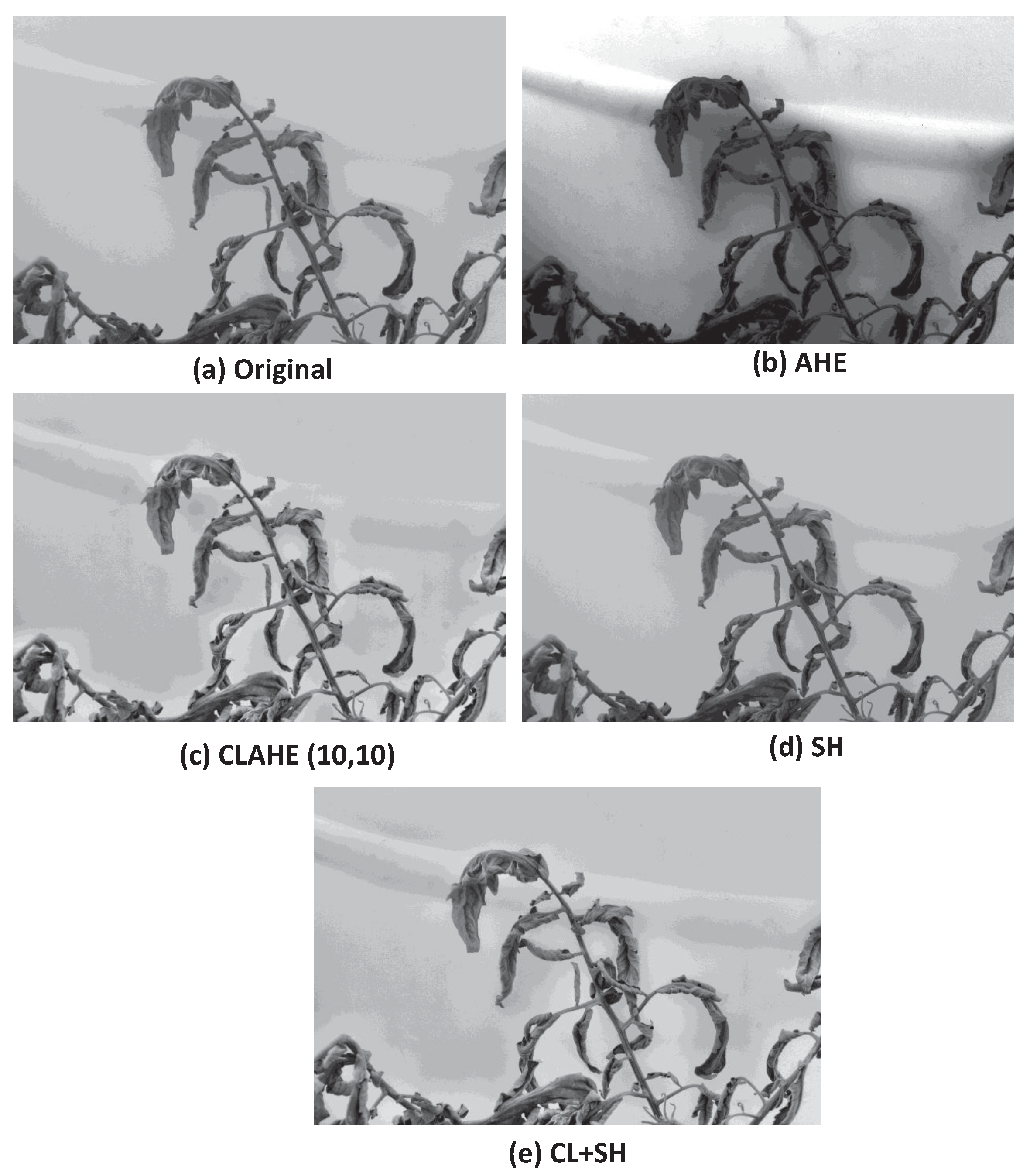
3.2. Qualitative Analysis of Image Preprocessing Techniques
3.2.1. AHE
3.2.2. CLAHE
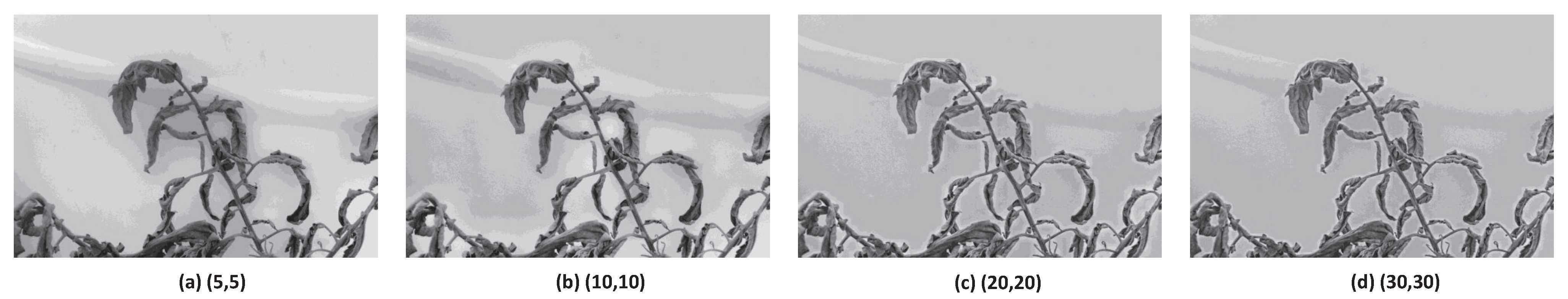
3.2.3. Image Sharpening
3.2.4. CLAHE + Sharpening (CL + SH)
3.2.5. Quality of the Augmented Data
3.3. Quantitative Analysis
3.3.1. Choosing the Base Classifier
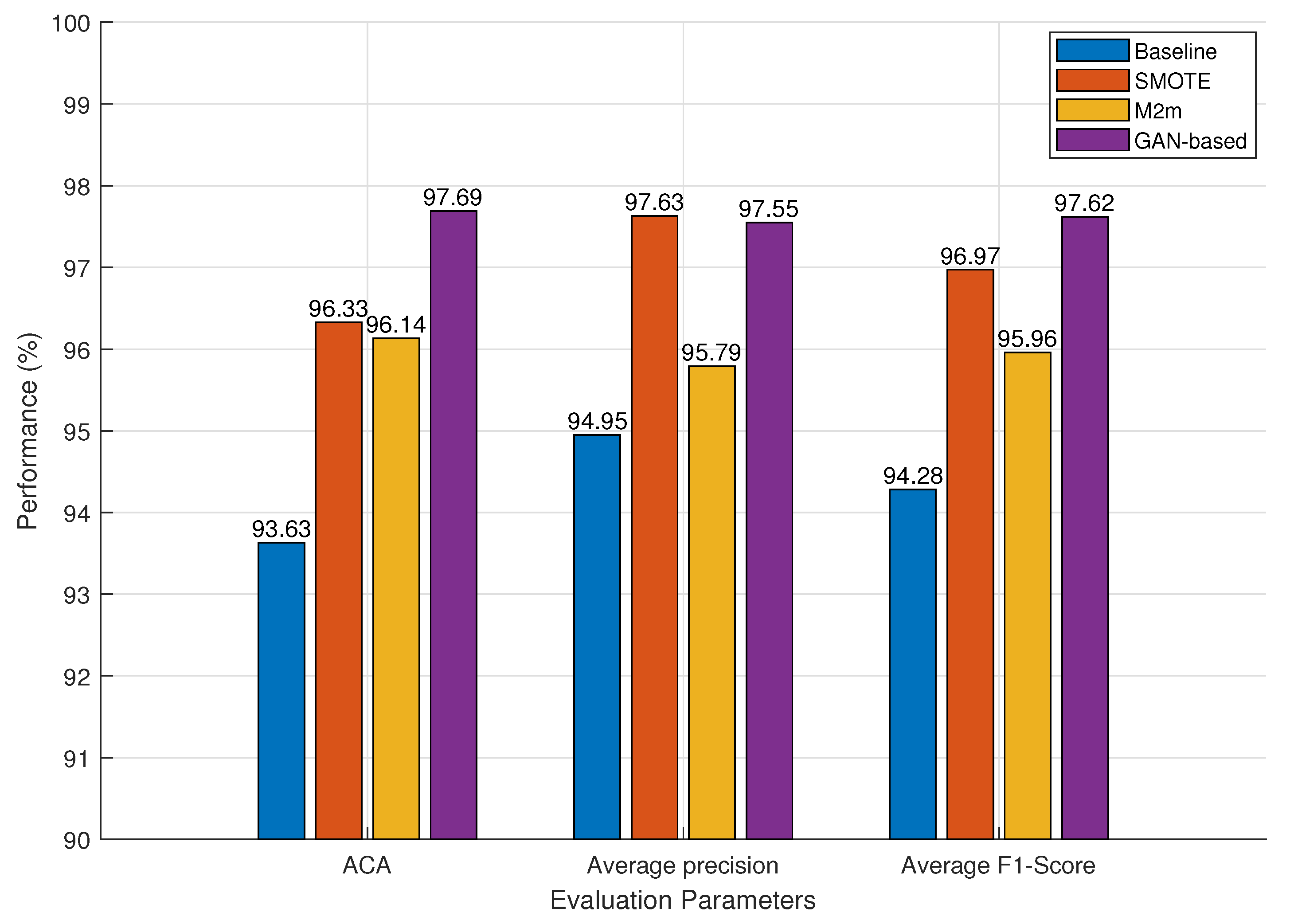
3.3.2. Impact of Class Handling Methods
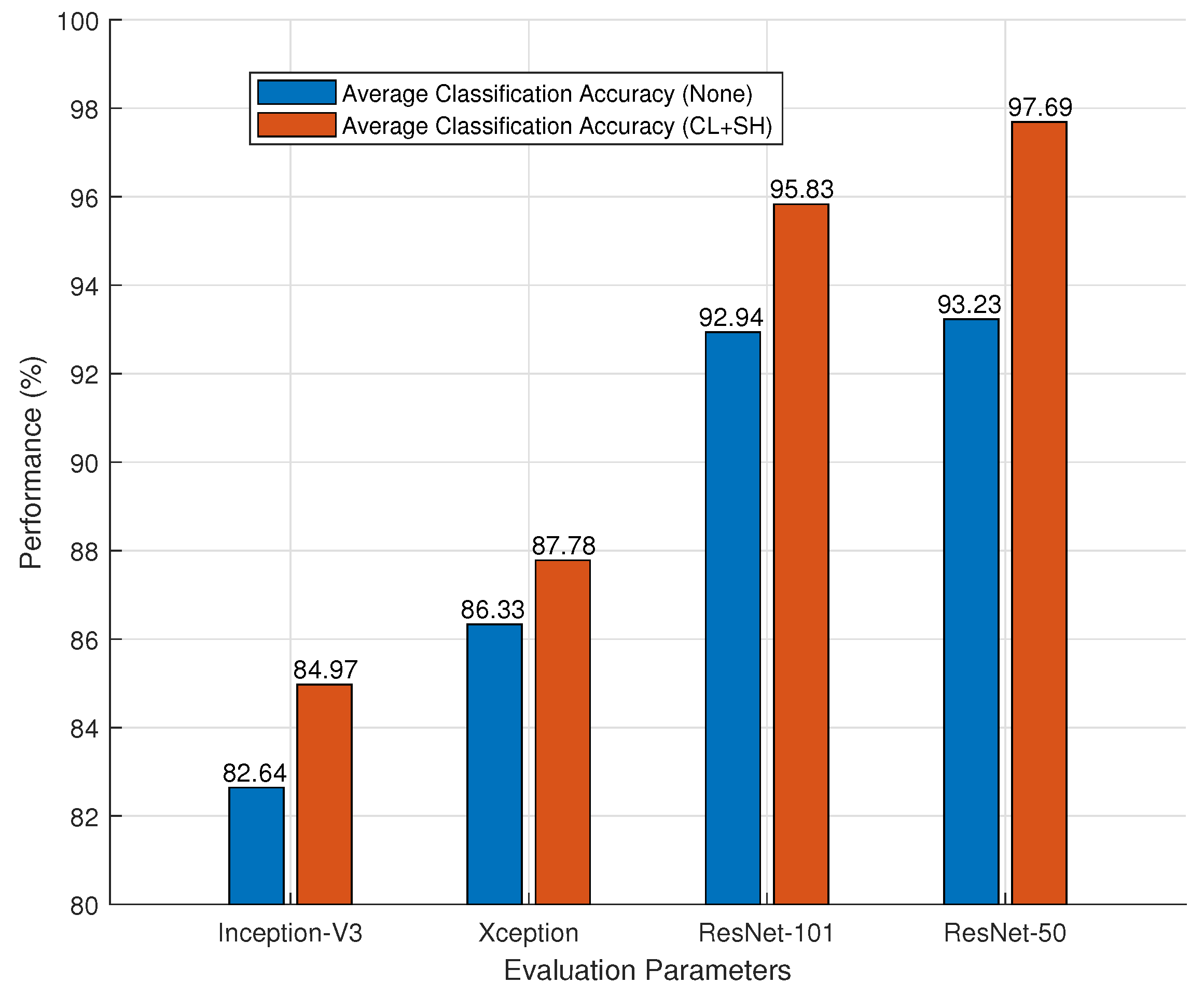
3.3.3. Impact of Preprocessing Techniques
3.3.4. Lessons Learned and Future Directions
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bai, Y.; Xiong, Y.; Huang, J.; Zhou, J.; Zhang, B. Accurate prediction of soluble solid content of apples from multiple geographical regions by combining deep learning with spectral fingerprint features. Postharvest Biol. Technol. 2019, 156, 110943. [Google Scholar] [CrossRef]
- World Health Organization. The State of Food Security and Nutrition in the World 2018: Building Climate Resilience for Food Security and Nutrition; Food & Agriculture Org: Rome, Italy, 2018.
- Ojo, M.O.; Zahid, A. Deep learning in controlled environment agriculture: A review of recent advancements, challenges and prospects. Sensors 2022, 22, 7965. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Ganapathysubramanian, B.; Sarkar, S.; Singh, A. Deep learning for plant stress phenotyping: Trends and future perspectives. Trends Plant Sci. 2018, 23, 883–898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohanty, S.P.; Hughes, D.P.; Salathé, M. Using deep learning for image-based plant disease detection. Front. Plant Sci. 2016, 7, 1419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferentinos, K.P. Deep learning models for plant disease detection and diagnosis. Comput. Electron. Agric. 2018, 145, 311–318. [Google Scholar] [CrossRef]
- Caldwell, D.; Kim, B.-S.; Iyer-Pascuzzi, A.S. Ralstonia solanacearum differentially colonizes roots of resistant and susceptible tomato plants. Phytopathology 2017, 107, 528–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Unay, D.; Gosselin, B.; Kleynen, O.; Leemans, V.; Destain, M.-F.; Debeir, O. Automatic grading of bi-colored apples by multispectral machine vision. Comput. Electron. Agric. 2011, 75, 204–212. [Google Scholar] [CrossRef] [Green Version]
- Gavhale, K.R.; Gawande, U.; Hajari, K.O. Unhealthy region of citrus leaf detection using image processing techniques. In Proceedings of the International Conference for Convergence for Technology-2014, Pune, India, 6–8 April 2014; pp. 1–6. [Google Scholar]
- Vetal, S.; Khule, R. Tomato plant disease detection using image processing. Int. J. Adv. Res. Comput. Commun. Eng. 2017, 6, 293–297. [Google Scholar] [CrossRef]
- Sun, Y.; Kamel, M.S.; Wong, A.K.; Wang, Y. Cost-sensitive boosting for classification of imbalanced data. Pattern Recognit. 2007, 40, 3358–3378. [Google Scholar] [CrossRef]
- Japkowicz, N. Supervised versus unsupervised binary-learning by feedforward neural networks. Mach. Learn. 2001, 42, 97–122. [Google Scholar] [CrossRef] [Green Version]
- Ling, C.X.; Sheng, V.S. Cost-sensitive learning and the class imbalance problem. Encycl. Mach. Learn. 2008, 2011, 231–235. [Google Scholar]
- Johnson, J.M.; Khoshgoftaar, T.M. Survey on deep learning with class imbalance. J. Big Data 2019, 6, 27. [Google Scholar] [CrossRef] [Green Version]
- Ling, C.X.; Li, C. Data mining for direct marketing: Problems and solutions. KDD 1998, 98, 73–79. [Google Scholar]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Janowczyk, A.; Madabhushi, A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J. Pathol. Inform. 2016, 7, 29. [Google Scholar] [CrossRef]
- Zhang, C.; Zhou, P.; Li, C.; Liu, L. A convolutional neural network for leaves recognition using data augmentation. In Proceedings of the 2015 IEEE International Conference on Computer and Information Technology; Ubiquitous Computing and Communications; Dependable, Autonomic and Secure Computing; Pervasive Intelligence and Computing, Liverpool, UK, 26–28 October 2015; pp. 2143–2150. [Google Scholar]
- Pan, Z.; Yu, W.; Yi, X.; Khan, A.; Yuan, F.; Zheng, Y. Recent progress on generative adversarial networks (gans): A survey. IEEE Access 2019, 7, 36322–36333. [Google Scholar] [CrossRef]
- Mai, G.; Cao, K.; Yuen, P.C.; Jain, A.K. On the reconstruction of face images from deep face templates. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 41, 1188–1202. [Google Scholar] [CrossRef] [Green Version]
- Hu, G.; Wu, H.; Zhang, Y.; Wan, M. A low shot learning method for tea leaf’s disease identification. Comput. Electron. Agric. 2019, 163, 104852. [Google Scholar] [CrossRef]
- Saikawa, T.; Cap, Q.H.; Kagiwada, S.; Uga, H.; Iyatomi, H. Aop: An anti-overfitting pretreatment for practical image-based plant diagnosis. In Proceedings of the 2019 IEEE International Conference on Big Data (Big Data), Los Angeles, CA, USA, 9–12 December 2019; pp. 5177–5182. [Google Scholar]
- Mirza, M.; Osindero, S. Conditional generative adversarial nets. arXiv 2014, arXiv:1411.1784. [Google Scholar]
- Tian, Y.; Yang, G.; Wang, Z.; Li, E.; Liang, Z. Detection of apple lesions in orchards based on deep learning methods of cyclegan and yolov3-dense. J. Sens. 2019, 2019, 7630926. [Google Scholar] [CrossRef]
- Cap, Q.H.; Uga, H.; Kagiwada, S.; Iyatomi, H. Leafgan: An effective data augmentation method for practical plant disease diagnosis. IEEE Trans. Autom. Sci. Eng. 2020, 1258–1267. [Google Scholar] [CrossRef]
- Kim, J.; Jeong, J.; Shin, J. M2m: Imbalanced classification via major-to-minor translation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 13896–13905. [Google Scholar]
- Radford, A.; Metz, L.; Chintala, S. Unsupervised representation learning with deep convolutional generative adversarial networks. arXiv 2015, arXiv:1511.06434. [Google Scholar]
- Pizer, S.M.; Amburn, E.P.; Austin, J.D.; Cromartie, R.; Geselowitz, A.; Greer, T.; ter Haar Romeny, B.; Zimmerman, J.B.; Zuiderveld, K. Adaptive histogram equalization and its variations. Comput. Vis. Graph. Image Process. 1987, 39, 355–368. [Google Scholar] [CrossRef]
- Reza, A.M. Realization of the contrast limited adaptive histogram equalization (clahe) for real-time image enhancement. J. VLSI Signal Process. Syst. Signal Image Video Technol. 2004, 38, 35–44. [Google Scholar] [CrossRef]
- Kubat, M.; Matwin, S. Addressing the curse of imbalanced training sets: One-sided selection. ICML 1997, 97, 179. [Google Scholar]
- Oksuz, K.; Cam, B.C.; Kalkan, S.; Akbas, E. Imbalance problems in object detection: A review. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 43, 3388–3415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buda, M.; Maki, A.; Mazurowski, M.A. A systematic study of the class imbalance problem in convolutional neural networks. Neural Netw. 2018, 106, 249–259. [Google Scholar] [CrossRef] [Green Version]
- Drummond, C.; Holte, R.C. C4. 5, class imbalance, and cost sensitivity: Why under-sampling beats over-sampling. Workshop Learn. Imbalanced Datasets II 2003, 11, 1–8. [Google Scholar]
- Nafi, N.M.; Hsu, W.H. Addressing class imbalance in image-based plant disease detection: Deep generative vs. sampling-based approaches. In Proceedings of the 2020 International Conference on Systems, Signals and Image Processing (IWSSIP), Niteroi, Brazil, 1–3 July 2020; pp. 243–248. [Google Scholar]
- Han, H.; Wang, W.-Y.; Mao, B.-H. Borderline-smote: A new over-sampling method in imbalanced data sets learning. In Proceedings of the International Conference on Intelligent Computing, Hefei, China, 23–26 August 2005; pp. 878–887. [Google Scholar]
- He, H.; Bai, Y.; Garcia, E.; Li, S.A. Adaptive synthetic sampling approach for imbalanced learning. In Proceedings of the 2008 IEEE International Joint Conference on Neural Networks (IEEE World Congress on Computational Intelligence), Hong Kong, China, 1–8 June 2008. [Google Scholar]
- Bang, S.; Baek, F.; Park, S.; Kim, W.; Kim, H. Image augmentation to improve construction resource detection using generative adversarial networks, cut-and-paste, and image transformation techniques. Autom. Constr. 2020, 115, 103198. [Google Scholar] [CrossRef]
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. Tensorflow: A system for large-scale machine learning. OSDI 2016, 16, 265–283. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Chao, X.; Hu, X.; Feng, J.; Zhang, Z.; Wang, M.; He, D. Construction of apple leaf diseases identification networks based on xception fused by se module. Appl. Sci. 2021, 11, 4614. [Google Scholar] [CrossRef]
- Wang, C.; Chen, D.; Hao, L.; Liu, X.; Zeng, Y.; Chen, J.; Zhang, G. Pulmonary image classification based on inception-v3 transfer learning model. IEEE Access 2019, 7, 146533–146541. [Google Scholar] [CrossRef]
- Singh, D.; Yadav, D.; Sinha, S.; Choudhary, G. Effect of temperature, cultivars, injury of root and inoculums load of ralstonia solanacearum to cause bacterial wilt of tomato. Arch. Phytopathol. Plant Prot. 2014, 47, 1574–1583. [Google Scholar] [CrossRef]
- Mullick, S.S.; Datta, S.; Dhekane, S.G.; Das, S. Appropriateness of performance indices for imbalanced data classification: An analysis. Pattern Recognit. 2020, 102, 107197. [Google Scholar] [CrossRef]
- Magalhães, S.A.; Castro, L.; Moreira, G.; Santos, F.N.D.; Cunha, M.; Dias, J.; Moreira, A.P. Evaluating the single-shot multibox detector and yolo deep learning models for the detection of tomatoes in a greenhouse. Sensors 2021, 21, 3569. [Google Scholar] [CrossRef]

| Classifier | ACA (%) |
|---|---|
| Inception-V3 | 81.73 ± 0.672 |
| Xception | 85.49 ± 0.231 |
| ResNet-101 | 92.02 ± 0.055 |
| ResNet-50 | 92.15 ± 0.04 |
| Method | Precision | Recall | F1-Score | AUC | Accuracy |
|---|---|---|---|---|---|
| Baseline | 84.39 ± 0.205 | 92.15 ± 0.04 | 88.23 ± 0.138 | 89.33 ± 0.095 | 90.03 ± 0.036 |
| SMOTE | 90.13 ± 0.017 | 92.87 ± 0.021 | 91.47 ± 0.016 | 94.69 ± 0.015 | 93.12 ± 0.022 |
| M2m | 89.72 ± 0.036 | 92.59 ± 0.029 | 91.12 ± 0.034 | 93.16 ± 0.037 | 92.55 ± 0.035 |
| GAN-based | 93.70 ± 0.017 | 93.23 ± 0.016 | 93.46 ± 0.014 | 96.12 ± 0.011 | 95.99 ± 0.013 |
| Method | Precision | Recall | F1-Score | AUC | Accuracy |
|---|---|---|---|---|---|
| Baseline | 84.39 ± 0.205 | 92.15 ± 0.04 | 88.23 ± 0.138 | 89.33 ± 0.095 | 90.03 ± 0.036 |
| SMOTE | 91.53 ± 0.026 | 92.73 ± 0.031 | 92.12 ± 0.027 | 92.22 ± 0.028 | 90.87 ± 0.028 |
| M2m | 88.15 ± 0.058 | 92.49 ± 0.041 | 90.24 ± 0.039 | 90.51 ± 0.031 | 86.91 ± 0.064 |
| GAN-based | 91.99 ± 0.019 | 92.97 ± 0.02 | 92.47 ± 0.019 | 93.28 ± 0.024 | 95.62 ± 0.021 |
| Preprocessing Techniques | Classes | Overall | |||
|---|---|---|---|---|---|
| Indices | Healthy | Disease | Indices | Performance (%) | |
| AHE | Precision (%) | 87.23 ± 0.092 | 83.97 ± 0.256 | Average Precision | 85.60 ± 0.101 |
| Recall (%) | 86.13 ± 0.116 | 84.09 ± 0.354 | ACA | 85.11 ± 0.158 | |
| F1-Score (%) | 86.67 ± 0.183 | 84.02 ± 0.392 | Average F1-score | 85.35 ± 0.279 | |
| CLAHE | Precision (%) | 96.85 ± 0.01 | 95.82 ± 0.011 | Average Precision | 96.33 ± 0.01 |
| Recall (%) | 95.59 ± 0.012 | 96.27 ± 0.009 | ACA | 95.93 ± 0.01 | |
| F1-score (%) | 96.21 ± 0.009 | 96.04 ± 0.009 | Average F1-score | 96.13 ± 0.009 | |
| SH | Precision (%) | 94.72 ± 0.019 | 93.86 ± 0.017 | Average Precision | 94.29 ± 0.025 |
| Recall (%) | 93.95 ± 0.021 | 93.32 ± 0.025 | ACA | 93.63 ± 0.022 | |
| F1-score (%) | 94.33 ± 0.023 | 93.59 ± 0.02 | Average F1-score | 93.96 ± 0.016 | |
| CL + SH | Precision (%) | 97.71 ± 0.009 | 97.39 ± 0.01 | Average Precision | 97.55 ± 0.009 |
| Recall (%) | 98.27 ± 0.007 | 97.11 ± 0.007 | ACA | 97.69 ± 0.008 | |
| F1-score (%) | 97.99 ± 0.007 | 97.24 ± 0.008 | Average F1-score | 97.62 ± 0.008 | |
| None | Precision (%) | 94.54 ± 0.011 | 92.86 ± 0.013 | Average Precision | 93.70 ± 0.017 |
| Recall (%) | 93.82 ± 0.011 | 92.64 ± 0.013 | ACA | 93.23 ± 0.016 | |
| F1-score (%) | 94.18 ± 0.012 | 92.75 ± 0.012 | Average F1-score | 93.46 ± 0.014 | |
| Classifier | ACA (CL + SH) (%) | ACA (None) (%) |
|---|---|---|
| Inception-V3 | 84.97 ± 0.342 | 82.64 ± 0.552 |
| Xception | 87.78 ± 0.132 | 86.33 ± 0.159 |
| ResNet-101 | 95.83 ± 0.019 | 92.94 ± 0.031 |
| ResNet-50 | 97.69 ± 0.008 | 93.23 ± 0.016 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ojo, M.O.; Zahid, A. Improving Deep Learning Classifiers Performance via Preprocessing and Class Imbalance Approaches in a Plant Disease Detection Pipeline. Agronomy 2023, 13, 887. https://doi.org/10.3390/agronomy13030887
Ojo MO, Zahid A. Improving Deep Learning Classifiers Performance via Preprocessing and Class Imbalance Approaches in a Plant Disease Detection Pipeline. Agronomy. 2023; 13(3):887. https://doi.org/10.3390/agronomy13030887
Chicago/Turabian StyleOjo, Mike O., and Azlan Zahid. 2023. "Improving Deep Learning Classifiers Performance via Preprocessing and Class Imbalance Approaches in a Plant Disease Detection Pipeline" Agronomy 13, no. 3: 887. https://doi.org/10.3390/agronomy13030887
APA StyleOjo, M. O., & Zahid, A. (2023). Improving Deep Learning Classifiers Performance via Preprocessing and Class Imbalance Approaches in a Plant Disease Detection Pipeline. Agronomy, 13(3), 887. https://doi.org/10.3390/agronomy13030887






