Identification and Analysis of the Catalase Gene Family Response to Abiotic Stress in Nicotiana tabacum L.
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Data Sources
2.2. Identification of CAT Members in Tobacco
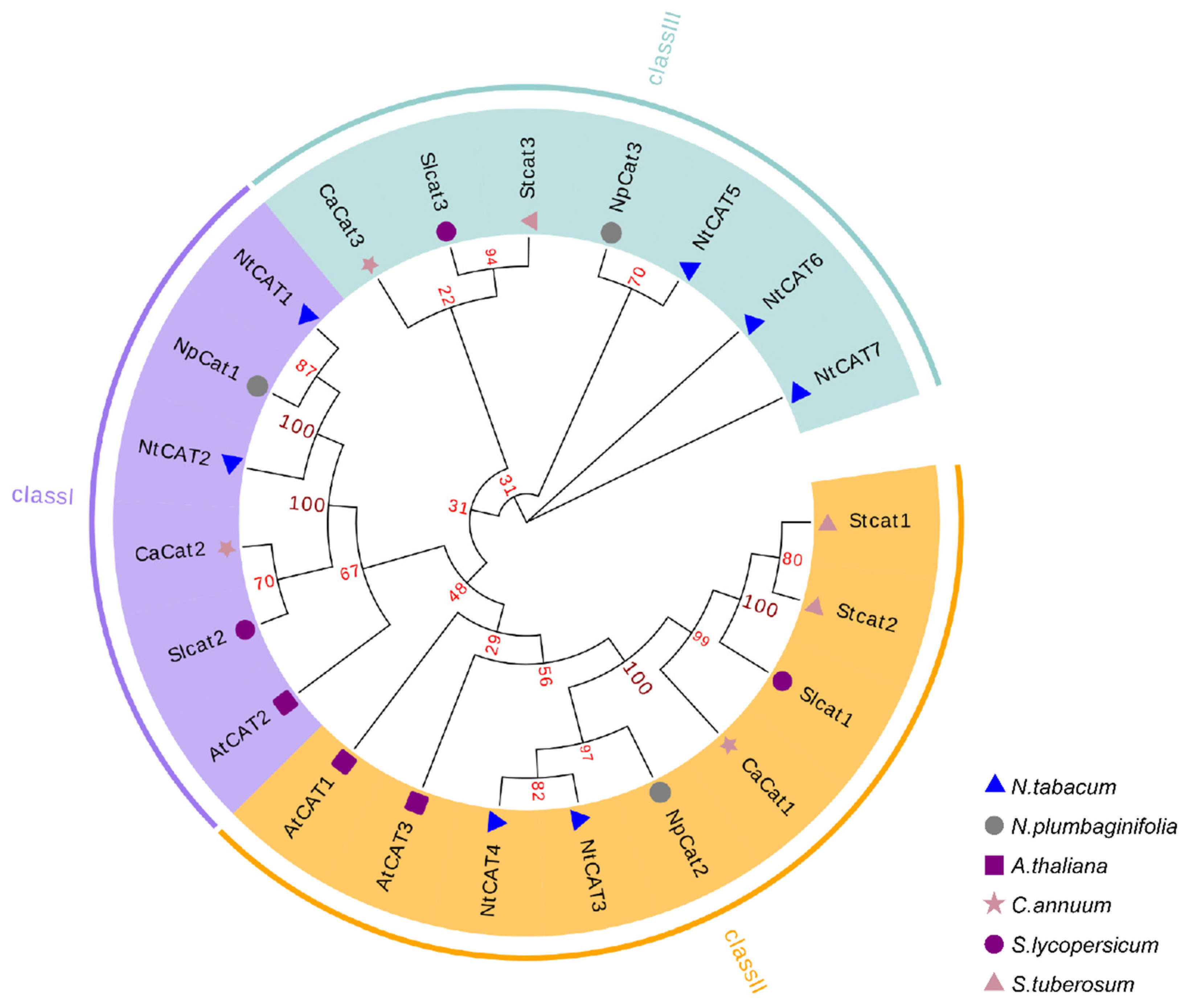
2.3. Phylogenetic Analysis
2.4. Gene Structure Analysis
2.5. Conserved Motif Prediction and Domain Presentation
2.6. Expression Profiling Analysis by Quantitative Real-Time PCR
2.7. Gene Ontology (GO) and KEGG Orthology (KO) Enrichment Analysis
2.8. Secondary and Tertiary Structures of CAT Proteins
3. Results
3.1. Identification of Tobacco CAT Genes
3.2. Phylogenetic Analysis of NtCATs
3.3. NtCAT Structure Analysis
3.4. NtCATs Conserved Motif Prediction
3.5. NtCAT Expression in Different Tissues and Times
3.6. Response of NtCATs to Various Abiotic Stresses
3.7. NtCAT Function Analysis
3.8. Secondary Structures of CAT Proteins
3.9. Tertiary Structures of CAT Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hasanuzzaman, M.; Bhuyan, M.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Mahmud, J.A.; Fujita, M.; Fotopoulos, V. Reactive Oxygen Species and Antioxidant Defense in Plants under Abiotic Stress: Revisiting the Crucial Role of a Universal Defense Regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef]
- Sofo, A.; Scopa, A.; Nuzzaci, M.; Vitti, A. Ascorbate Peroxidase and Catalase Activities and Their Genetic Regulation in Plants Subjected to Drought and Salinity Stresses. Int. J. Mol. Sci. 2015, 16, 13561–13578. [Google Scholar] [CrossRef] [Green Version]
- Mhamdi, A.; Queval, G.; Chaouch, S.; Vanderauwera, S.; Van Breusegem, F.; Noctor, G. Catalase function in plants: A focus on Arabidopsis mutants as stress-mimic models. J. Exp. Bot. 2010, 61, 4197–4220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Cheng, Y.; Chen, D.; Liu, D.; Hu, M.; Dong, J.; Zhang, X.; Song, L.; Shen, F. The Catalase Gene Family in Cotton: Genome-Wide Characterization and Bioinformatics Analysis. Cells 2019, 8, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, L.; Yang, Y.; Jiang, L.; Liu, S. The catalase gene family in cucumber: Genome-wide identification and organization. Genet. Mol. Biol. 2016, 39, 408–415. [Google Scholar] [CrossRef] [Green Version]
- Frugoli, J.A.; Zhong, H.H.; Nuccio, M.L.; McCourt, P.; McPeek, M.A.; Thomas, T.L.; McClung, C.R. Catalase is encoded by a multigene family in Arabidopsis thaliana (L.). Heynh. Plant Physiol. 1996, 112, 327–336. [Google Scholar] [CrossRef] [Green Version]
- Esaka, M.; Yamada, N.; Kitabayashi, M.; Setoguchi, Y.; Tsugeki, R.; Kondo, M.; Nishimura, M. cDNA cloning and differential gene expression of three catalases in pumpkin. Plant Mol. Biol. 1997, 33, 141–155. [Google Scholar] [CrossRef] [PubMed]
- Vuosku, J.; Sutela, S.; Kestilä, J.; Jokela, A.; Sarjala, T.; Häggman, H. Expression of catalase and retinoblastoma-related protein genes associates with cell death processes in Scots pine zygotic embryogenesis. BMC Plant Biol. 2015, 15, 88. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.J.; Wu, S.D.; Huang, G.J.; Shen, C.Y.; Afiyanti, M.; Li, W.J.; Lin, Y.H. Expression of a cloned sweet potato catalase SPCAT1 alleviates ethephon-mediated leaf senescence and H(2)O(2) elevation. J. Plant Physiol. 2012, 169, 86–97. [Google Scholar] [CrossRef]
- Zámocký, M.; Koller, F. Understanding the structure and function of catalases: Clues from molecular evolution and in vitro mutagenesis. Prog. Biophys. Mol. Biol. 1999, 72, 19–66. [Google Scholar] [CrossRef]
- Zhou, Y.B.; Liu, C.; Tang, D.Y.; Yan, L.; Wang, D.; Yang, Y.Z.; Gui, J.S.; Zhao, X.Y.; Li, L.G.; Tang, X.D.; et al. The Receptor-Like Cytoplasmic Kinase STRK1 Phosphorylates and Activates CatC, Thereby Regulating H2O2 Homeostasis and Improving Salt Tolerance in Rice. Plant Cell 2018, 30, 1100–1118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willekens, H.; Inzé, D.; Van Montagu, M.; Van Camp, W. Catalases in plants. Mol. Breed. 1995, 1, 207–228. [Google Scholar] [CrossRef]
- Willekens, H.; Langebartels, C.; Tire, C.; Van Montagu, M.; Inze, D.; Van Camp, W. Differential expression of catalase genes in Nicotiana plumbaginifolia (L.). Proc. Natl. Acad. Sci. 1994, 91, 10450–10454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwamoto, M.; Higo, H.; Higo, K. Differential diurnal expression of rice catalase genes: The 5′-flanking region of CatA is not sufficient for circadian control. Plant Sci. 2000, 151, 39–46. [Google Scholar] [CrossRef]
- Jiao, Z.; Wang, J.; Tian, Y.; Wang, S.; Sun, X.; Li, S.; Ma, W.; Zhou, T.; Fan, Z. Maize catalases localized in peroxisomes support the replication of maize chlorotic mottle virus. Phytopathol. Res. 2021, 3, 17. [Google Scholar] [CrossRef]
- Lee, S.H.; An, C.S. Differential expression of three catalase genes in hot pepper (Capsicum annuum L.). Mol. Cells 2005, 20, 247–255. [Google Scholar]
- Willekens, H.; Villarroel, R.; Van Montagu, M.; Inzé, D.; Van Camp, W. Molecular identification of catalases from Nicotiana plumbaginifolia (L.). FEBS Lett. 1994, 352, 79–83. [Google Scholar] [CrossRef] [Green Version]
- Zhong, H.H.; Mcclung, C.R. The circadian clock gates expression of two Arabidopsis catalase genes to distinct and opposite circadian phases. Mol. Gen. Genet. MGG 1996, 251, 196–203. [Google Scholar]
- Joo, J.; Lee, Y.H.; Song, S.I. Rice CatA, CatB, and CatC are involved in environmental stress response, root growth, and photorespiration, respectively. J. Plant Biol. 2014, 57, 375–382. [Google Scholar] [CrossRef]
- Du, Y.Y.; Wang, P.C.; Chen, J.; Song, C.P. Comprehensive functional analysis of the catalase gene family in Arabidopsis thaliana. J. Integr. Plant Biol. 2008, 50, 1318–1326. [Google Scholar] [CrossRef]
- Li, Y.; Chen, L.; Mu, J.; Zuo, J. LESION SIMULATING DISEASE1 interacts with catalases to regulate hypersensitive cell death in Arabidopsis. Plant Physiol. 2013, 163, 1059–1070. [Google Scholar] [CrossRef] [Green Version]
- Zimmermann, P.; Heinlein, C.; Orendi, G.; Zentgraf, U. Senescence-specific regulation of catalases in Arab. thaliana (L.). Heynh. Plant Cell Environ. 2006, 29, 1049–1060. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.J.; Li, X.D.; Ratnasekera, D.; Wang, C.; Liu, W.X.; Song, L.F.; Zhang, W.Z.; Wu, W.H. Arabidopsis CALCIUM-DEPENDENT PROTEIN KINASE8 and CATALASE3 Function in Abscisic Acid-Mediated Signaling and H2O2 Homeostasis in Stomatal Guard Cells under Drought Stress. Plant Cell 2015, 27, 1445–1460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shikanai, T.; Takeda, T.; Yamauchi, H.; Sano, S.; Tomizawa, K.-I.; Yokota, A.; Shigeoka, S. Inhibition of ascorbate peroxidase under oxidative stress in tobacco having bacterial catalase in chloroplasts. FEBS Lett. 1998, 428, 47–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, H.; Chen, Z.; Du, H.; Klessig, D.F. Development of necrosis and activation of disease resistance in transgenic tobacco plants with severely reduced catalase levels. Plant J. 1997, 11, 993–1005. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wei, X.; Wang, M.; Zhu, X.; Zhao, Y.; Wei, F.; Xia, Z. Overexpression of NtabDOG1L promotes plant growth and enhances drought tolerance in Nicotiana tabacum. Plant Sci. 2019, 287, 110186. [Google Scholar] [CrossRef]
- Fernandez-Pozo, N.; Menda, N.; Edwards, J.D.; Saha, S.; Tecle, I.Y.; Strickler, S.R.; Bombarely, A.; Fisher-York, T.; Pujar, A.; Foerster, H. The Sol Genomics Network (SGN)—From genotype to phenotype to breeding. Nucleic Acids Res. 2014, 43, D1036–D1041. [Google Scholar] [CrossRef]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2011, 40, D1202–D1210. [Google Scholar] [CrossRef]
- Edwards, K.D.; Fernandez-Pozo, N.; Drake-Stowe, K.; Humphry, M.; Evans, A.D.; Bombarely, A.; Allen, F.; Hurst, R.; White, B.; Kernodle, S.P.; et al. A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genom. 2017, 18, 448. [Google Scholar] [CrossRef] [Green Version]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2015, 44, D279–D285. [Google Scholar] [CrossRef]
- Sayers, E.W.; Agarwala, R.; Bolton, E.E.; Brister, J.R.; Canese, K.; Clark, K.; Connor, R.; Fiorini, N.; Funk, K.; Hefferon, T. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2019, 47, D23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, A.L.; Attwood, T.K.; Babbitt, P.C.; Blum, M.; Bork, P.; Bridge, A.; Brown, S.D.; Chang, H.-Y.; El-Gebali, S.; Fraser, M.I. InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2018, 47, D351–D360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Xie, Y.; Ma, J.; Luo, X.; Nie, P.; Zuo, Z.; Lahrmann, U.; Zhao, Q.; Zheng, Y.; Zhao, Y. IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics 2015, 31, 3359–3361. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef] [Green Version]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, 182–185. [Google Scholar] [CrossRef] [Green Version]
- Alam, N.B.; Ghosh, A. Comprehensive analysis and transcript profiling of Arabidopsis thaliana and Oryza sativa catalase gene family suggests their specific roles in development and stress responses. Plant Physiol. Biochem. 2018, 123, 54–64. [Google Scholar] [CrossRef]






| Gene Name | Gene ID | Length (aa) | MW (kDa) | pI | GRAVY | Subcellular Localization |
|---|---|---|---|---|---|---|
| NtCAT1 | Nitab4.5_0000702g0130.1 | 492 | 56.824 | 6.70 | −0.568 | pero, chlo, cyto |
| NtCAT2 | Nitab4.5_0006960g0010.1 | 492 | 56.822 | 6.86 | −0.564 | pero, chlo, cyto |
| NtCAT3 | Nitab4.5_0009821g0010.1 | 476 | 55.254 | 6.50 | −0.531 | cyto, pero, plas, cysk_plas, nucl |
| NtCAT4 | Nitab4.5_0000407g0130.1 | 492 | 56.824 | 6.60 | −0.518 | cyto, pero, mito, plas, cysk_plas |
| NtCAT5 | Nitab4.5_0000588g0030.1 | 492 | 56.947 | 6.95 | −0.569 | pero, mito, chlo, cyto |
| NtCAT6 | Nitab4.5_0011405g0020.1 | 490 | 56.673 | 6.75 | −0.555 | pero, mito, chlo, cyto |
| NtCAT7 | Nitab4.5_0009395g0030.1 | 91 | 10.249 | 9.25 | 0.349 | plas, golg_plas, golg |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Wang, D.; Tang, H.; Li, H.; Zhang, X.; Dong, S.; Zhang, L.; Yang, L. Identification and Analysis of the Catalase Gene Family Response to Abiotic Stress in Nicotiana tabacum L. Agronomy 2023, 13, 936. https://doi.org/10.3390/agronomy13030936
Liu Z, Wang D, Tang H, Li H, Zhang X, Dong S, Zhang L, Yang L. Identification and Analysis of the Catalase Gene Family Response to Abiotic Stress in Nicotiana tabacum L. Agronomy. 2023; 13(3):936. https://doi.org/10.3390/agronomy13030936
Chicago/Turabian StyleLiu, Zhonghui, Di Wang, Heng Tang, Haozhen Li, Xiaohua Zhang, Shaolin Dong, Li Zhang, and Long Yang. 2023. "Identification and Analysis of the Catalase Gene Family Response to Abiotic Stress in Nicotiana tabacum L." Agronomy 13, no. 3: 936. https://doi.org/10.3390/agronomy13030936
APA StyleLiu, Z., Wang, D., Tang, H., Li, H., Zhang, X., Dong, S., Zhang, L., & Yang, L. (2023). Identification and Analysis of the Catalase Gene Family Response to Abiotic Stress in Nicotiana tabacum L. Agronomy, 13(3), 936. https://doi.org/10.3390/agronomy13030936





