Relationships between Rhizosphere Environments and Growth of 10-Year-Old Wild-Simulated Ginseng
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Soil Samples Cultivating WSG
2.2. Soil Analysis
2.3. Growth Characteristics of WSG Cultivated in Different Sites
2.4. Extraction of Soil DNA and Polymerase Chain Reaction (PCR) Amplification
2.5. Pyrosequencing and Data Processing
2.6. Data Analysis and Principal Coordinate Analysis
3. Results and Discussion
3.1. Information of Various WSG Cultivation Sites
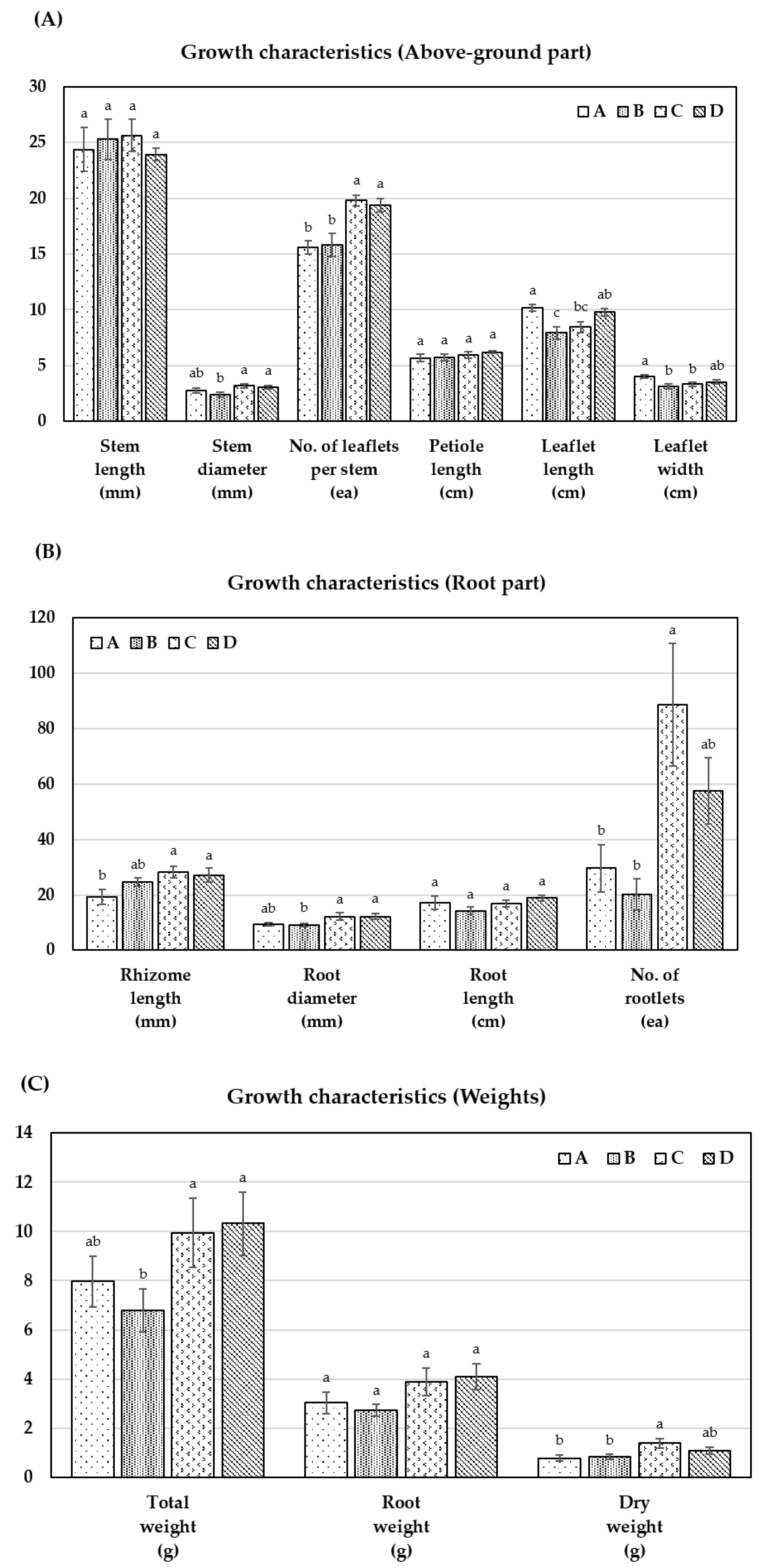
3.2. Growth Characteristics of 10-Year-Old WSG Cultivated in Different Sites
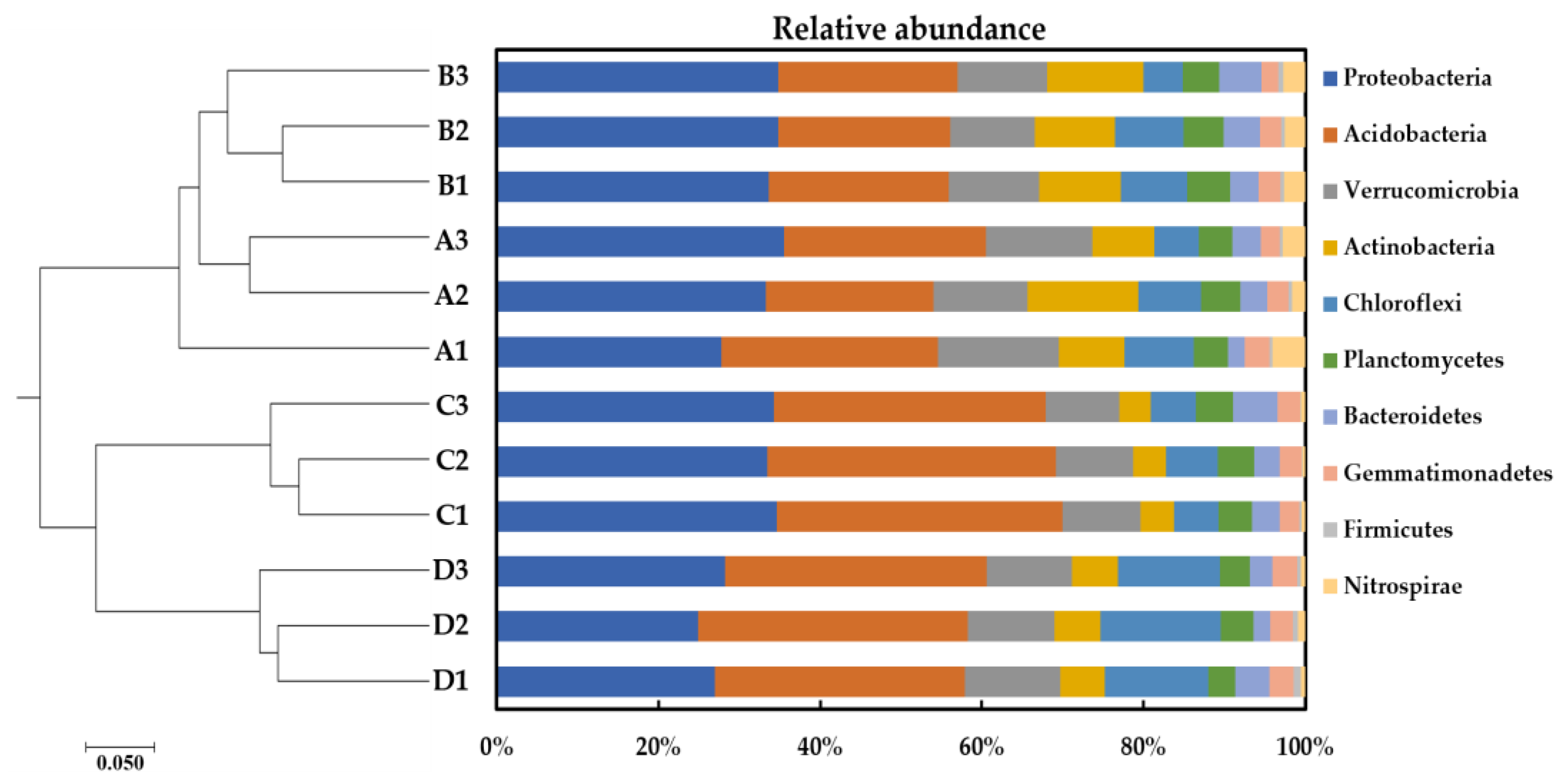
3.3. Bacterial Community Profiles
3.4. Correlation Analyses between Soil Chemical Properties and Bacterial Community
3.5. Correlation Analysis between Growth Characteristics of WSG and Soil Bacterial Community
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kim, K.Y.; Kim, H.J.; Um, Y.R.; Jeon, K.S. Effect of soil properties and soil bacterial community on early growth characteristics of wild-simulated ginseng (Panax ginseng C.A. Meyer) in coniferous and mixed forest. Korean J. Med. Crop Sci. 2020, 28, 183–194. [Google Scholar] [CrossRef]
- Lee, D.S. Weather characteristics and growth of a forest ginseng cultivation site. J. Korean Soc. For. Sci. 2011, 99, 863–870. [Google Scholar]
- Ramegowda, V.; Senthil-Kumar, M. The interactive effects of simultaneous biotic and abiotic stresses on plants: Mechanistic understanding from drought and pathogen combination. J. Plant Physiol. 2015, 176, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.; Irulappan, V.; Bagavathiannan, M.V.; Senthil-Kumar, M. Impact of combined abiotic and biotic stresses on plant growth and avenues for crop improvement by exploiting physio-morphological traits. Front. Plant Sci. 2017, 8, 537. [Google Scholar] [CrossRef]
- Hatfield, J.L.; Prueger, J.H. Temperature extremes: Effect on plant growth and development. Weather Clim. Extrem. 2015, 10, 4–10. [Google Scholar] [CrossRef]
- Ahluwalia, O.; Singh, P.C.; Bhatia, R. A review on drought stress in plants: Implications, mitigation and the role of plant growth promoting rhizobacteria. Resour. Environ. Sustain. 2021, 5, 100032. [Google Scholar] [CrossRef]
- Tekaya, N.; Saiapina, O.; Ouada, H.B.; Lagarde, F.; Namour, P.; Ouada, H.B.; Jaffrezic-Renault, N. Bi-enzymatic conductometric biosensor for detection of heavy metal ions and pesticides in water samples based on enzymatic inhibition in Arthrospira platensis. J. Environ. Protect. 2014, 5, 441–453. [Google Scholar] [CrossRef]
- Denance, N.; Sanchez-Vellet, A.; Goffner, D.; Molilna, A. Disease resistance or growth: The role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 2013, 4, 155. [Google Scholar] [CrossRef]
- Selvakumar, G.; Kim, K.; Hu, S.; Sa, T. Effect of salinity on plants and the role of arbuscular mycorrhizal fungi and plant growth-promoting rhizobacteria in alleviation of salt stress. In Physiological Mechanisms and Adaptation Strategies in Plants under Changing Environment; Ahmad, P., Wani, M.R., Eds.; Springer: New York, NY, USA, 2014; Volume 1, pp. 115–144. [Google Scholar]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef]
- Prasad, R.; Kamal, S.; Sharma, P.K.; Oelmueller, R.; Varma, A. Root endophte Piriformospora indica DSM 11827 alters plants morphology, enhances biomass and antioxidant activity of medicinal plant Bacopa monniera. J. Basic Microbiol. 2013, 52, 1016–1024. [Google Scholar] [CrossRef]
- Pei, Z.; Eichenberg, D.; Bruelheide, H.; Krober, W.; Kuhn, P.; Li, Y.; von Oheimb, G.; Purschke, O.; Scholten, T.; Buscot, F.; et al. Soil and tree species traits both shape soil microbial communities during early growth of Chinese subtropical forests. Soil Biol. Biochem. 2016, 96, 180–190. [Google Scholar] [CrossRef]
- Llado, S.; Lopez-Mondejar, R.; Baldrian, P. Drivers of microbial community structure in forest soils. Appl. Microbiol. Biotechnol. 2018, 102, 4331–4338. [Google Scholar] [CrossRef]
- Sogin, M.L.; Morrison, H.G.; Huber, J.A.; Welch, D.M.; Huse, S.M.; Neal, P.R.; Arrieta, J.M.; Herndl, G.J. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. USA 2006, 103, 12115–12120. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Tao, B.; Guo, J.; Li, J.; Chen, G. Changes in the microbial community structure and soil chemical properties of vertisols under different cropping systems in Northern China. Front. Environ. Sci. 2018, 6, 132. [Google Scholar] [CrossRef]
- Wu, Z.; Liu, Q.; Li, Z.; Cheng, W.; Sun, J.; Guo, Z.; Li, Y.; Zhou, J.; Meng, D.; Li, H.; et al. Environmental factors shaping the diversity of bacterial communities that promote rice production. BMC Microbiol. 2018, 18, 51. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Xu, J.; Li, Y.; Fang, H.; Niu, W.; Li, X.; Zhang, Y.; Ding, W.; Chen, S. Manipulation of microbial community in the rhizosphere alleviates the replanting issues in Panax ginseng. Soil Biol. Biochem. 2018, 125, 64–74. [Google Scholar] [CrossRef]
- Lee, B.J.; Eo, S.H. Soil bacterial community in red pine forest of Mt. Janggunbong, Bonghwa-gun, Gyeongbuk, Korea, using next generation sequencing. J. Korean For. Soc. 2017, 106, 121–129. [Google Scholar]
- Yun, Y.B.; Huh, J.H.; Jeong, D.H.; Kim, J.; Um, Y. Correlation analysis between growth characteristics and ginsenoside contents of 4-year-old wild-simulated ginseng (Panax ginseng C.A. Meyer) with different cultivation sites. J. Appl. Biol. Chem. 2022, 65, 253–259. [Google Scholar] [CrossRef]
- Korean Soil Information. Available online: http://soil.rda.go.kr/geoweb/soilmain.do (accessed on 7 May 2011).
- Rural Development Administration (RDA). Analysis Manual of Comprehensive Examination Laboratory (Soil, Plant, Water and Liquid Manure); Rural Development Administration: Suwon, Republic of Korea, 2013; pp. 31–53. [Google Scholar]
- Korea Seed and Variety Service(KSVS). Know-How of Characteristics Investigation of the Crops: Ginseng (Panax ginseng Meyer); Korea Seed and Variety Service: Gimcheon, Republic of Korea, 2014. [Google Scholar]
- Canfora, L.; Vendramin, E.; Felici, B.; Tarricone, L.; Florio, A.; Benedetti, A. Vineyard microbiome variations during different fertilization practices revealed by 16S rRNA gene sequencing. Appl. Soil Ecol. 2018, 123, 71–80. [Google Scholar] [CrossRef]
- Schloss, P.D. A high-throughput DNA sequence aligner for microbial ecology studies. PLoS ONE 2009, 4, e8230. [Google Scholar] [CrossRef]
- Nurek, T.; Gendek, A.; Roman, K. Forest residues as a renewable source of energy: Elemental composition and physical properties. BioResources 2019, 14, 6–20. [Google Scholar] [CrossRef]
- Scholer, A.; Jacquiod, S.; Vestergaard, G.; Schulz, S.; Schloter, M. Analysis of soil microbial communities based on amplicon sequencing of marker genes. Biol. Fertil. Soils 2017, 53, 485–489. [Google Scholar] [CrossRef]
- Anderson, M.J. DISTLM v. 5: A FORTRAN Computer Program to Calculate a Distance-Based Multivariate Analysis for a Linear Model; Department of Statistics, University of Auckland: Auckland, New Zealand, 2004; p. 10. [Google Scholar]
- Gruba, P.; Mulder, J. Tree species affect cation exchange capacity (CEC) and cation binding properties of organic matter in acid forest soils. Sci. Total Environ. 2015, 511, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.; Ota, A.T.; Kabeya, D.; Okamoto, T.; Saitoh, T.; Nishiyama, Y. The effects of mixed broad-leaved trees on the collembolan community in larch plantations of central Japan. Appl. Soil Ecol. 2014, 83, 125–132. [Google Scholar] [CrossRef]
- Bu, W.S.; Gu, H.J.; Zhang, C.C.; Zhang, Y.; Singh, A.N.; Fang, X.M.; Fan, J.; Wang, H.M.; Chen, F.S. Mixed broadleaved tree species increases soil phosphorus availability but decreases coniferous tree nutrient concentration in subtropical China. Forests 2020, 11, 461. [Google Scholar] [CrossRef]
- Gruba, P.; Mulder, J. Relationship between aluminum in soils and soil water in mineral horizons of a range of acid forest soils. Soil Sci. Soc. Am. J. 2008, 72, 1150–1157. [Google Scholar] [CrossRef]
- Mueller, K.E.; Eissenstat, D.M.; Hobbie, S.E.; Oleksyn, J.; Jagodzinski, A.M.; Reich, P.B.; Chadwick, O.A.; Chorover, J. Tree species effects on coupled cycles of carbon, nitrogen, and acidity in mineral soils at a common garden experiment. Biogeochemistry 2012, 111, 601–614. [Google Scholar] [CrossRef]
- Chamberlain, J.L.; Prisley, S.; McGuffin, M. Understanding the relationship between American ginseng harvest and hardwood forest inventory and timber harvest to improve co-management of the forest of Eastern United States. J. Sustain. For. 2013, 32, 605–624. [Google Scholar] [CrossRef]
- Chung, J.M.; Moon, H.S. Soil characteristics by the site types around Nari Sasin in Ulleung island. J. Agric. Life Sci. 2011, 44, 44–50. [Google Scholar]
- Kim, C.; Choo, G.C.; Cho, H.S.; Lim, J.T. Soil properties of cultivation sites for mountain-cultivated ginseng at local level. J. Ginseng Res. 2015, 39, 76–80. [Google Scholar] [CrossRef]
- Binkley, D.; Fisher, R.F. Ecology and Management of Forest Soils, 4th ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2013; pp. 39–57. [Google Scholar]
- Xia, Z.; Bai, E.; Wang, Q.; Gao, D.; Zhou, J.; Jiang, P.; Wu, J. Biogeographic distribution patterns of bacteria in typical Chinese forest soils. Front. Microbiol. 2016, 7, 1106. [Google Scholar] [CrossRef] [PubMed]
- Yin, N.; Koide, R.T. Microbial activity, microarthropods and the phenomenon of positive, non-additive decomposition of mixed litter. J. Soil Ecol. 2019, 76, 150570. [Google Scholar] [CrossRef]
- Apsley, D.; Caroll, C. Growing American Ginseng in Ohio: Selecting a Site; Extension Factsheet; The Ohio State University: Columbus, OH, USA, 2004; p. 4. [Google Scholar]
- Seo, S.M.; Woo, S.Y.; Lee, D.S. A study on the photosynthetic rates of Panax ginseng in the different age and provinces. J. Korean For. Soc. 2007, 96, 357–361. [Google Scholar]
- Woo, S.Y.; Lee, D.S. A study on the growth and environments of Panax ginseng in the different forest stands(I). Korean J. Agric. For. Meteorol. 2002, 4, 65–71. [Google Scholar]
- Borges, C.V.; Minatel, I.O.; Gomez-Gomez, H.A.; Lima, P.P. Medicinal plants: Influence of environmental factors on the content of secondary metabolites. In Medicinal Plants and Environmental Challenges. Ghorbanpour, M.; Varma, A., Ed.; Springer International Publishing: New York, NY, USA, 2017; pp. 259–277. [Google Scholar]
- Kim, K.; Um, Y.; Jeong, D.H.; Kim, H.J.; Kim, M.J.; Jeon, K.S. Study on the correlation between the soil bacterial community and growth characteristics of wild-simulated ginseng(Panax ginseng C.A. Meyer). Korean J. Environ. Biol. 2019, 37, 380–388. [Google Scholar] [CrossRef]
- Morales-Cedeno, L.; del Orozco-Mosqueda, M.C.; Loeza-Lara, P.; Parra-Cota, F.I.; de los Santos-Villalobos, S.; Santoyo, G. Plant growth-promoting bacterial endophytes as biocontrol agents of pre- and post-harvest diseases: Fundamentals, methods of application and future perspectives. Microbiol. Res. 2021, 242, 126612. [Google Scholar] [CrossRef]
- Lugtenberg, G.; Kamilova, F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef]
- Glick, B.R. Plant growth-promoting bacteria: Mechanisms and applications. Scientifica 2012, 2012, 963401. [Google Scholar] [CrossRef]
- Miransari, M. soil microbes and plant fertilization. Appl. Microbiol Biotechnol. 2011, 92, 875–885. [Google Scholar] [CrossRef]
- Sharifi, R.; Ryu, C.M. Chatting with a tiny belowground member of the holobiome: Communication between plants and growth-promoting rhizobacteria. Adv. Bot. Res. 2017, 82, 135–160. [Google Scholar]
- Schnitzer, S.A.; Klironomos, J.N.; HilleRisLambers, J.; Kinkel, L.L.; Reich, P.B.; Xiao, K.; Rillig, M.C.; Sikes, B.A.; Callaway, R.M.; Mangan, S.A. Soil microbes drive the classic plant diversity-productivity pattern. Ecology 2011, 92, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Sun, H.; Xu, C.; Ma, L.; Li, M.; Shao, C.; Guan, Y.; Liu, N.; Liu, Z.; Zhang, S.; et al. Analysis of rhizosphere bacterial and fungal communities associated with rusty root disease of Panax ginseng. Appl. Soil Ecol. 2019, 138, 245–252. [Google Scholar] [CrossRef]
- Dong, L.; Li, Y.; Xu, J.; Yang, J.; Wei, G.; Shen, L.; Ding, W.; Chen, S. Biofertilizer regulate the soil microbial community and enhance Panax ginseng yields. Chin. Med. 2019, 13, 20. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; Roh, A.S.; Choi, S.C.; Kim, E.J.; Choi, M.T.; Ahn, B.K.; Kim, S.K.; Lee, Y.H.; Joa, J.H.; Kang, S.S.; et al. Soil pH and electrical conductivity are key edaphic factors shaping bacterial communities of greenhouse soils in Korea. J. Microbiol. 2016, 54, 838–845. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; Del Rio, T.G. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef]
- Zuo, J.; Zu, M.; Liu, L.; Song, X.; Yuan, Y. Composition and diversity of bacterial communities in the rhizosphere of the Chinese medicinal herb Dendrobium. BMC Plant Biol. 2021, 21, 127. [Google Scholar] [CrossRef]
- Su, J.F.; Wang, Z.; Huang, T.L.; Zhang, H.; Zhang, H. Simultaneous removal of nitrate, phosphorus and cadmium using a novel multifunctional biomaterial immobilized aerobic strain Proteobacteria Cupriavidus H29. Bioresour. Technol. 2020, 307, 123196. [Google Scholar] [CrossRef]
- Spain, A.M.; Krumholz, L.R.; Elshahed, M.S. Abundance, composition, diversity and novelty of soil Proteobacteria. Int. Soc. Microb. Ecol. J. 2009, 3, 992–1000. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, J. Evaluating bacterial community structures in oil collected from the sea surface and sediment in the northern Gulf of Mexico after the deepwater horizon oil spill. Microbiol. Open 2013, 2, 492–504. [Google Scholar] [CrossRef]
- He, Z.; Hu, Y.; Yin, Z.; Hu, Y.; Zhong, H. Microbial diversity of chromium-contaminated soils and characterization of six chromium-removing bacteria. Environ. Manag. 2016, 57, 1319–1328. [Google Scholar] [CrossRef]
- Lee, D.G.; Cho, K.C.; Chu, K.H. Identification of triclosan-degrading bacteria in a triclosan enrichment culture using stable isotope probing. Biodegradation 2014, 25, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Xie, H.J.; Xu, J.; Xu, X.; Zhang, J.; Hu, Z.; Liu, C.; Liang, S.; Wang, Q.; Wang, J. Bacterial community variation and microbial mechanism of triclosan(TCS) removal by constructed wetlands with different types of plants. Sci. Total Environ. 2015, 505, 633–639. [Google Scholar] [CrossRef]
- Kielak, A.M.; Cipriano, M.A.P.; Kuramae, E.E. Acidobacteria strains from subdivision 1 act as plant growth-promoting bacteria. Arch. Microbiol. 2016, 198, 987–993. [Google Scholar] [CrossRef] [PubMed]
- Kalam, S.; Basu, A.; Ahmad, I.; Sayyed, R.Z.; El-Enshasy, H.A.; Dailin, D.J.; Suriani, N.L. Recent understanding of soil acidobacteria and their ecological significance: A critical review. Front. Microbiol. 2020, 11, 580024. [Google Scholar] [CrossRef]
- Yoneda, Y.; Yamamoto, K.; Makino, A.; Tanaka, Y.; Meng, X.Y.; Hashimoto, J.; Shin-ya, K.; Satoh, N.; Fujie, M.; Toyama, T.; et al. Novel plant-associated Acidobacteria promotes growth of common floating aquatic plants, duckweeds. Microorganisms 2021, 9, 1133. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.; Wu, X.; Qiu, Q.; Duan, C.; Jones, D.L. Seasonal variations in soil microbial communities under different land restoration types in a subtropical mountains region, Southwest China. Appl. Soil Ecol. 2020, 153, 103634. [Google Scholar] [CrossRef]
- Mechri, B.; Mariem, F.B.; Baham, M.; Elhadj, S.B.; Hammami, M. Change in soil properties and the soil microbial community following land spreading of olive mill wastewater affects olive trees key physiological parameters and the abundance of arbuscular mycorrhizal fungi. Soil Biol. Biochem. 2008, 40, 152–161. [Google Scholar] [CrossRef]
- Preem, J.K.; Truu, J.; Truu, M.; Mander, Ü.; Oopkaup, K.; Lõhmus, K.; Helmisaari, H.S.; Uri, V.; Zobel, M. Bacterial community structure and its relationship to soil physico-chemical characteristics in alder stands with different management histories. Ecol. Eng. 2012, 49, 10–17. [Google Scholar] [CrossRef]
- Kim, K.; Kim, H.J.; Jeong, D.H.; Huh, J.H.; Jeon, K.S.; Um, Y. Correlation between soil bacterial community structure and soil properties in cultivation sites of 13-year-old wild-simulated ginseng (Panax ginseng C.A. Meyer). Appl. Sci. 2021, 11, 937. [Google Scholar] [CrossRef]
- Lim, S.U. Plant Growth and Nutrients. In Fertilizer; Ilsin: Seoul, Republic of Korea, 2005; pp. 38–45. [Google Scholar]
- Altomare, C.; Tringovska, I. Beneficial Soil Microorganisms, an Ecological Alternative for Soil Fertility Management. In Genetics, Biofuels and Local Farming Systems. Sustainable Agriculture Reviews; Lichtfouse, E., Ed.; Springer: Dordrecht, The Netherlands; Cham, Switzerland, 2011; pp. 161–214. [Google Scholar]
- Bapiri, A.; Bååth, E.; Rousk, J. Drying-rewetting cycles affect fungal and bacterial growth differently in an arable soil. Microb. Ecol. 2010, 60, 419–428. [Google Scholar] [CrossRef]
- Joa, J.H.; Moon, K.H.; Kim, S.C.; Moon, D.G.; Koh, S.W. Effect of Temperature Condition on Nitrogen Mineralization of Organic Matter and Soil Microbial Community Structure in non-Volcanic Ash Soil. Korean J. Soil Sci. Fertil. 2012, 45, 377–384. [Google Scholar] [CrossRef]
- Paz-González, A.; Vieira, S.R.; Taboada Castro, M.T. The effect of cultivation on the spatial variability of selected properties of an umbric horizon. Geoderma 2000, 37, 273–292. [Google Scholar] [CrossRef]
- Le, X.H.; Franco, C.M.M.; Ballard, R.A.; Drew, E.A. Isolation and characterization of endophytic actinobacteria and their effect on the early growth and nodulation of lucerne (Medicago sativa L.). Plant Soil. 2016, 405, 13–24. [Google Scholar] [CrossRef]
- Gohel, S.D.; Sharma, A.K.; Thakrar, F.J.; Singh, S.P. Endophytic actinobacteria and their interactions with plant host systems. In Understanding Host-Microbiome Interaction-An Omics Approach; Singh, R.P., Ed.; Springer Nature Singapore Pte Ltd.: Singapore, 2017; pp. 247–266. [Google Scholar]
- Purushotham, N.; Jones, E.; Monk, J.; Ridgway, H. Community structure of endophytic actinobacteria in a New Zealand native medicinal plant Pseudowintera colorata (horopito) and their influence on plant growth. Microbiol. Ecol. 2018, 76, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Li, J.; Zhang, X.; Chen, Q.; Liu, M.; Ao, X.; Gu, Y.; Liao, D.; Xu, K.; Ma, M.; et al. Actinobacteria associated with Glycyrrhiza inflata Bat. are diverse and have plant growth promoting and antimicrobial activity. Sci. Rep. 2018, 8, 13661. [Google Scholar] [CrossRef]
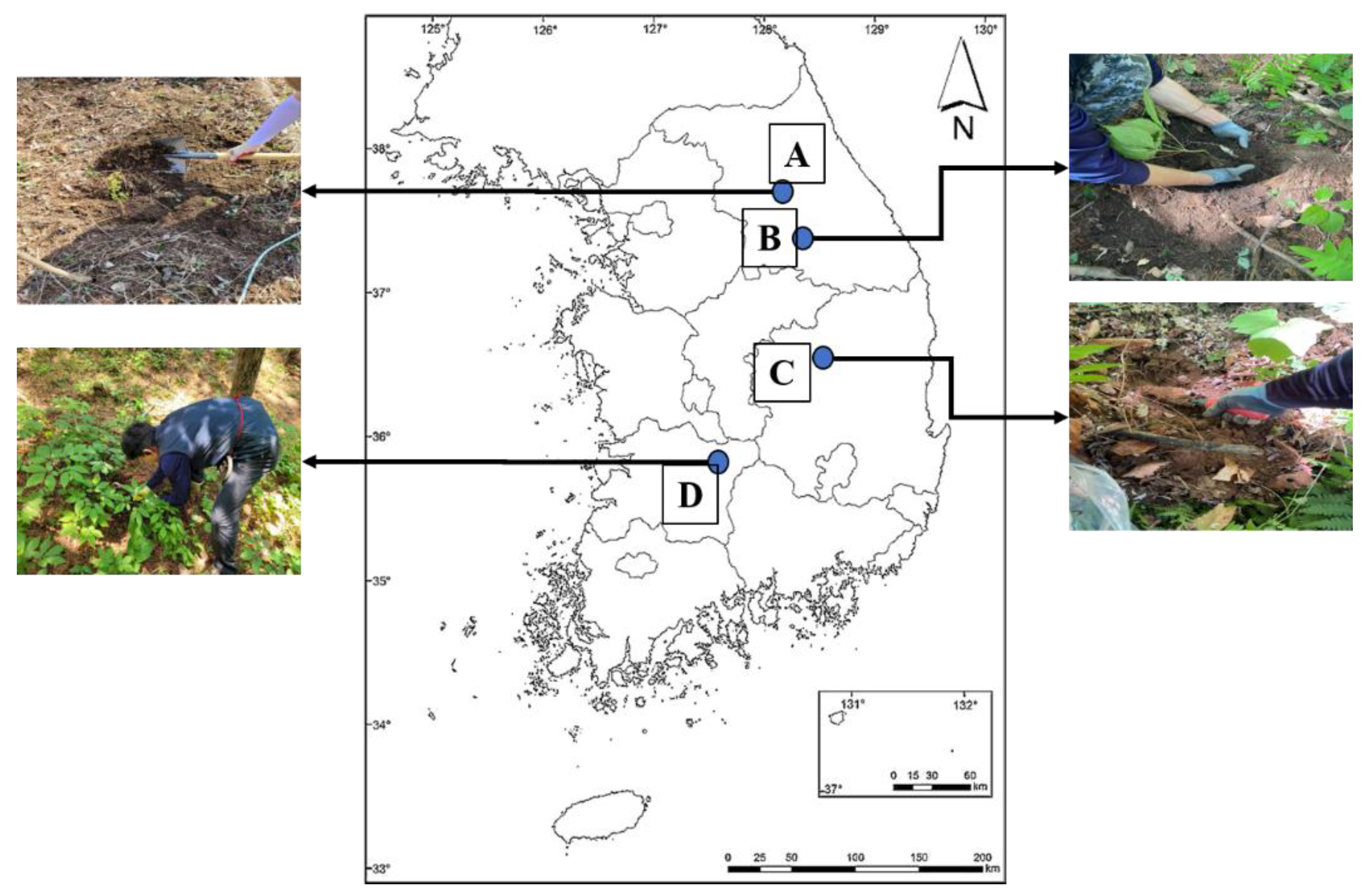

| Cultivation Site | Sand (%) | Loam (%) | Clay (%) | Soil Texture | Topography | Species of Tree | Forest Physiognomy | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Slope | HASL a | Average | Percentage | |||||||||
| TH b | DBH c | |||||||||||
| ° | Direction | m | m | cm | % | |||||||
| A | 67.5 | 18.0 | 14.5 | Sandy loam | 25 | Southwest | 931 | Conifer | Pinus koraiensis | 18.5 | 29.0 | 15.4 |
| Broad-leaved | Betula dahurica Pall. Ulmus davidiana var. japonica | 17.4 | 20.3 | 84.6 | ||||||||
| Total | 17.95 | 24.65 | 100 | |||||||||
| B | 35.5 | 32.4 | 32.1 | Loamy sand | 22 | Southwest | 615 | Conifer | Larix kaempferi | 10.0 | 26.0 | 6.3 |
| Broad-leaved | Prunus sect. Cerasus | 11.26 | 20.36 | 93.7 | ||||||||
| Total | 10.63 | 23.18 | 100 | |||||||||
| C | 65.9 | 12.6 | 21.5 | Sandy clay loam | 8 | Southeast | 391 | Conifer | - | - | - | - |
| Broad-leaved | Robinia pusedoacacia | 9.57 | 13.57 | 100 | ||||||||
| Total | 9.57 | 13.57 | 100 | |||||||||
| D | 84.8 | 5.1 | 10.1 | Clay loam | 18 | Northeast | 717 | Conifer | Larix kaempferi | 21.6 | 28.4 | 55.6 |
| Broad-leaved | Fraxinus rhynchophylla Morus alba | 8.0 | 9.0 | 44.4 | ||||||||
| Total | 14.8 | 18.7 | 100 | |||||||||
| pH | EC | OM z | TN y | Avail. P2O5 x | Exchangable Cations | CEC s | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| K w | Ca v | Mg u | Na t | |||||||
| (1:5) | dS/m (1:5) | % | % | mg/kg | -------------------------------------- cmol+/kg -------------------------------------- | |||||
| A | 4.57 ± 0.04b | 0.31 ± 0.02a | 20.25 ± 0.69a | 0.65 ± 0.02a | 231.55 ± 11.36a | 0.46 ± 0.04a | 7.12 ± 0.45a | 0.93 ± 0.09a | 0.07 ± 0.02a | 46.31 ± 0.31a |
| B | 4.84 ± 0.04a | 0.21 ± 0.02b | 9.81 ± 0.95b | 0.35 ± 0.04b | 134 ± 15b | 0.22 ± 0.02bc | 5.88 ± 0.84a | 0.77 ± 0.10a | 0.04 ± 0.01ab | 27.14 ± 2.25b |
| C | 4.25 ± 0.03c | 0.15 ± 0.01c | 5.27 ± 0.23c | 0.23 ± 0.02c | 144.8 ± 44.4b | 0.26 ± 0.02b | 0.60 ± 0.09b | 0.16 ± 0.02b | 0.03 ± 0.00b | 18.02 ± 1.54c |
| D | 4.18 ± 0.02c | 0.17 ± 0.01bc | 10.63 ± 0.47b | 0.39 ± 0.03b | 232.18 ± 14.59a | 0.17 ± 0.01c | 0.88 ± 0.11b | 0.21 ± 0.02b | 0.02 ± 0.00b | 26.59 ± 1.25b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yun, Y.-B.; Kim, K.; Huh, J.-H.; Um, Y. Relationships between Rhizosphere Environments and Growth of 10-Year-Old Wild-Simulated Ginseng. Agronomy 2023, 13, 1313. https://doi.org/10.3390/agronomy13051313
Yun Y-B, Kim K, Huh J-H, Um Y. Relationships between Rhizosphere Environments and Growth of 10-Year-Old Wild-Simulated Ginseng. Agronomy. 2023; 13(5):1313. https://doi.org/10.3390/agronomy13051313
Chicago/Turabian StyleYun, Yeong-Bae, Kiyoon Kim, Jeong-Hoon Huh, and Yurry Um. 2023. "Relationships between Rhizosphere Environments and Growth of 10-Year-Old Wild-Simulated Ginseng" Agronomy 13, no. 5: 1313. https://doi.org/10.3390/agronomy13051313
APA StyleYun, Y.-B., Kim, K., Huh, J.-H., & Um, Y. (2023). Relationships between Rhizosphere Environments and Growth of 10-Year-Old Wild-Simulated Ginseng. Agronomy, 13(5), 1313. https://doi.org/10.3390/agronomy13051313






