Microbial Mechanisms of the Priming Effect over 12 Years of Different Amounts of Nitrogen Management
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site and N Application
2.2. Maize Residue Preparation, Return, and Sampling
2.3. Total CO2-C Mineralization, Native SOC Mineralization, Residue-Derived C Mineralization, and Priming Effect (PE) Analyses
2.4. Microbial Biomass Carbon (MBC) and Microbial Carbon Use Efficiency (CUE)
2.5. High-Throughput Sequencing
2.6. Statistical Analysis
3. Results
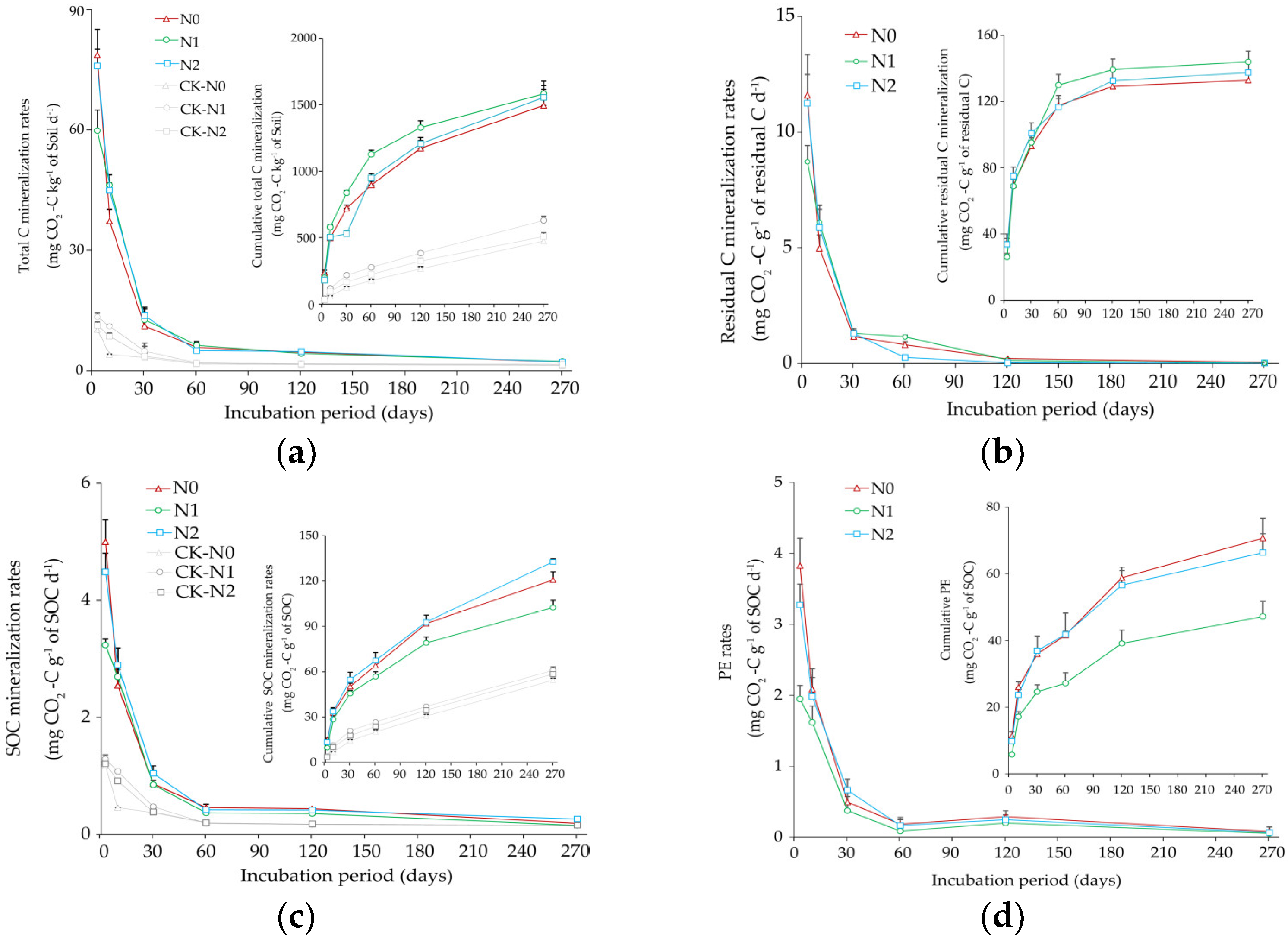
3.1. Total C, Residual C, Soil Organic C (SOC) Mineralization, and Priming Effect (PE)
3.2. Microbial Biomass C (MBC) and Residue-Derived MBC
3.3. Microbial C Use Efficiency (CUE)
3.4. Soil Microbial Communities’ Structure and Co-Occurrence Networks
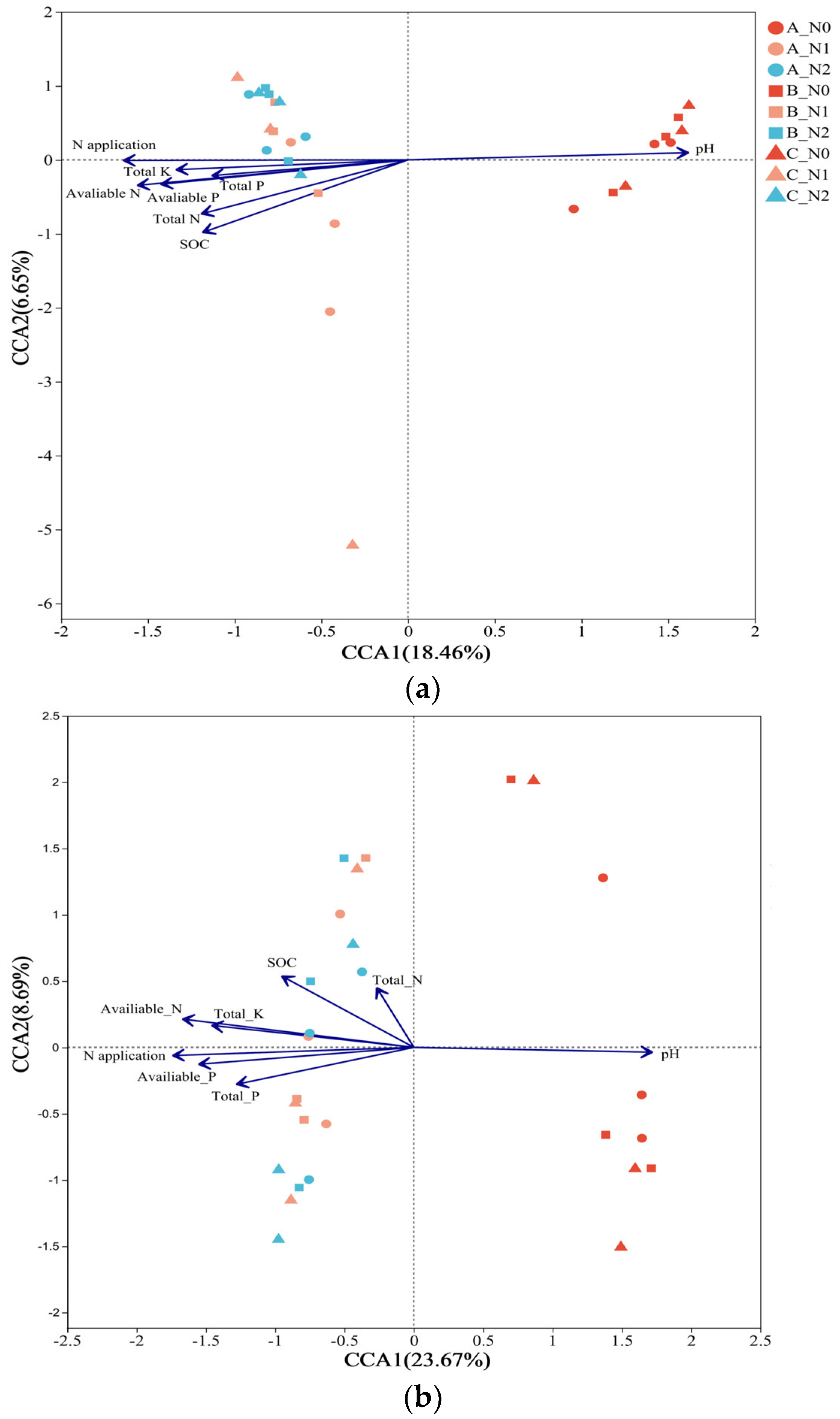
3.5. The Effects of Soil Chemical Properties on Assessments of Microbial Communities and Keystone Taxa
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blaud, A.; Menon, M.; Van, D.; Lair, G.J.; Banwart, S. Effects of Dry and Wet Sieving of Soil on Identification and Interpretation of Microbial Community Composition. Adv. Agron. 2017, 142, 119–142. [Google Scholar]
- Pankhurst, C.; Kirkby, C.; Hawke, B.; Harch, B. Impact of a change in tillage and crop residue management practice on soil chemical and microbiological properties in a cereal-producing red duplex soil in NSW, Australia. Biol. Fertil. Soils 2002, 35, 189–196. [Google Scholar]
- Song, X.J.; Li, J.Y.; Liu, X.T.; Liang, G.P.; Li, S.P.; Zhang, M.N.; Zheng, F.J.; Wang, B.S.; Wu, X.P.; Wu, H.J. Altered microbial resource limitation regulates soil organic carbon sequestration based on ecoenzyme stoichiometry under long-term tillage systems. Land Degrad. Dev. 2022, 33, 2795–2808. [Google Scholar] [CrossRef]
- Najera, F.; Dippold, M.A.; Boy, J.; Seguel, O.; Koester, M.; Stock, S.; Merino, C.; Kuzyakov, Y.; Matus, F. Effects of drying/rewetting on soil aggregate dynamics and implications for organic matter turnover. Biol. Fertil. Soils 2020, 56, 893–905. [Google Scholar] [CrossRef]
- Kok, D.D.; Scherer, L.; Vries, D.W.; Trimbos, P.M.; Bodegom, V. Relationships of priming effects with organic amendment composition and soil microbial properties. Geoderma 2022, 422, 115951. [Google Scholar] [CrossRef]
- Jackson, R.B.; Lajtha, K.; Crow, S.E.; Hugelius, G.; Kramer, M.G.; Piñeiro, G. The ecology of soil carbon: Pools, vulnerabilities, and biotic and abiotic controls. Annu. Rev. Ecol. Evol. Syst. 2020, 48, 419–445. [Google Scholar] [CrossRef]
- Chen, H.; Li, W.J.; Zheng, W.N.; Yin, C.; Fan, X.P.; Ye, M.J.; Gao, Z.X.; Wu, C.Y.; Liang, Y.C. Linking nitrogen- and straw-sensitive indicator species and their co-occurrences to priming effect in agricultural soil exposed to long-term nitrogen fertilization. Soil Biol. Biochem. 2023, 176, 108881. [Google Scholar] [CrossRef]
- Fang, Y.Y.; Nazaries, L.; Singh, B.K.; Singh, B.P. Microbial mechanisms of carbon priming effects revealed during the interaction of crop residue and nutrient inputs in contrasting soils. Glob. Chang. Biol. 2018, 24, 2775–2790. [Google Scholar] [CrossRef]
- Song, X.J.; Liu, X.T.; Liang, G.P.; Li, S.P.; Li, J.Y.; Zhang, M.N.; Zheng, F.J.; Ding, W.T.; Wu, X.P.; Wu, H.J. Positive priming effect explained by microbial nitrogen mining and stoichiometric decomposition at different stages. Soil Biol. Biochem. 2022, 175, 108852. [Google Scholar] [CrossRef]
- Wang, X.H.; Li, S.Y.; Zhu, B.; Peter, M.H.; Chen, G.S.; Yao, X.D.; Wu, D.M.; Yang, Z.J.; Lyu, M.K. Yusheng YangLong-term nitrogen deposition inhibits soil priming effects by enhancing phosphorus limitation in a subtropical forest. Glob. Change Biol. 2023, 29, 4081–4093. [Google Scholar] [CrossRef]
- Marie, B.; Tabata, B.; Jens, D.; Christine, W.; Rainer, G.J. Microbial carbon use efficiency of litter with distinct C/N ratios in soil at different temperatures, including microbial necromass as growth component. Biol. Fertil. Soils 2022, 58, 761–770. [Google Scholar]
- Wang, X.L.; Chen, Y.; Yang, K.P.; Duan, F.Y.; Liu, P.; Wang, Z.G.; Wang, J.W. Effects of legume intercropping and nitrogen input on net greenhouse gas balances, intensity, carbon footprint and crop productivity in sweet maize cropland in South China. J. Clean. Prod. 2021, 314, 127997. [Google Scholar] [CrossRef]
- Fang, Y.; Singh, B.P.; Collins, D.; Li, B.; Zhu, J.; Tavakkoli, E. Nutrient supply enhanced wheat residue Carbon mineralization, microbial growth, and microbial carbon-use efficiency when residues were supplied at high rate in contrasting soils. Soil Biol. Biochem. 2018, 126, 168–178. [Google Scholar] [CrossRef]
- Cui, H.; Chen, P.F.; He, C.; Jiang, Z.H.; Lan, R.; Yang, J.P. Soil microbial community structure dynamics shape the rhizosphere priming effect patterns in the paddy soil. Sci. Total Environ. 2023, 857, 159459. [Google Scholar] [CrossRef]
- Moran-Rodas, V.E.; Joergensen, R.G.; Wachendorf, C. Does liming improve microbial carbon use efficiency after maize litter addition in a tropical acidic soil? Biol. Fertil. Soils 2023, 1–9. [Google Scholar] [CrossRef]
- Schimel, J. Modeling ecosystem-scale carbon dynamics in soil: The microbial dimension. Soil Biol. Biochem. 2023, 178, 108948. [Google Scholar] [CrossRef]
- Aamer, M.; Chattha, M.B.; Mahmood, A.; Naqve, M.; Hassan, M.U.; Shaaban, M.; Rasul, F.; Batool, M.; Rasheed, A.; Tang, H.Y. Rice Residue-Based Biochar Mitigates N2O Emission from Acid Red Soil. Agronomy 2021, 11, 2462. [Google Scholar] [CrossRef]
- Dai, S.S.; He, P.; You, M.Y.; Li, L.J. The presence of soybean, but not soybean cropping frequency has influence on SOM priming in crop rotation systems. Plant Soil 2023, 487, 511–520. [Google Scholar] [CrossRef]
- Li, S.; Liu, J.J.; Yao, Q.; Yu, Z.H.; Li, Y.S.; Jin, J.; Liu, X.B.; Wang, G.H. Short-term lime application impacts microbial community composition and potential function in an acid black soil. Plant Soil 2022, 470, 35–50. [Google Scholar] [CrossRef]
- Zhang, C.; Zhao, X.; Liang, A.; Li, Y.Y.; Song, Q.; Li, X.Y.; Li, D.P.; Hou, N. Insight into the soil aggregate-mediated restoration mechanism of degraded black soil via biochar addition: Emphasizing the driving role of core microbial communities and nutrient cycling. Environ. Res. 2023, 228, 115895. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.X.; Pan, W.K.; Sheng, T.; Sun, X.D.; Xie, Y.N.; Chadwick, D.R.; Hill, P.W.; Si, L.L.; Wu, L.G.; Jones, D.L. Maize and soybean experience fierce competition from soil microorganisms for the uptake of organic and inorganic nitrogen and sulphur: A pot test using 13 C, 15 N, 14 C, and 35 S labelling. Soil Biol. Biochem. 2021, 157, 108260. [Google Scholar] [CrossRef]
- Feng, J.; Tang, M.; Zhu, B. Soil priming effect and its responses to nutrient addition along a tropical forest elevation gradient. Glob. Chang. Biol. 2021, 27, 2793–2806. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Lu, C.; Luo, J.; Gong, X.; Guo, D.; Ma, Y. Matured compost amendment improves compost nutrient content by changing the bacterial community during the composting of Chinese herb residues. Front. Microbiol. 2023, 14, 1146546. [Google Scholar] [CrossRef]
- Wang, X.J.; Clayton, R.; Butterly, C.R.; Baldock, J.F.; Tang, C.C. Long-term stabilization of crop residues and soil organic carbon affected by residue quality and initial soil pH. Sci. Total Environ. 2017, 587, 502–509. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Chen, M.; Yang, Z.L.; Cong, M.F.; Zhu, X.P.; Jia, H.T. Soil Microbial Community Response to Nitrogen Application on a Swamp Meadow in the Arid Region of Central Asia. Front. Microbiol. 2022, 12, 797306. [Google Scholar] [CrossRef]
- Goldstein, A.; Turner, W.R.; Spawn, S.A.; Anderson-Teixeira, K.J.; Cook-Patton, S.; Fargione, J.; Gibbs, H.K.; Griscom, B.; Hewson, J.H.; Howard, J.F.; et al. Protecting irrecoverable carbon in Earth’s ecosystems. Nat. Clim. Chang. 2020, 10, 287–295. [Google Scholar] [CrossRef]
- Chen, R.R.; Senbayram, M.; Blagodatsky, S.; Myachina, O.; Dittert, K.; Lin, X.G.; Blagodatskaya, E.; Kuzyakov, Y. Soil C and N availability determine the priming effect: Microbial N mining and stoichiometric decomposition theories. Glob. Chang. Biol. 2014, 20, 2356–2367. [Google Scholar] [CrossRef]
- Ramirez, K.S.; Craine, J.M.; Fierer, N. Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Glob. Change Biol. 2012, 18, 1918–1927. [Google Scholar] [CrossRef]
- Bai, L.F.; Zhang, X.Q.; Li, B.Z.; Sun, F.C.; Zhao, X.Q.; Wang, Y.F.; Lu, Z.Y.; Zhang, D.J.; Fang, J. Fungal communities are more sensitive to nitrogen fertilization than bacteria in different spatial structures of silage maize under short-term nitrogen fertilization. Appl. Soil Ecol. 2022, 170, 104275. [Google Scholar] [CrossRef]
- Autret, B.; Guillier, H.; Pouteau, V.; Mary, B.; Chenu, C. Similar specific mineralization rates of organic carbon and nitrogen in incubated soils under contrasted arable cropping systems. Soil Tillage Res. 2020, 204, 104712. [Google Scholar] [CrossRef]
- Fu, X.H.; Li, S.Q.; Shen, Y.F.; Yue, S.C. Dynamic changes in bacterial community structure are associated with distinct priming effect patterns. Soil Biol. Biochem. 2022, 169, 108671. [Google Scholar] [CrossRef]
- Rottjers, L.; Faust, K. From hairballs to hypotheses-biological insights from microbial networks. FEMS Microbiol. Rev. 2018, 42, 761–780. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Lv, J.; Yu, M.; Ma, Z.; Xi, H.; Kou, C.; He, Z.; Shen, A. Long-term decomposed straw return positively affects the soil microbial community. J. Appl. Microbiol. 2020, 128, 138–150. [Google Scholar] [CrossRef]
- Xiao, D.; He, X.Y.; Wang, G.H.; Xu, X.C.; Hu, Y.J.; Chen, X.B.; Zhang, W.; Su, Y.R.; Wang, K.L.; Soromotin, A.V.; et al. Network analysis reveals bacterial and fungal keystone taxa involved in straw and soil organic matter mineralization. Appl. Soil Ecol. 2022, 173, 104395. [Google Scholar] [CrossRef]
- Banerjee, S.; Schlaeppi, K.; Van der Heijden, M.G.A. Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 2018, 16, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Li, L.J.; Xia, Z.B.; Ye, R.Z.; Doane, T.A.; Horwath, W.R. Soil microbial biomass size and soil carbon influence the priming effect from carbon inputs depending on nitrogen availability. Soil Biol. Biochem. 2018, 119, 41–49. [Google Scholar] [CrossRef]
- Fang, Y.; Singh, B.P.; Badgery, W.; He, X. In situ assessment of new carbon and nitrogen assimilation and allocation in contrastingly managed dryland wheat crop–soil systems. Agric. Ecosyst. Environ. 2016, 235, 80–90. [Google Scholar] [CrossRef]
- Mo, F.; Zhang, Y.Y.; Liu, Y.; Liao, Y.C. Microbial carbon-use efficiency and straw-induced priming effect within soil aggregates are regulated by tillage history and balanced nutrient supply. Biol. Fertil. Soils 2021, 57, 409–420. [Google Scholar] [CrossRef]
- Zhuang, G.; Shuangyi, L.; Roland, B.; Zhu, P.; Peng, C.; An, T.T.; Cheng, N.; Liu, X.; Li, T.G.; Xu, Z.Q.; et al. Differential long-term fertilization alters residue-derived labile organic carbon fractions and microbial community during straw residue decomposition. Soil Tillage Res. 2021, 213, 105120. [Google Scholar]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Johan, B.P.; Bengtsson, P.J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2018, 47, 259–264. [Google Scholar] [CrossRef]
- Qiu, Q.Y.; Wang, H.; Zhang, Q.F.; Mgelwa, A.S.; Zhu, B.; Hu, Y.L. Negative priming effect from tree leaf and root residues with contrasting chemical composition. Geoderma 2022, 427, 116118. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mah, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, 2584. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Q.; Song, M.; Li, D.D.; Ma, L.; Zhao, B.Z.; Zhang, J.B. Effect of long-term fertilization on decomposition of crop residues and their incorporation into microbial communities of 6-year stored soils. Biol. Fertil. Soils 2020, 56, 25–37. [Google Scholar] [CrossRef]
- Sarker, J.R.; Singh, B.P.; Fang, Y.; Cowie, A.L.; Dougherty, W.J.; Collins, D.; Dalal, R.C.; Singh, B.K. Tillage history and crop residue input enhanced native carbon mineralisation and nutrient supply in contrasting soils under long-term farming systems. Soil Tillage Res. 2019, 193, 71–84. [Google Scholar] [CrossRef]
- Liu, X.A.; Sun, J.; Mau, R.L.; Finley, B.K.; Compson, Z.G.; Gestel, N.V.; Brown, J.R.; Schwartz, E.; Dijkstra, P.; Hungate, B.A. Labile carbon input determines the direction and magnitude of the priming effect. Appl. Soil Ecol. 2017, 109, 7–13. [Google Scholar] [CrossRef]
- Li, J.N.; Zhao, J.; Liao, X.H.; Yi, Q.; Zhang, W.; Lin, H.F.; Liu, K.P.; Peng, P.Q.; Wang, K.L. Long-term returning agricultural residues increases soil microbe-nematode network complexity and ecosystem multifunctionality. Geoderma 2023, 430, 116340. [Google Scholar] [CrossRef]
- Zhang, S.B.; Fang, Y.Y.; Kawasaki, A.; Tavakkoli, E.; Cai, Y.J.; Wang, H.L.; Ge, T.D.; Zhou, J.S.; Yu, B.; Li, Y.F. Biochar significantly reduced nutrient-induced positive priming in a subtropical forest soil. Biol. Fertil. Soils 2023, 1–19. [Google Scholar] [CrossRef]
- Arcand, M.M.; Levy, B.D.J.; Helgason, B.L. Resource legacies of organic and conventional management differentiate soil microbial carbon use. Front. Microbiol. 2017, 8, 02293. [Google Scholar] [CrossRef]
- Naether, A.; Foesel, B.U.; Naegele, V.; Wust, P.K.; Weinert, J.; Bonkowski, M.; Alt, F.; Oelmann, Y.; Polle, A.; Lohaus, G. Environmental factors affect acidobacterial communities below the subgroup level in grassland and forest soils. Appl. Environ. Microbiol. 2012, 78, 7398–7406. [Google Scholar] [CrossRef]
- Xie, N.; An, T.; Zhuang, J.; Mark, R.; Sean, S.; Li, S.; Wang, J.K. High initial soil organic matter level combined with aboveground plant residues increased microbial carbon use efficiency but accelerated soil priming effect. Biogeochemistry 2022, 160, 1–15. [Google Scholar] [CrossRef]
- Chen, X.B.; Xia, Y.H.; Rui, Y.C.; Ning, Z.; Hu, Y.J.; Tang, H.M.; He, H.B.; Li, H.X.; Kuzyakov, Y.K.; Ge, T.D.; et al. Microbial carbon use efficiency, biomass turnover, and necromass accumulation in paddy soil depending on fertilization. Agric. Ecosyst. Environ. 2020, 106, 816. [Google Scholar] [CrossRef]
- Wang, C.; Xiao, R.; Guo, Y.; Wang, Q.; Zhang, M. Changes in soil microbial community composition during phragmites australis straw decomposition in salt marshes with freshwater pumping. Sci. Total Environ. 2020, 762, 143996. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.Y.; Zhang, D.Q.; Yan, Y.Q.; Yang, C.; Fang, B.T.; Li, X.D.; Shao, Y.H.; Wang, H.F.; Yue, J.Q.; Wang, Y.J. Short-term application of chicken manure under different nitrogen rates alters structure and co-occurrence pattern but not diversity of soil microbial community in wheat field. Front. Microbiol. 2022, 13, 975571. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, J.; Mi, W.T.; Mu, W.Y.; Tao, W.H.; Chen, C.; Mi, W.H.; Su, L.J. Effects of slow- and controlled-release nitrogen fertilizers on bacterial and fungal community composition in a double-cropping rice system. Soil Tillage Res. 2023, 230, 105714. [Google Scholar] [CrossRef]
- Hui, C.; Jiang, H.; Liu, B.; Wei, R.; Zhang, Y.P.; Zhang, Q.C.; Liang, Y.C.; Zhao, Y.H. Chitin degradation and the temporary response of bacterial chitinolytic communities to chitin amendment in soil under different fertilization regime. Sci. Total Environ. 2020, 705, 136003. [Google Scholar] [CrossRef] [PubMed]
- Wagg, C.; Schlaeppi, K.; Banerjee, S.; Kuramae, E.E.; Heijden, M.G.A.V.D. Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nat. Commun. 2019, 10, 4841. [Google Scholar] [CrossRef]
- Liu, S.W.; Yu, H.; Yu, Y.H.; Huang, J.; Zhou, Z.Y.; Zeng, J.X.; Chen, P.B.; Xiao, F.S.; He, Z.L.; Yan, Q.Y. Ecological stability of microbial communities in Lake Donghu regulated by keystone taxa. Ecol. Indic. 2022, 136, 108695. [Google Scholar] [CrossRef]
- Wei, X.; Zhu, Z.; Liu, Y.; Luo, Y.; Deng, Y.; Xu, X.; Liu, S.; Richter, A.; Shibistova, O.; Guggenberger, G.; et al. C:N:P stoichiometry regulates soil organic carbon mineralization and concomitant shifts in microbial community composition in paddy soil. Biol. Fertil. Soils 2020, 56, 1093–1107. [Google Scholar] [CrossRef]
- Zhong, Y.; Liu, J.; Jia, X.; Shan, G.Z.; Wang, R.; Yan, W. Microbial community assembly and metabolic function during wheat straw decomposition under different nitrogen fertilization treatments. Biol. Fertil. Soils 2020, 14, 697–710. [Google Scholar] [CrossRef]
- Yan, F.; Wang, S.Q.; Huang, Z.H.; Liu, Y.R.; He, L.L.; Qian, F.Y. Microbial ecological responses of partial nitritation/anammox granular sludge to real water matrices and its potential application. Environ. Res. 2023, 266, 115701. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.D.; Tan, X.F.; Ye, Z.R.; Li, B.J.; Zhang, Y.; Chen, X.T.; Kong, D.D. Soil bacterial community composition and diversity respond to soil environment in rooftop agricultural system. Environ. Technol. Innov. 2023, 30, 103042. [Google Scholar] [CrossRef]
- Fei, Y.M.; Zhang, B.G.; He, J.X.; Chen, C.B.; Liu, H. Dynamics of vertical vanadium migration in soil and interactions with indigenous microorganisms adjacent to tailing reservoir. J. Hazard Mater. 2022, 424, 127608. [Google Scholar] [CrossRef] [PubMed]

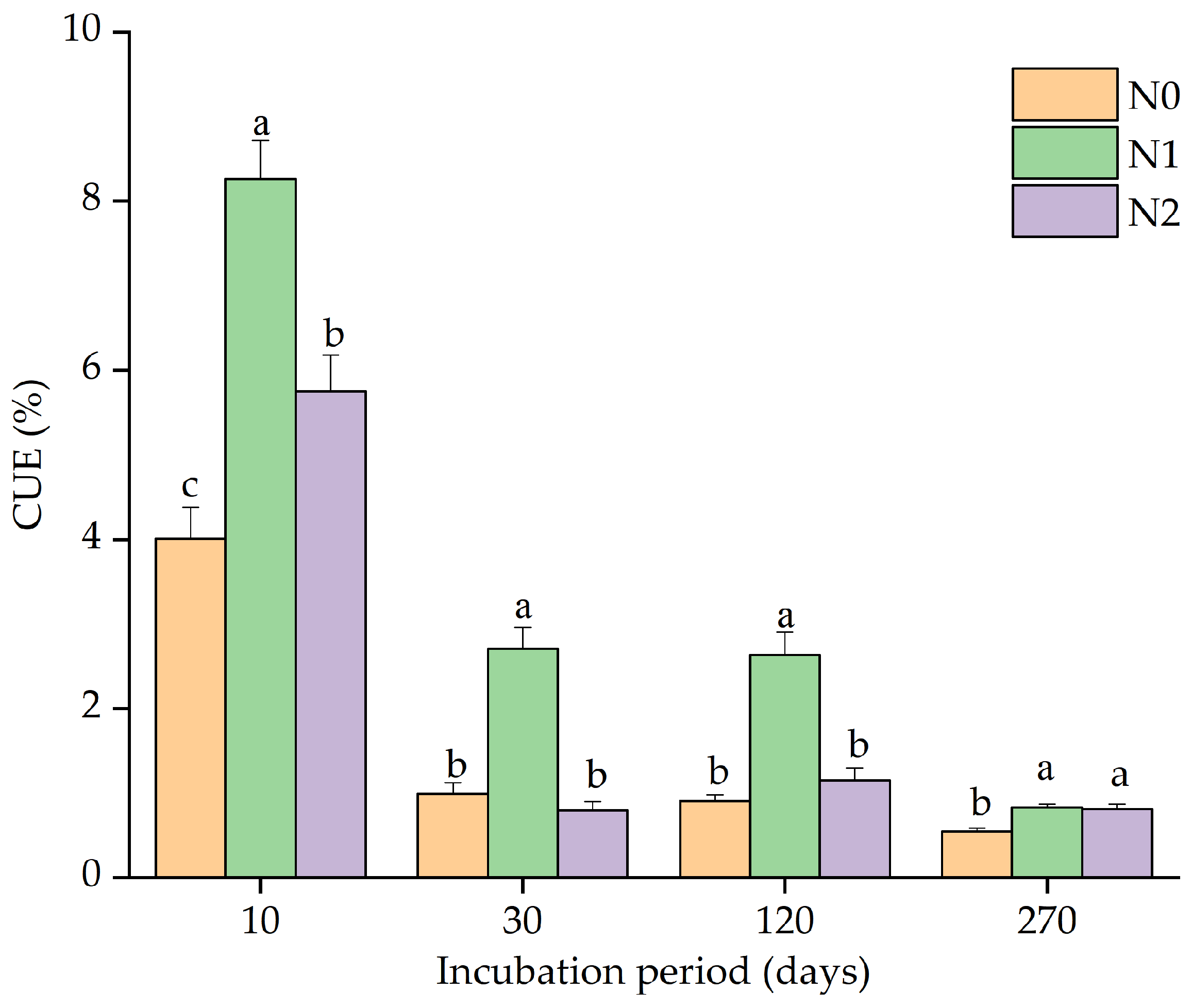

| Treatments | pH | SOC | TN | TP | TK | AN | AP | AK | Annual CO2 Emissions |
|---|---|---|---|---|---|---|---|---|---|
| g/kg | g/kg | g/kg | g/kg | mg/kg | mg/kg | mg/kg | kg/hm2 | ||
| N0 | 6.763 ± 0.108 a | 15.042 ± 0.338 b | 0.604 ± 0.012 c | 0.969 ± 0.012 b | 17.497 ± 0.971 b | 70.741 ± 3.739 b | 251.734 ± 13.435 c | 307.040 ± 2.323 b | 472.279 ± 23.76 a |
| N1 | 4.71 ± 0.155 b | 17.774 ± 0.320 a | 0.807 ± 0.014 a | 1.152 ± 0.014 a | 21.104 ± 0.792 a | 93.445 ± 1.575 a | 372.309 ± 5.404 a | 304.765 ± 4.684 a | 361.774 ± 18.87 b |
| N2 | 4.15 ± 0.023 c | 16.134 ± 0.293 b | 0.680 ± 0.015 b | 1.062 ± 0.015 ab | 19.383 ± 0.977 a | 87.323 ± 3.436 a | 333.077 ± 14.173 b | 395.305 ± 14.903 a | 378.424 ± 17.233 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, K.; Peng, P.; Duan, F.; Tang, H.; Wu, K.; Wu, Z.; Li, F.; Chen, Y.; Zou, C.; Liu, L.; et al. Microbial Mechanisms of the Priming Effect over 12 Years of Different Amounts of Nitrogen Management. Agronomy 2023, 13, 1783. https://doi.org/10.3390/agronomy13071783
Yang K, Peng P, Duan F, Tang H, Wu K, Wu Z, Li F, Chen Y, Zou C, Liu L, et al. Microbial Mechanisms of the Priming Effect over 12 Years of Different Amounts of Nitrogen Management. Agronomy. 2023; 13(7):1783. https://doi.org/10.3390/agronomy13071783
Chicago/Turabian StyleYang, Kepan, Peng Peng, Fuyuan Duan, Hu Tang, Kaixuan Wu, Zhenying Wu, Fan Li, Yong Chen, Chaoqun Zou, Lei Liu, and et al. 2023. "Microbial Mechanisms of the Priming Effect over 12 Years of Different Amounts of Nitrogen Management" Agronomy 13, no. 7: 1783. https://doi.org/10.3390/agronomy13071783





