Multi-Omics Approaches in Plant–Microbe Interactions Hold Enormous Promise for Sustainable Agriculture
Abstract
1. Introduction
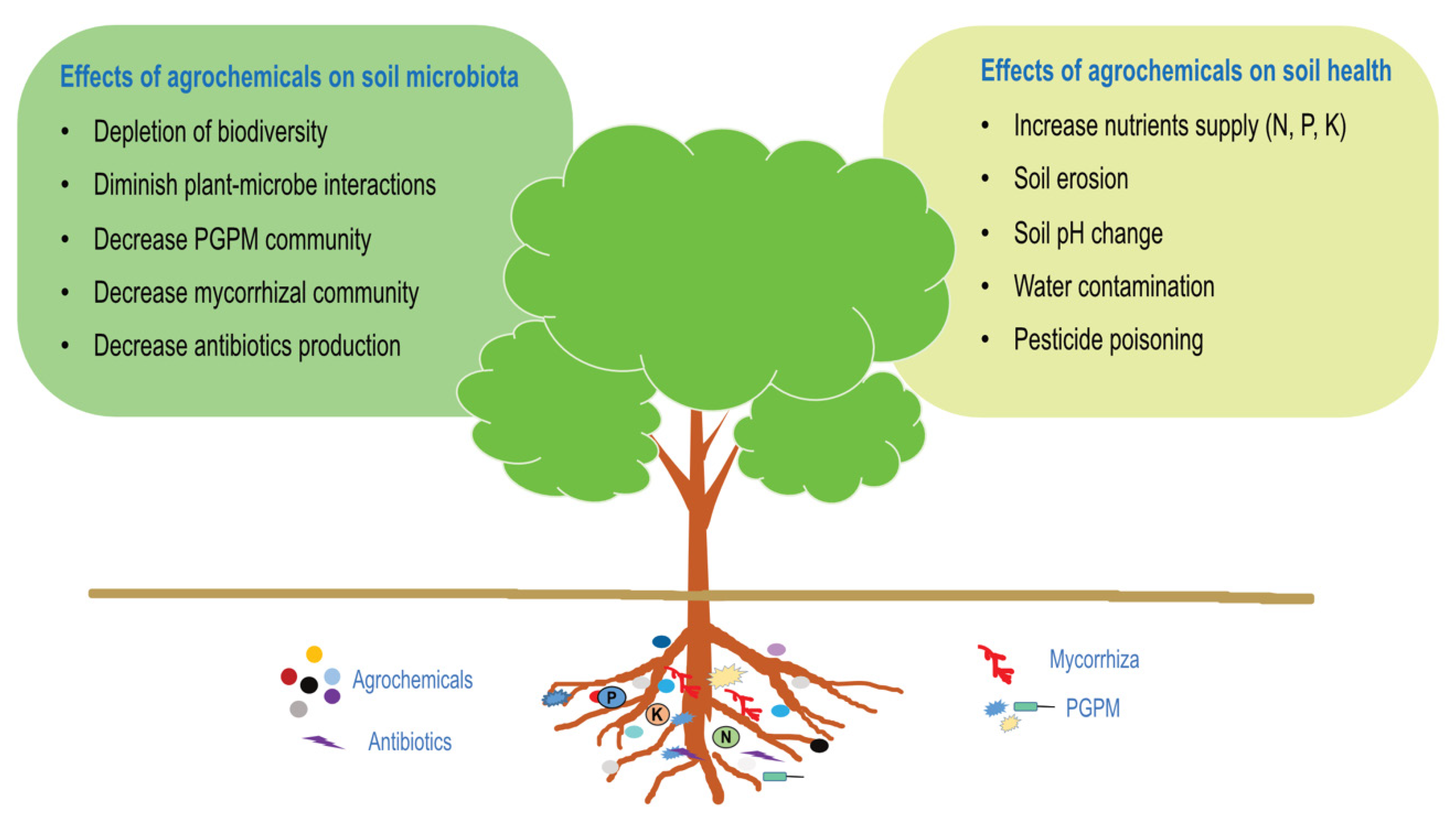
2. Impact of Excessive Consumption of Agrochemicals and Abiotic Parameters on Soil Microbiota–Plant Interactions
3. Soil Microbiota—An Ecological Engineer
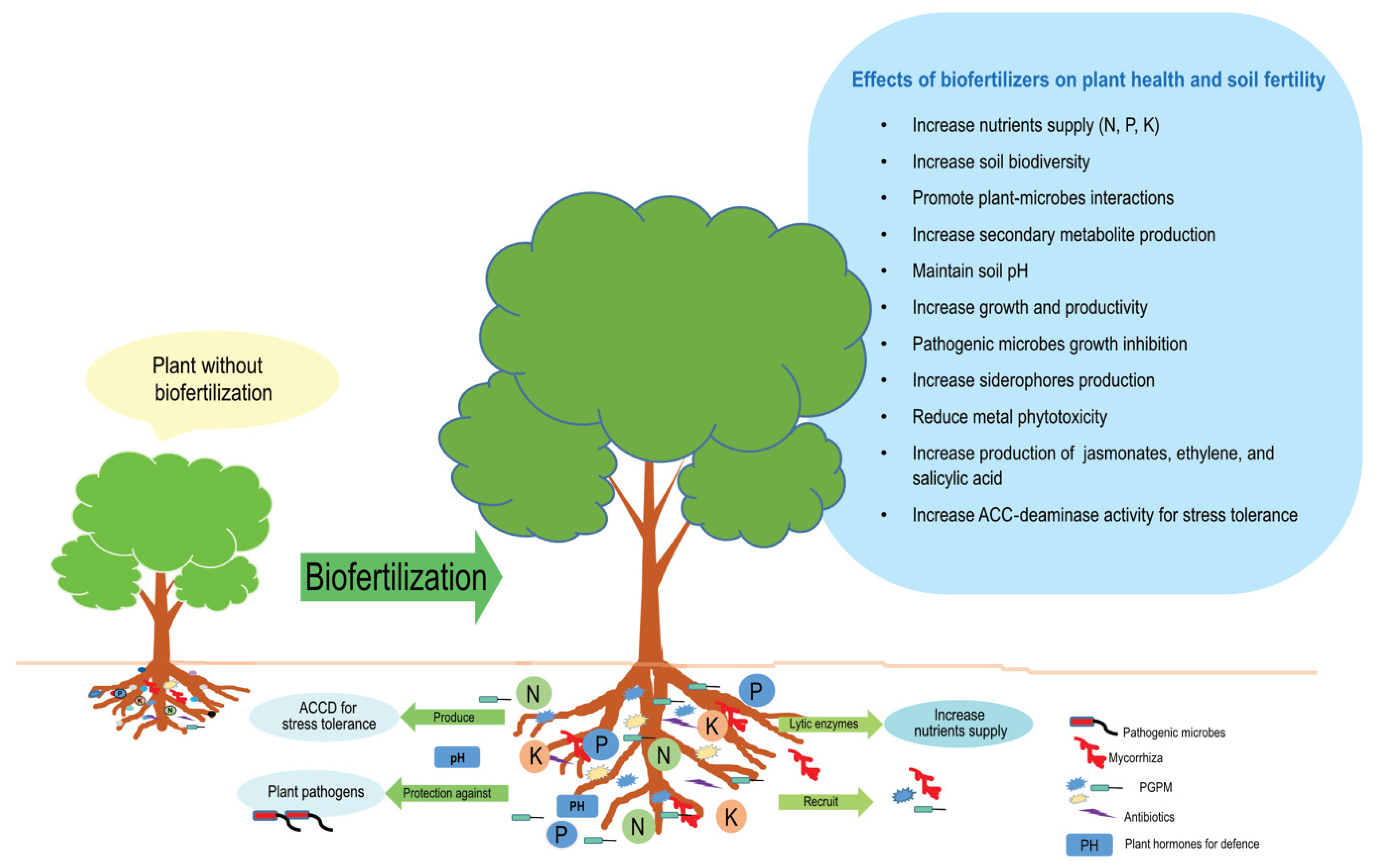
4. PGPM Boost Plant Growth and Productivity
5. Mycorrhizal Fungi Boost Plant Growth and Productivity
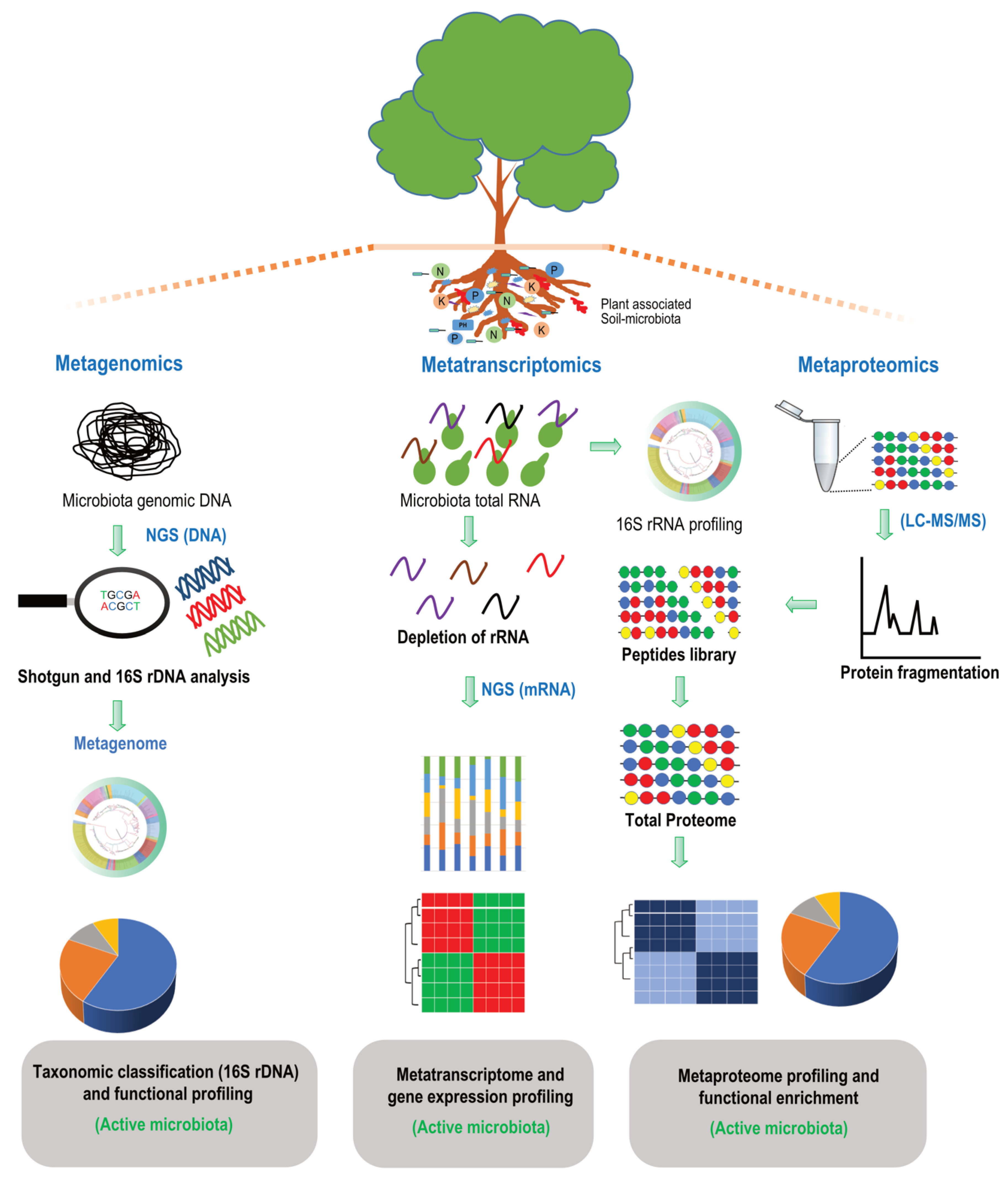
6. Multi-Omics—An Integrated Approach for a Better Insight into Soil-Microbiota–Plant Interactions
7. Conclusions and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berg, G.; Rybakova, D.; Grube, M.; Köberl, M. The plant microbiome explored: Implications for experimental botany. J. Exp. Bot. 2016, 67, 995–1002. [Google Scholar] [CrossRef] [PubMed]
- Berg, G. Plant-microbe interactions promoting plant growth and health: Perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 2009, 84, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Gunnell, D.; Eddleston, M.; Phillips, M.R.; Konradsen, F. The global distribution of fatal pesticide self-poisoning: Systematic review. BMC Public Health 2007, 7, 357. [Google Scholar] [CrossRef] [PubMed]
- Carrión, V.J.; Perez-Jaramillo, J.; Cordovez, V.; Tracanna, V.; de Hollander, M.; Ruiz-Buck, D.; Mendes, L.W.; van Ijcken, W.F.J.; Gomez-Exposito, R.; Raaijmakers, J.M.; et al. Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science 2019, 366, 606–612. [Google Scholar] [CrossRef]
- Vimal, S.R.; Singh, J.S.; Arora, N.K.; Singh, S. Soil-Plant-Microbe Interactions in Stressed Agriculture Management: A Review. Pedosphere 2017, 27, 177–192. [Google Scholar] [CrossRef]
- Glick, B.R. Introduction to Plant Growth-promoting Bacteria. In Beneficial Plant-Bacterial Interactions; Springer International Publishing: Cham, Switzerland, 2015; pp. 1–28. [Google Scholar] [CrossRef]
- Weyens, N.; van der Lelie, D.; Taghavi, S.; Newman, L.; Vangronsveld, J. Exploiting plant–microbe partnerships to improve biomass production and remediation. Trends Biotechnol. 2009, 27, 591–598. [Google Scholar] [CrossRef]
- Rajkumar, M.; Sandhya, S.; Prasad, M.N.V.; Freitas, H. Perspectives of plant-associated microbes in heavy metal phytoremediation. Biotechnol. Adv. 2012, 30, 1562–1574. [Google Scholar] [CrossRef]
- Pilon-Smits, E. Phytoremediation. Annu. Rev. Plant Biol. 2005, 56, 15–39. [Google Scholar] [CrossRef]
- Khan, A.G. Mycorrhizoremediation—An enhanced form of phytoremediation. J. Zhejiang Univ. Sci. B 2006, 7, 503–514. [Google Scholar] [CrossRef]
- Lin, X.; Feng, Y.; Zhang, H.; Chen, R.; Wang, J.; Zhang, J.; Chu, H. Long-Term Balanced Fertilization Decreases Arbuscular Mycorrhizal Fungal Diversity in an Arable Soil in North China Revealed by 454 Pyrosequencing. Environ. Sci. Technol. 2012, 46, 5764–5771. [Google Scholar] [CrossRef]
- Shen, J.P.; Zhang, L.M.; Guo, J.F.; Ray, J.L.; He, J.Z. Impact of long-term fertilization practices on the abundance and composition of soil bacterial communities in Northeast China. Appl. Soil Ecol. 2010, 46, 119–124. [Google Scholar] [CrossRef]
- Suding, K.N.; Collins, S.L.; Gough, L.; Clark, C.; Cleland, E.E.; Gross, K.L.; Milchunas, D.G.; Pennings, S. Functional- and abundance-based mechanisms explain diversity loss due to N fertilization. Proc. Natl. Acad. Sci. USA 2005, 102, 4387–4392. [Google Scholar] [CrossRef] [PubMed]
- Hautier, Y.; Niklaus, P.A.; Hector, A. Competition for Light Causes Plant Biodiversity Loss After Eutrophication. Science 2009, 324, 636–638. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Cernava, T. Overhauling the assessment of agrochemical-driven interferences with microbial communities for improved global ecosystem integrity. Environ. Sci. Ecotechnol. 2020, 4, 100061. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Singh, S.K.; Modi, A.; Singh, P.K.; Yeka Zhimo, V.; Kumar, A. Impacts of agrochemicals on soil microbiology and food quality. In Agrochemicals Detection, Treatment and Remediation; Elsevier: Amsterdam, The Netherlands, 2020; pp. 101–116. [Google Scholar] [CrossRef]
- Lamb, E.G.; Kembel, S.W.; Cahill, J.F. Shoot, but not root, competition reduces community diversity in experimental mesocosms. J. Ecol. 2009, 97, 155–163. [Google Scholar] [CrossRef]
- Harpole, W.S.; Tilman, D. Grassland species loss resulting from reduced niche dimension. Nature 2007, 446, 791–793. [Google Scholar] [CrossRef]
- Crawley, M.J.; Johnston, A.E.; Silvertown, J.; Dodd, M.; de Mazancourt, C.; Heard, M.S.; Henman, D.F.; Edwards, G.R. Determinants of Species Richness in the Park Grass Experiment. Am. Nat. 2005, 165, 179–192. [Google Scholar] [CrossRef]
- Li, F.; Liu, M.; Li, Z.; Jiang, C.; Han, F.; Che, Y. Changes in soil microbial biomass and functional diversity with a nitrogen gradient in soil columns. Appl. Soil Ecol. 2013, 64, 1–6. [Google Scholar] [CrossRef]
- Pii, Y.; Mimmo, T.; Tomasi, N.; Terzano, R.; Cesco, S.; Crecchio, C. Microbial interactions in the rhizosphere: Beneficial influences of plant growth-promoting rhizobacteria on nutrient acquisition process. A review. Biol. Fertil. Soils. 2015, 51, 403–415. [Google Scholar] [CrossRef]
- Ai, C.; Liang, G.; Sun, J.; Wang, X.; He, P.; Zhou, W.; He, X. Reduced dependence of rhizosphere microbiome on plant-derived carbon in 32-year long-term inorganic and organic fertilized soils. Soil. Biol. Biochem. 2015, 80, 70–78. [Google Scholar] [CrossRef]
- Lambers, H.; Mougel, C.; Jaillard, B.; Hinsinger, P. Plant-microbe-soil interactions in the rhizosphere: An evolutionary perspective. Plant Soil. 2009, 321, 83–115. [Google Scholar] [CrossRef]
- Egerton-Warburton, L.M.; Johnson, N.C.; Allen, E.B. Mycorrhizal Community Dynamics Following Nitrogen Fertilization: A Cross-Site Test in Five Grasslands. Ecol. Monogr. 2007, 77, 527–544. [Google Scholar] [CrossRef]
- Ling, N.; Zhu, C.; Xue, C.; Chen, H.; Duan, Y.; Peng, C.; Guo, S.; Shen, Q. Insight into how organic amendments can shape the soil microbiome in long-term field experiments as revealed by network analysis. Soil. Biol. Biochem. 2016, 99, 137–149. [Google Scholar] [CrossRef]
- Wang, H.; Wei, Z.; Mei, L.; Gu, J.; Yin, S.; Faust, K.; Raes, J.; Deng, Y.; Wang, Y.; Shen, Q.; et al. Combined use of network inference tools identifies ecologically meaningful bacterial associations in a paddy soil. Soil. Biol. Biochem. 2017, 105, 227–235. [Google Scholar] [CrossRef]
- Wang, J.; Song, Y.; Ma, T.; Raza, W.; Li, J.; Howland, J.G.; Huang, Q.; Shen, Q. Impacts of inorganic and organic fertilization treatments on bacterial and fungal communities in a paddy soil. Appl. Soil. Ecol. 2017, 112, 42–50. [Google Scholar] [CrossRef]
- Nosheen, S.; Ajmal, I.; Song, Y. Microbes as Biofertilizers, a Potential Approach for Sustainable Crop Production. Sustainability 2021, 13, 1868. [Google Scholar] [CrossRef]
- Kumar, R.; Saurabh, K.; Kumawat, N.; Sundaram, P.K.; Mishra, J.S.; Singh, D.K.; Hans, H.; Krishna, B.; Bhatt, B.P. Sustaining Productivity Through Integrated Use of Microbes in Agriculture. In Role of Microbial Communities for Sustainability; Springer: Singapore, 2021; pp. 109–145. [Google Scholar] [CrossRef]
- Fadiji, A.E.; Yadav, A.N.; Santoyo, G.; Babalola, O.O. Understanding the plant-microbe interactions in environments exposed to abiotic stresses: An overview. Microbiol. Res. 2023, 271, 127368. [Google Scholar] [CrossRef]
- Sharma, S.; Chandra, D.; Sharma, A.K. Rhizosphere Plant–Microbe Interactions under Abiotic Stress. In Rhizosphere Biology: Interactions between Microbes and Plant; Springer: Singapore, 2021; pp. 195–216. [Google Scholar] [CrossRef]
- Etesami, H.; Beattie, G.A. Plant-Microbe Interactions in Adaptation of Agricultural Crops to Abiotic Stress Conditions. In Probiotics and Plant Health; Springer: Singapore, 2017; pp. 163–200. [Google Scholar] [CrossRef]
- Berens, M.L.; Wolinska, K.W.; Spaepen, S.; Ziegler, J.; Nobori, T.; Nair, A.; Krüler, V.; Winkelmüller, T.M.; Wang, Y.; Mine, A.; et al. Balancing trade-offs between biotic and abiotic stress responses through leaf age-dependent variation in stress hormone cross-talk. Proc. Natl. Acad. Sci. USA 2019, 116, 2364–2373. [Google Scholar] [CrossRef]
- Li, H.; La, S.; Zhang, X.; Gao, L.; Tian, Y. Salt-induced recruitment of specific root-associated bacterial consortium capable of enhancing plant adaptability to salt stress. ISME J. 2021, 15, 2865–2882. [Google Scholar] [CrossRef]
- Osman, K.T. Saline and Sodic Soils. In Management of Soil Problems; Springer International Publishing: Cham, Switzerland, 2018; pp. 255–298. [Google Scholar] [CrossRef]
- Xu, L.; Naylor, D.; Dong, Z.; Simmons, T.; Pierroz, G.; Hixson, K.K.; Kim, Y.-M.; Zink, E.M.; Engbrecht, K.M.; Wang, Y.; et al. Drought delays development of the sorghum root microbiome and enriches for monoderm bacteria. Proc. Natl. Acad. Sci. USA 2018, 115, E4284–E4293. [Google Scholar] [CrossRef]
- Furze, J.R.; Martin, A.R.; Nasielski, J.; Thevathasan, N.V.; Gordon, A.M.; Isaac, M.E. Resistance and resilience of root fungal communities to water limitation in a temperate agroecosystem. Ecol. Evol. 2017, 7, 3443–3454. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, C.R.; Copeland, J.; Wang, P.W.; Guttman, D.S.; Kotanen, P.M.; Johnson, M.T.J. Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl. Acad. Sci. USA 2018, 115, E1157–E1165. [Google Scholar] [CrossRef] [PubMed]
- Brunner, I.; Herzog, C.; Dawes, M.A.; Arend, M.; Sperisen, C. How tree roots respond to drought. Front Plant Sci. 2015, 6, 547. [Google Scholar] [CrossRef]
- Talaat, N.B.; Shawky, B.T. Microbe-Mediated Induced Abiotic Stress Tolerance Responses in Plants. In Plant-Microbe Interactions in Agro-Ecological Perspectives; Springer International Publishing: Cham, Switzerland, 2017; pp. 101–133. [Google Scholar] [CrossRef]
- Shekhawat, K.; Saad, M.M.; Sheikh, A.; Mariappan, K.; Al-Mahmoudi, H.; Abdulhakim, F.; Eida, A.A.; Jalal, R.; Masmoudi, K.; Hirt, H. Root endophyte induced plant thermotolerance by constitutive chromatin modification at heat stress memory gene loci. EMBO Rep. 2021, 22, e51049. [Google Scholar] [CrossRef]
- Khan, M.A.; Asaf, S.; Khan, A.L.; Jan, R.; Kang, S.-M.; Kim, K.-M.; Lee, I.-J. Thermotolerance effect of plant growth-promoting Bacillus cereus SA1 on soybean during heat stress. BMC Microbiol. 2020, 20, 175. [Google Scholar] [CrossRef] [PubMed]
- Wipf, H.M.L.; Bùi, T.N.; Coleman-Derr, D. Distinguishing Between the Impacts of Heat and Drought Stress on the Root Microbiome of Sorghum bicolor. Phytobiomes J. 2021, 5, 166–176. [Google Scholar] [CrossRef]
- Theocharis, A.; Bordiec, S.; Fernandez, O.; Paquis, S.; Dhondt-Cordelier, S.; Baillieul, F.; Clément, C.; Barka, E.A. Burkholderia phytofirmans PsJN Primes Vitis vinifera L. and Confers a Better Tolerance to Low Nonfreezing Temperatures. Mol. Plant-Microbe Interact. 2012, 25, 241–249. [Google Scholar] [CrossRef]
- Fernandez, O.; Theocharis, A.; Bordiec, S.; Feil, R.; Jacquens, L.; Clément, C.; Fontaine, F.; Barka, E.A. Burkholderia phytofirmans PsJN Acclimates Grapevine to Cold by Modulating Carbohydrate Metabolism. Mol. Plant-Microbe Interact. 2012, 25, 496–504. [Google Scholar] [CrossRef]
- Masciarelli, O.; Llanes, A.; Luna, V. A new PGPR co-inoculated with Bradyrhizobium japonicum enhances soybean nodulation. Microbiol. Res. 2014, 169, 609–615. [Google Scholar] [CrossRef]
- Nacoon, S.; Jogloy, S.; Riddech, N.; Mongkolthanaruk, W.; Ekprasert, J.; Cooper, J.; Boonlue, S. Combination of arbuscular mycorrhizal fungi and phosphate solubilizing bacteria on growth and production of Helianthus tuberosus under field condition. Sci. Rep. 2021, 11, 6501. [Google Scholar] [CrossRef]
- Singh, S.; Kapoor, K.K. Inoculation with phosphate-solubilizing microorganisms and a vesicular-arbuscular mycorrhizal fungus improves dry matter yield and nutrient uptake by wheat grown in a sandy soil. Biol. Fertil. Soils. 1998, 28, 139–144. [Google Scholar] [CrossRef]
- Diwan, D.; Rashid, M.M.; Vaishnav, A. Current understanding of plant-microbe interaction through the lenses of multi-omics approaches and their benefits in sustainable agriculture. Microbiol. Res. 2022, 265, 127180. [Google Scholar] [CrossRef] [PubMed]
- Schirawski, J.; Perlin, M. Plant–Microbe Interaction 2017—The Good, the Bad and the Diverse. Int. J. Mol. Sci. 2018, 19, 1374. [Google Scholar] [CrossRef] [PubMed]
- Dohroo, A.D.R. The role of plant growth promoting rhizobacteria, arbuscular mycorrhizal fungi and their helper bacteria on growth parameters and root rot of apple. Walailak J. Sci Technol. 2012, 2, 35–38. [Google Scholar]
- Bender, S.F.; Wagg, C.; van der Heijden, M.G.A. An Underground Revolution: Biodiversity and Soil Ecological Engineering for Agricultural Sustainability. Trends Ecol. Evol. 2016, 31, 440–452. [Google Scholar] [CrossRef]
- Agoussar, A.; Yergeau, E. Engineering the plant microbiota in the context of the theory of ecological communities. Curr. Opin. Biotechnol. 2021, 70, 220–225. [Google Scholar] [CrossRef]
- Foo, J.L.; Ling, H.; Lee, Y.S.; Chang, M.W. Microbiome engineering: Current applications and its future. Biotechnol. J. 2017, 12, 1600099. [Google Scholar] [CrossRef]
- Verhage, A.; van Wees, S.C.M.; Pieterse, C.M.J. Plant Immunity: It’s the Hormones Talking, But What Do They Say? Figure 1. Plant Physiol. 2010, 154, 536–540. [Google Scholar] [CrossRef]
- Bari, R.; Jones, J.D.G. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef]
- Lopes, M.J.S.; Dias-Filho, M.B.; Gurgel, E.S.C. Successful Plant Growth-Promoting Microbes: Inoculation Methods and Abiotic Factors. Front. Sustain. Food Syst. 2021, 5, 606454. [Google Scholar] [CrossRef]
- Ahmad, M.; Zahir, Z.A.; Asghar, H.N.; Asghar, M. Inducing salt tolerance in mung bean through coinoculation with rhizobia and plant-growth-promoting rhizobacteria containing 1-aminocyclopropane-1-carboxylate deaminase. Can. J. Microbiol. 2011, 57, 578–589. [Google Scholar] [CrossRef] [PubMed]
- Gahlot, D.; Kukreja, K.; Suneja, S.; Dudeja, S. Effect of digested distillery spent wash on nodulation, nutrient uptake and photosynthetic activity in chickpea (Cicer arietinum). Acta Agron. Hung. 2011, 59, 73–85. [Google Scholar] [CrossRef]
- Naamala, J.; Smith, D.L. Relevance of Plant Growth Promoting Microorganisms and Their Derived Compounds, in the Face of Climate Change. Agronomy 2020, 10, 1179. [Google Scholar] [CrossRef]
- Bhardwaj, D.; Ansari, M.W.; Sahoo, R.K.; Tuteja, N. Biofertilizers function as key player in sustainable agriculture by improving soil fertility, plant tolerance and crop productivity. Microb. Cell Factories 2014, 13, 66. [Google Scholar] [CrossRef]
- Sharma, P.; Kapoor, K. Problems and prospective of nitrogen fixation in agroforestry systems. Proc Indian Natl Sci Acad. 2005, B71, 145–161. [Google Scholar]
- Ahmed, E.A.; Hassan, E.A.; El Tobgy, K.; Ramadan, E. Evaluation of rhizobacteria of some medicinal plants for plant growth promotion and biological control. Ann. Agric. Sci. 2014, 59, 273–280. [Google Scholar] [CrossRef]
- Gupta, S.; Dangayach, S.; Shukla, K.; Agrawal, N.; Sundari, K.S. Investigating the role of PGPM in assisting plant growth under stress caused by organophosphate pesticide-phorate. Indo Glob. J. Pharm. Sci. 2014, 4, 142. [Google Scholar] [CrossRef]
- Parmar, P.; Sindhu, S.S. Potassium Solubilization by Rhizosphere Bacteria: Influence of Nutritional and Environmental Conditions. J. Microbial. Res. 2013, 3, 25–31. [Google Scholar]
- Ulloa-Ogaz, A.L.; Muñoz-Castellanos, L.N.; Nevárez-Moorillón, G.V. Biocontrol of phytopathogens: Antibiotic production as mechanism of control. Battle Against Microb. Pathog. Basic Sci. Technol. Adv. Educ. Programs 2015, 1, 305–309. [Google Scholar]
- Johansson, J.F.; Paul, L.R.; Finlay, R.D. Microbial interactions in the mycorrhizosphere and their significance for sustainable agriculture. FEMS Microbiol Ecol. 2004, 48, 1–13. [Google Scholar] [CrossRef]
- Ruiz-Lozano, J.M.; Aroca, R.; Zamarreño, M.; Molina, S.; Andreo-Jiménez, B.; Porcel, R.; García-Mina, J.M.; Ruyter-Spira, C.; López-Ráez, J.A. Arbuscular mycorrhizal symbiosis induces strigolactone biosynthesis under drought and improves drought tolerance in lettuce and tomato. Plant Cell Environ. 2016, 39, 441–452. [Google Scholar] [CrossRef]
- Shukla, N.; Awasthi, R.P.; Rawat, L.; Kumar, J. Biochemical and physiological responses of rice (Oryza sativa L.) as influenced by Trichoderma harzianum under drought stress. Plant Physiol. Biochem. 2012, 54, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Shukla, P.S.; Agarwal, P.K.; Jha, B. Improved Salinity Tolerance of Arachis hypogaea (L.) by the Interaction of Halotolerant Plant-Growth-Promoting Rhizobacteria. J. Plant Growth Regul. 2012, 31, 195–206. [Google Scholar] [CrossRef]
- Offre, P.; Pivato, B.; Mazurier, S.; Siblot, S.; Berta, G.; Lemanceau, P.; Mougel, C. Microdiversity of Burkholderiales associated with mycorrhizal and nonmycorrhizal roots of Medicago truncatula. FEMS Microbiol. Ecol. 2008, 65, 180–192. [Google Scholar] [CrossRef]
- Offre, P.; Pivato, B.; Siblot, S.; Gamalero, E.; Corberand, T.; Lemanceau, P.; Mougel, C. Identification of Bacterial Groups Preferentially Associated with Mycorrhizal Roots of Medicago truncatula. Appl. Environ. Microbiol. 2007, 73, 913–921. [Google Scholar] [CrossRef] [PubMed]
- Bakker, P.A.H.M.; Pieterse, C.M.J.; de Jonge, R.; Berendsen, R.L. The Soil-Borne Legacy. Cell 2018, 172, 1178–1180. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Vismans, G.; Yu, K.; Song, Y.; de Jonge, R.; Burgman, W.P.; Burmølle, M.; Herschend, J.; Bakker, P.A.H.M.; Pieterse, C.M.J. Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 2018, 12, 1496–1507. [Google Scholar] [CrossRef]
- Li, X.; Jousset, A.; de Boer, W.; Carrion, V.J.; Zhang, T.; Wang, X.; Kuramae, E.E. Legacy of land use history determines reprogramming of plant physiology by soil microbiome. ISME J. 2019, 13, 738–751. [Google Scholar] [CrossRef]
- Rolfe, S.A.; Griffiths, J.; Ton, J. Crying out for help with root exudates: Adaptive mechanisms by which stressed plants assemble health-promoting soil microbiomes. Curr. Opin. Microbiol. 2019, 49, 73–82. [Google Scholar] [CrossRef]
- Evelin, H.; Kapoor, R.; Giri, B. Arbuscular mycorrhizal fungi in alleviation of salt stress: A review. Ann. Bot. 2009, 104, 1263–1280. [Google Scholar] [CrossRef]
- Laranjeira, S.; Fernandes-Silva, A.; Reis, S.; Torcato, C.; Raimundo, F.; Ferreira, L.; Carnide, V.; Marques, G. Inoculation of plant growth promoting bacteria and arbuscular mycorrhizal fungi improve chickpea performance under water deficit conditions. Appl. Soil Ecol. 2021, 164, 103927. [Google Scholar] [CrossRef]
- Hussain, S.S. Microbe-Mediated Tolerance in Plants Against Biotic and Abiotic Stresses. In Microbial Interventions in Agriculture and Environment; Springer: Singapore, 2019; pp. 173–217. [Google Scholar] [CrossRef]
- Timmusk, S.; Behers, L.; Muthoni, J.; Muraya, A.; Aronsson, A.C. Perspectives and Challenges of Microbial Application for Crop Improvement. Front. Plant Sci. 2017, 8, 49. [Google Scholar] [CrossRef] [PubMed]
- Sheth, R.U.; Cabral, V.; Chen, S.P.; Wang, H.H. Manipulating Bacterial Communities by in situ Microbiome Engineering. Trends Genet. 2016, 32, 189–200. [Google Scholar] [CrossRef]
- Mueller, U.G.; Sachs, J.L. Engineering Microbiomes to Improve Plant and Animal Health. Trends Microbiol. 2015, 23, 606–617. [Google Scholar] [CrossRef]
- Trivedi, P.; Schenk, P.M.; Wallenstein, M.D.; Singh, B.K. Tiny Microbes, Big Yields: Enhancing food crop production with biological solutions. Microb. Biotechnol. 2017, 10, 999–1003. [Google Scholar] [CrossRef] [PubMed]
- Doni, F.; Miranti, M.; Mispan, M.S.; Mohamed, Z.; Uphoff, N. Multi-omics approaches for deciphering the microbial modulation of plants’ genetic potentials: What’s known and what’s next? Rhizosphere 2022, 24, 100613. [Google Scholar] [CrossRef]
- López-Mondéjar, R.; Kostovčík, M.; Lladó, S.; Carro, L.; García-Fraile, P. Exploring the plant microbiome through multi-omics approaches. In Probiotics in Agroecosystem; Springer: Berlin/Heidelberg, Germany, 2017; pp. 233–268. [Google Scholar] [CrossRef]
- Saini, D.K.; Chopra, Y.; Singh, J.; Sandhu, K.S.; Kumar, A.; Bazzer, S.; Srivastava, P. Comprehensive evaluation of mapping complex traits in wheat using genome-wide association studies. Mol. Breed. 2022, 42, 1. [Google Scholar] [CrossRef]
- Scossa, F.; Alseekh, S.; Fernie, A.R. Integrating multi-omics data for crop improvement. J. Plant Physiol. 2021, 257, 153352. [Google Scholar] [CrossRef]
- Bertola, M.; Ferrarini, A.; Visioli, G. Improvement of Soil Microbial Diversity through Sustainable Agricultural Practices and Its Evaluation by—Omics Approaches: A Perspective for the Environment, Food Quality and Human Safety. Microorganisms 2021, 9, 1400. [Google Scholar] [CrossRef]
- Bell, T.H.; Joly, S.; Pitre, F.E.; Yergeau, E. Increasing phytoremediation efficiency and reliability using novel omics approaches. Trends Biotechnol. 2014, 32, 271–280. [Google Scholar] [CrossRef]
- Braga, R.M.; Dourado, M.N.; Araújo, W.L. Microbial interactions: Ecology in a molecular perspective. Braz. J. Microbiol. 2016, 47, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, G.; Sbodio, A.; Tech, J.J.; Suslow, T.V.; Coaker, G.L.; Leveau, J.H.J. Leaf microbiota in an agroecosystem: Spatiotemporal variation in bacterial community composition on field-grown lettuce. ISME J. 2012, 6, 1812–1822. [Google Scholar] [CrossRef] [PubMed]
- Malla, M.A.; Dubey, A.; Yadav, S.; Kumar, A.; Hashem, A.; Abd Allah, E.F. Understanding and Designing the Strategies for the Microbe-Mediated Remediation of Environmental Contaminants Using Omics Approaches. Front. Microbiol. 2018, 9, 1132. [Google Scholar] [CrossRef] [PubMed]
- Roossinck, M.J. Plant Virus Metagenomics: Biodiversity and Ecology. Annu. Rev. Genet. 2012, 46, 359–369. [Google Scholar] [CrossRef]
- White, R.A.; Rivas-Ubach, A.; Borkum, M.I.; Köberl, M.; Bilbao, A.; Colby, S.M.; Hoyt, D.W.; Bingol, K.; Kim, Y.-M.; Wendler, J.P.; et al. The state of rhizospheric science in the era of multi-omics: A practical guide to omics technologies. Rhizosphere 2017, 3, 212–221. [Google Scholar] [CrossRef]
- Trivedi, P.; Mattupalli, C.; Eversole, K.; Leach, J.E. Enabling sustainable agriculture through understanding and enhancement of microbiomes. New Phytol. 2021, 230, 2129–2147. [Google Scholar] [CrossRef]
- Saraiva, J.P.; Worrich, A.; Karakoç, C.; Kallies, R.; Chatzinotas, A.; Centler, F.; da Rocha, U.N. Mining Synergistic Microbial Interactions: A Roadmap on How to Integrate Multi-Omics Data. Microorganisms 2021, 9, 840. [Google Scholar] [CrossRef]
- Xu, L.; Pierroz, G.; Wipf, H.M.-L.; Gao, C.; Taylor, J.W.; Lemaux, P.G.; Coleman-Derr, D. Holo-omics for deciphering plant-microbiome interactions. Microbiome 2021, 9, 69. [Google Scholar] [CrossRef]
- Crandall, S.G.; Gold, K.M.; Jiménez-Gasco, M.M.; Filgueiras, C.C.; Willett, D.S. A multi-omics approach to solving problems in plant disease ecology. PLoS ONE 2020, 15, e0237975. [Google Scholar] [CrossRef]
- Yamazaki, S.; Mardani-Korrani, H.; Kaida, R.; Ochiai, K.; Kobayashi, M.; Nagano, A.J.; Fujii, Y.; Sugiyama, A.; Aoki, Y. Field multi-omics analysis reveals a close association between bacterial communities and mineral properties in the soybean rhizosphere. Sci. Rep. 2021, 11, 8878. [Google Scholar] [CrossRef]
- Igiehon, N.O.; Babalola, O.O. Below-ground-above-ground Plant-microbial Interactions: Focusing on Soybean, Rhizobacteria and Mycorrhizal Fungi. Open Microbiol. J. 2018, 12, 261–279. [Google Scholar] [CrossRef] [PubMed]
- Sessitsch, A.; Hardoim, P.; Doering, J.; Weilharter, A.; Krause, A.; Woyke, T.; Mitter, B.; Hauberg-Lotte, L.; Friedrich, F.; Rahalkar, M.; et al. Functional Characteristics of an Endophyte Community Colonizing Rice Roots as Revealed by Metagenomic Analysis. Mol. Plant Microbe Interact. 2012, 25, 28–36. [Google Scholar] [CrossRef]
- Higdon, S.M.; Pozzo, T.; Kong, N.; Huang, B.C.; Yang, M.L.; Jeannotte, R.; Brown, C.T.; Bennett, A.B.; Weimer, B.C. Genomic characterization of a diazotrophic microbiota associated with maize aerial root mucilage. PLoS ONE 2020, 15, e0239677. [Google Scholar] [CrossRef] [PubMed]
- Schauer, N.; Fernie, A.R. Plant metabolomics: Towards biological function and mechanism. Trends Plant Sci. 2006, 11, 508–516. [Google Scholar] [CrossRef]
- Maggini, V.; Mengoni, A.; Bogani, P.; Firenzuoli, F.; Fani, R. Promoting model systems of microbiota-medicinal plant interactions. Trends Plant Sci. 2020, 25, 223–225. [Google Scholar] [CrossRef] [PubMed]
- Pascale, A.; Proietti, S.; Pantelides, I.S.; Stringlis, I.A. Modulation of the root microbiome by plant molecules: The basis for targeted disease suppression and plant growth promotion. Front. Plant Sci. 2020, 10, 1741. [Google Scholar] [CrossRef]
- Wang, P.; Niu, B. Plant specialized metabolites modulate root microbiomes. Sci. China Life Sci. 2019, 62, 1111–1113. [Google Scholar] [CrossRef]
- Xu, L.; Dong, Z.; Chiniquy, D.; Pierroz, G.; Deng, S.; Gao, C.; Diamond, S.; Simmons, T.; Wipf, H.M.-L.; Caddell, D.; et al. Genome-resolved metagenomics reveals role of iron metabolism in drought-induced rhizosphere microbiome dynamics. Nat. Commun. 2021, 12, 3209. [Google Scholar] [CrossRef]
- Narsing Rao, M.P.; Lohmaneeratana, K.; Bunyoo, C.; Thamchaipenet, A. Actinobacteria–Plant Interactions in Alleviating Abiotic Stress. Plants 2022, 11, 2976. [Google Scholar] [CrossRef]
- Sakurai, N. Recent applications of metabolomics in plant breeding. Breed. Sci. 2022, 72, 21065. [Google Scholar] [CrossRef]
- Ellis, D.I.; Goodacre, R. Metabolic fingerprinting in disease diagnosis: Biomedical applications of infrared and Raman spectroscopy. Analyst 2006, 131, 875–885. [Google Scholar] [CrossRef] [PubMed]
- Knief, C.; Delmotte, N.; Chaffron, S.; Stark, M.; Innerebner, G.; Wassmann, R.; von Mering, C.; A Vorholt, J. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 2012, 6, 1378–1390. [Google Scholar] [CrossRef] [PubMed]
- Medina-Cordoba, L.K.; Chande, A.T.; Rishishwar, L.; Mayer, L.W.; Valderrama-Aguirre, L.C.; Valderrama-Aguirre, A.; Gaby, J.C.; Kostka, J.E.; Jordan, I.K. Genomic characterization and computational phenotyping of nitrogen-fixing bacteria isolated from Colombian sugarcane fields. Sci. Rep. 2021, 11, 9187. [Google Scholar] [CrossRef] [PubMed]
- Ke, J.; Wang, B.; Yoshikuni, Y. Microbiome Engineering: Synthetic Biology of Plant-Associated Microbiomes in Sustainable Agriculture. Trends Biotechnol. 2021, 39, 244–261. [Google Scholar] [CrossRef] [PubMed]
| Biofertiliser and Their Biochemical Property | Enriched Microorganism(s) | Reference(s) |
|---|---|---|
| Nitrogen fixing | Rhizobium spp. and Frankia spp. | [1] |
| Phosphorus solubilizing | Pseudomonas striata and Penicillium spp. | [2,3] |
| Zinc solubilizing | Pseudomonas and Bacillus spp. | [4,5] |
| Sulphur oxidising | Acidothiobacillus spp., Xanthobacter and Pseudomonas spp. | [6] |
| Potassium mobilizing | Aspergillus niger, Bacillus spp., Acidothiobacillus spp., Burkholderia spp., Ferrooxidans spp., Paenibacillus spp. and Pseudomonas spp. | [7,8] |
| Other plant-growth-promoting microbes | Agrobacterium spp., Enterobacter spp., Streptomyces spp., Pseudomonas fluorescens and Xanthomonas spp. | [9] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, U.; Raj, S.; Sreenikethanam, A.; Maddheshiya, R.; Kumari, S.; Han, S.; Kapoor, K.K.; Bhaskar, R.; Bajhaiya, A.K.; Gahlot, D.K. Multi-Omics Approaches in Plant–Microbe Interactions Hold Enormous Promise for Sustainable Agriculture. Agronomy 2023, 13, 1804. https://doi.org/10.3390/agronomy13071804
Kumar U, Raj S, Sreenikethanam A, Maddheshiya R, Kumari S, Han S, Kapoor KK, Bhaskar R, Bajhaiya AK, Gahlot DK. Multi-Omics Approaches in Plant–Microbe Interactions Hold Enormous Promise for Sustainable Agriculture. Agronomy. 2023; 13(7):1804. https://doi.org/10.3390/agronomy13071804
Chicago/Turabian StyleKumar, Umesh, Subhisha Raj, Arathi Sreenikethanam, Rahul Maddheshiya, Seema Kumari, Sungsoo Han, Krishan K. Kapoor, Rakesh Bhaskar, Amit K. Bajhaiya, and Dharmender K. Gahlot. 2023. "Multi-Omics Approaches in Plant–Microbe Interactions Hold Enormous Promise for Sustainable Agriculture" Agronomy 13, no. 7: 1804. https://doi.org/10.3390/agronomy13071804
APA StyleKumar, U., Raj, S., Sreenikethanam, A., Maddheshiya, R., Kumari, S., Han, S., Kapoor, K. K., Bhaskar, R., Bajhaiya, A. K., & Gahlot, D. K. (2023). Multi-Omics Approaches in Plant–Microbe Interactions Hold Enormous Promise for Sustainable Agriculture. Agronomy, 13(7), 1804. https://doi.org/10.3390/agronomy13071804









