Weed Identification in Maize Fields Based on Improved Swin-Unet
Abstract
1. Introduction
2. Data Processing
2.1. Data Acquisition
2.2. Data Enhancement
3. Model Construction
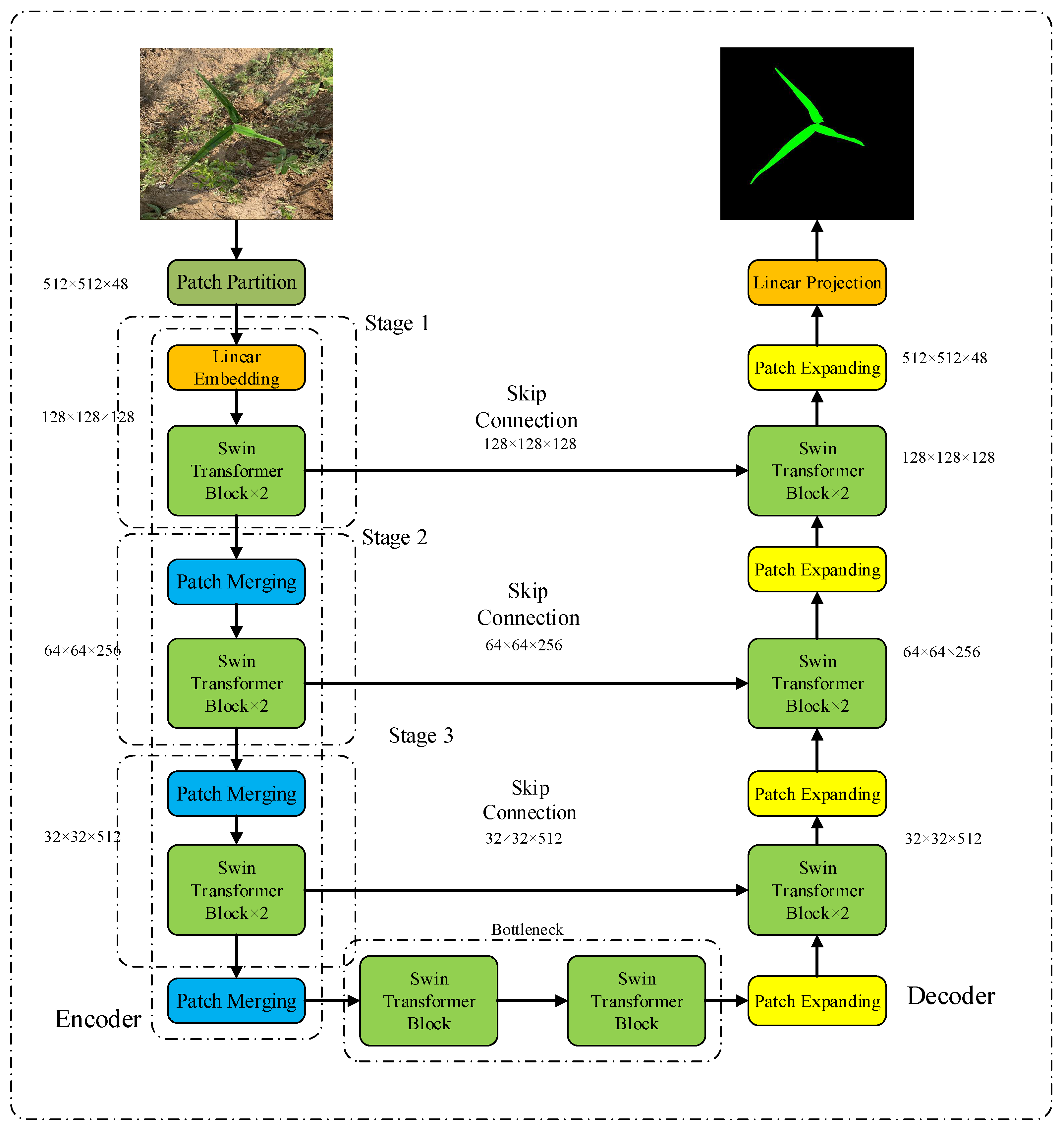
3.1. Swin-Unet Semantic Segmentation Model
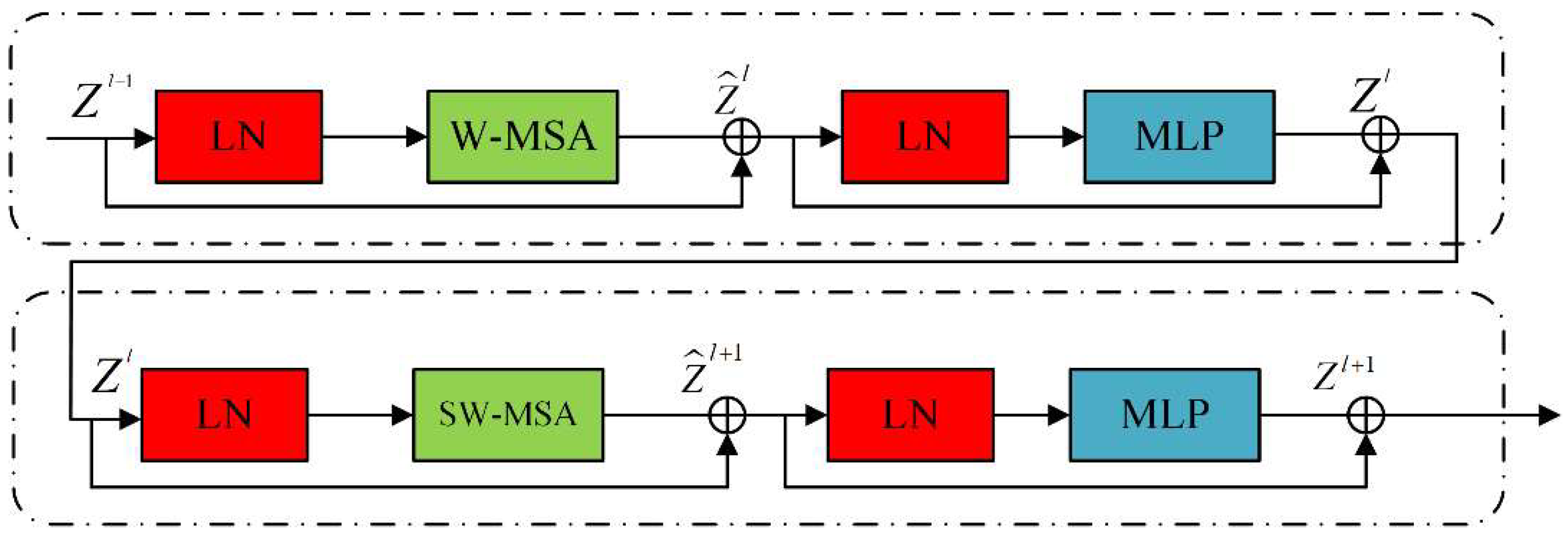
3.2. Swin Transformer Block
3.3. DropBlock Regularization
3.4. Weed Identification Model Based on Improved Swin-Unet
4. Weed Recognition Test
4.1. Test Environment
4.2. Parameter Settings
4.3. Model Evaluation Metrics
5. Results and Analysis
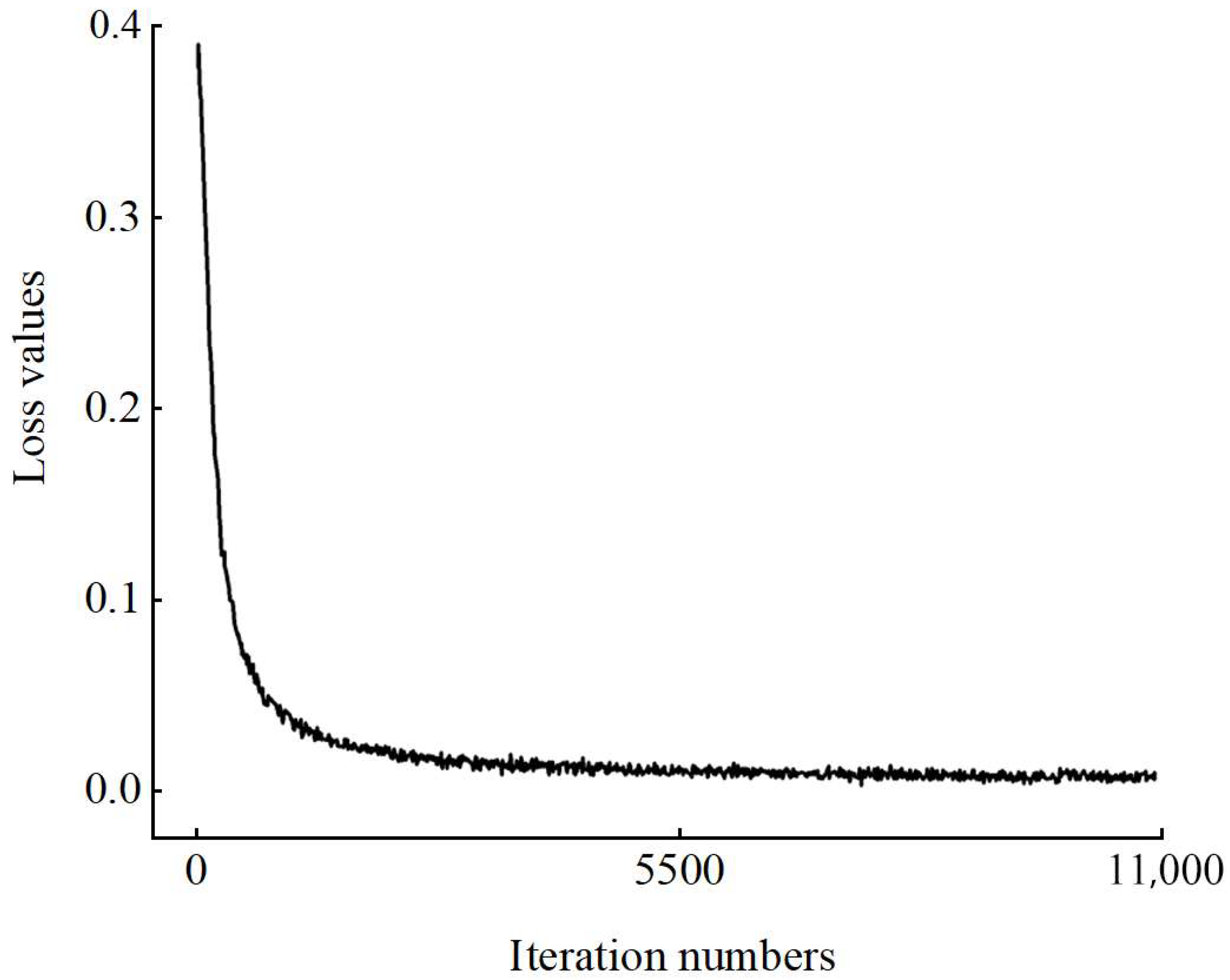
5.1. Training Error
5.2. Model Comparison
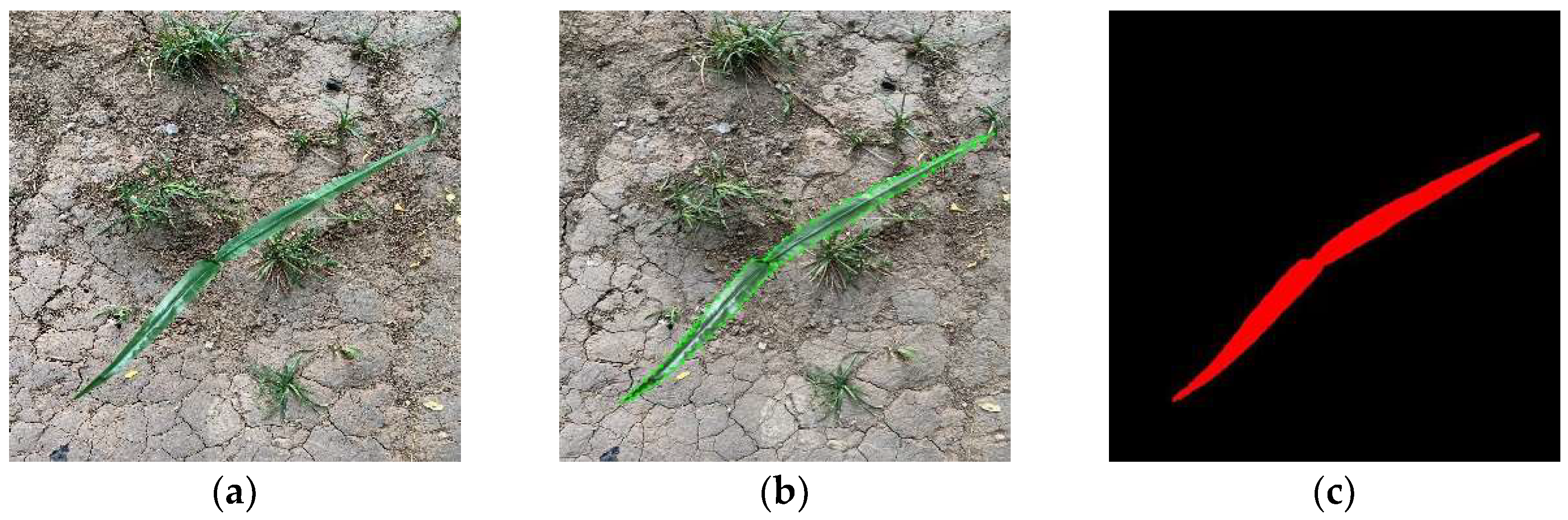
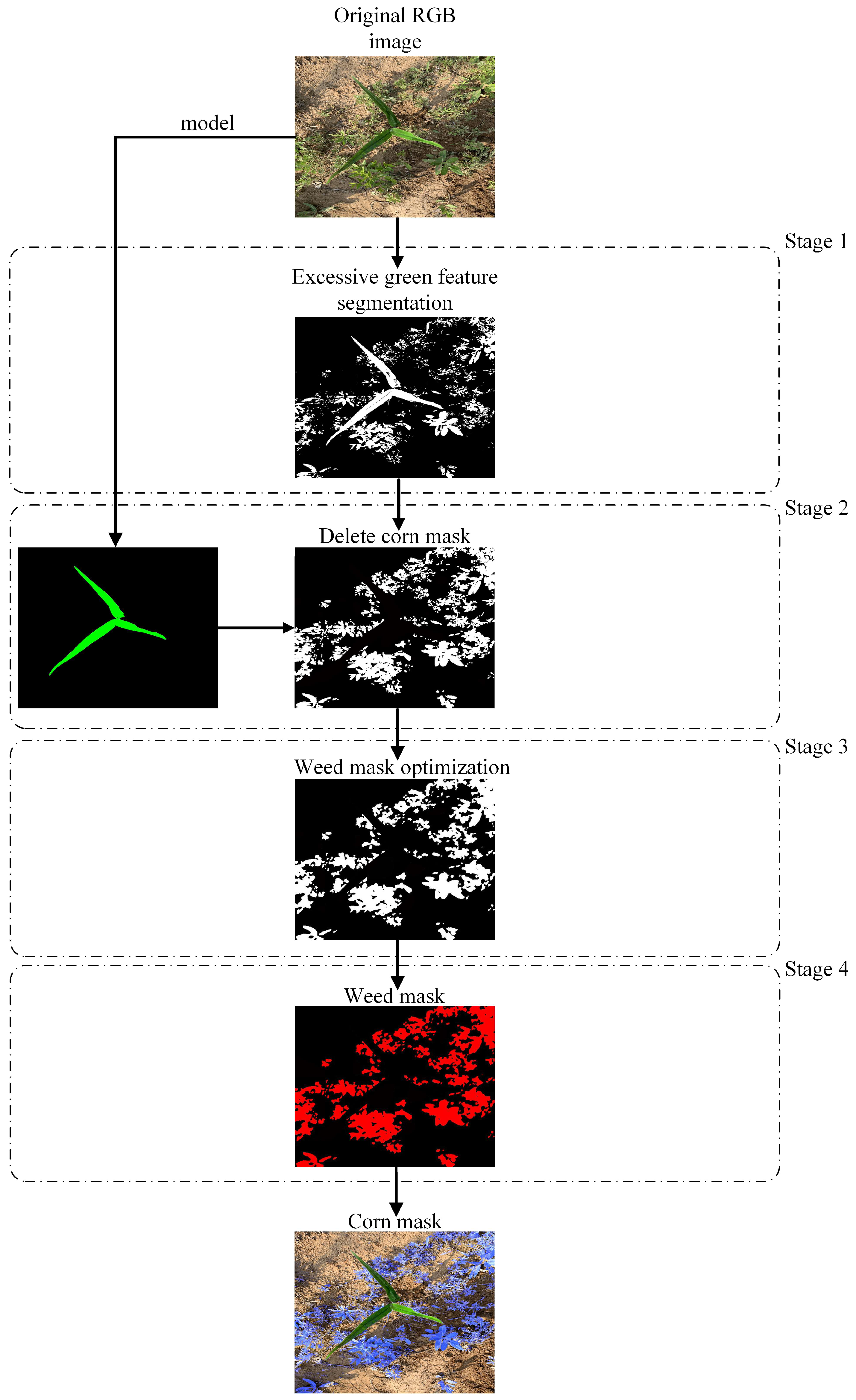
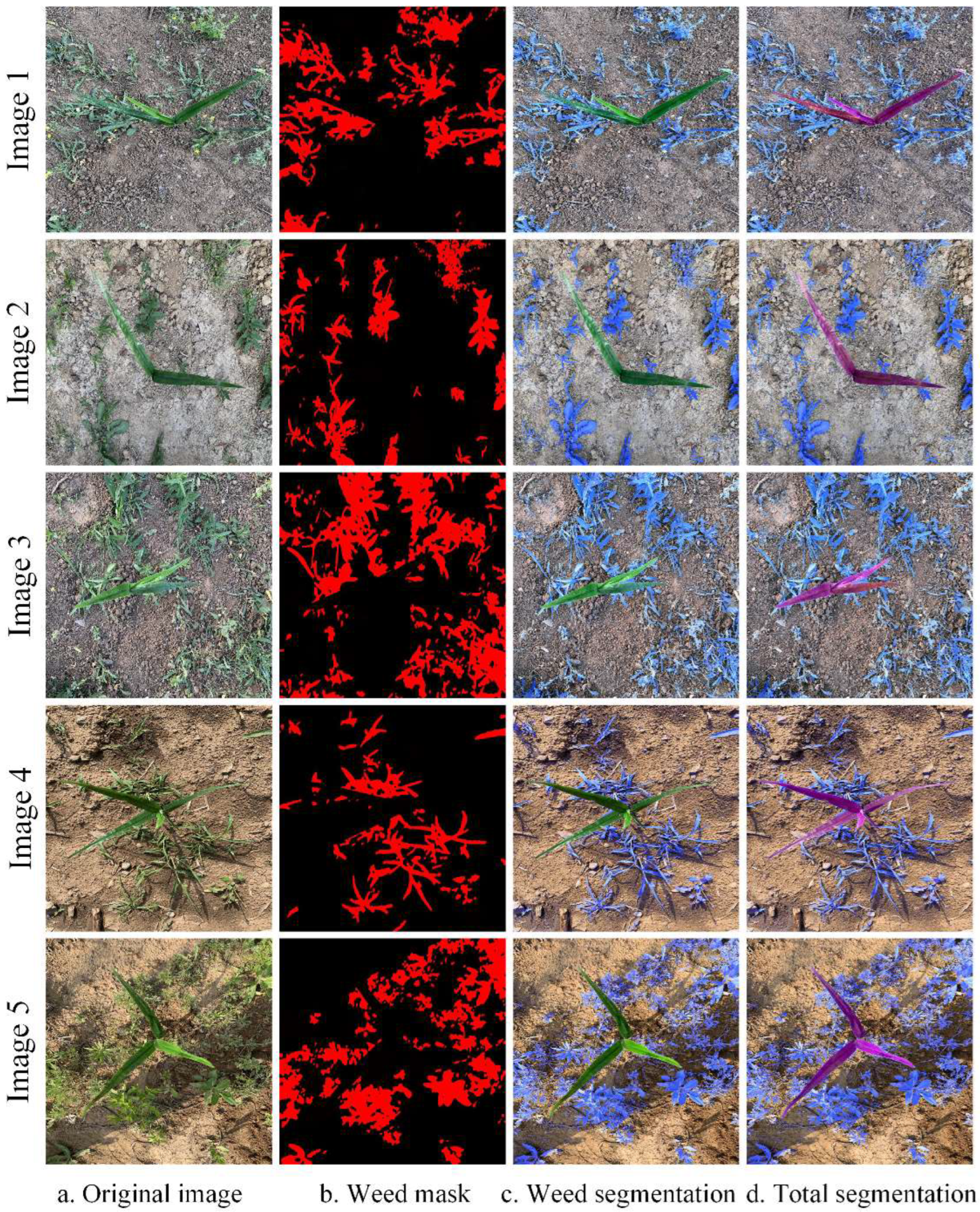
5.3. Maize Identification and Segmentation
6. Conclusions
- (1)
- The proposed model achieved up to 96.52% mPA and 93.71% mIoU, superior to those achieved by the DeepLabv3+, PSANet, Mask R-CNN, and original Swin-Unet models, indicating its effectiveness for target recognition and segmentation. The crop masks obtained through segmentation are used to obtain a weed mask through a morphological processing algorithm. Because the weed region can be obtained directly from the maize mask, only the maize seedlings must be labeled in the training set, greatly reducing the labor required. The method can efficiently and accurately identify and segment maize and weeds in a complex maize field environments, even where crops and weeds in some overlap.
- (2)
- The proposed model exhibited a higher inference speed than the original Swin-Unet, DeepLabv3+, and PSANet models; the processing time for each frame was 5.28 × 10−2 s. Hence, the proposed method is sufficiently fast for real-time data processing in applications such as vision for a weeding robot.
- (3)
- The proposed model in this paper, compared to similar studies, shows a slight improvement but lacks significant advantages. In future research, we will further enhance the model structure to improve the practicality of the method and develop a more efficient and accurate weed identification approach.
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Z.P. Development of chemical weed control and integrated weed management in China. Weed Biol. Manag. 2003, 3, 197–203. [Google Scholar] [CrossRef]
- Li, X. Outstanding problems and management countermeasures in weed control on farmland in China in recent years. Plant Prot. 2018, 44, 77–84. [Google Scholar]
- Machleb, J.; Peteinatos, G.G.; Kollenda, B.L.; Andújar, D.; Gerhards, R. Sensor-based mechanical weed control: Present state and prospects. Comput. Electron. Agric. 2020, 176, 105638. [Google Scholar] [CrossRef]
- Zhang, S. Weed control technologies in major crop fields in China. J. Weed Sci. 2020, 38, 50–55. [Google Scholar]
- Duan, X.; Han, J.; Ba, J.; Shi, J.; Zhang, Y.; Kang, L.; Wen, Y. Current status and development trend of chemical weed control in corn 417 fields. Hortic. Seedl. 2019, 39, 54–56. [Google Scholar]
- Saha, D.; Cregg, B.M.; Sidhu, M.K. A review of non-chemical weed control practices in Christmas tree production. Forests 2020, 11, 554. [Google Scholar] [CrossRef]
- Muola, A.; Fuchs, B.; Laihonen, M.; Rainio, K.; Heikkonen, L.; Ruuskanen, S.; Saikkonen, K.; Helander, M. Risk in the circular food economy: Glyphosate-based herbicide residues in manure fertilizers decrease crop yield. Sci. Total Environ. 2021, 750, 141422. [Google Scholar] [CrossRef]
- Yuan, H.; Zhao, N.; Cheng, M. Research progress and prospect of weed identification in the field based on image processing. J. Agric. Mach. 2020, 51 (Suppl. S2), 323–334. [Google Scholar]
- Utstumo, T.; Urdal, F.; Brevik, A.; Dørum, J.; Netland, J.; Overskeid, Ø.; Berge, T.W.; Gravdahl, J.T. Robotic in-row weed control in vegetables. Comput. Electron. Agric. 2018, 154, 36–45. [Google Scholar] [CrossRef]
- Huang, L.; Liu, W.; Huang, W.; Zhao, J.; Song, F. Remote sensing monitoring of powdery mildew in winter wheat by combining wavelet analysis and support vector machine. J. Agric. Eng. 2017, 33, 188–195. [Google Scholar]
- Zhai, Z.; Xu, Z.; Zhou, X.; Wang, L.; Zhang, J. Identification of cotton blind toon weevil hazard classes based on plain Bayesian classifier. J. Agric. Eng. 2015, 31, 204–211. [Google Scholar]
- Liang, X.; Chen, B.; Li, M.; Wei, C.; Feng, J. A dynamic counting method for cotton rows based on HOG features and SVM. J. Agric. Eng. 2020, 36, 173–181. [Google Scholar]
- Chen, S.; Wu, S.; Yu, X. Identification of buckwheat diseases based on convolutional neural networks combined with image processing techniques. J. Agric. Eng. 2021, 37, 155–163. [Google Scholar]
- Jiang, H.; Zhang, C.; Qiao, Y.; Zhang, Z.; Zhang, W.; Song, C. CNN feature based graph convolutional network for weed and crop recognition in smart farming. Comput. Electron. Agric. 2020, 174, 105450. [Google Scholar] [CrossRef]
- Zhou, L.; Mu, H.; Ma, H.; Chen, G. Remote sensing yield estimation of winter wheat in northern China based on convolutional neural network. J. Agric. Eng. 2019, 35, 119–128. [Google Scholar]
- Peng, W.; Lan, Y.; Yue, X.; Cheng, Z.; Wang, L.; Cen, Z.; Lu, Y.; Hong, J. Research on weed identification in rice fields based on deep convolutional neural network. J. South China Agric. Univ. 2020, 41, 75–81. [Google Scholar]
- Meng, Q.; Zhang, M.; Yang, X.; Liu, Y.; Zhang, Z. Identification of corn seedlings and weeds based on lightweight convolution combined with feature information fusion. J. Agric. Mach. 2020, 51, 238–245, 303. [Google Scholar]
- Wang, C.; Wu, X.; Zhang, Y.; Wang, W. Weed identification in corn field scenes based on shift window Transformer network. J. Agric. Eng. 2022, 38, 133–142. [Google Scholar]
- Takahashi, R.; Matsubara, T.; Uehara, K. Data augmentation using random image cropping and patching for deep CNNs. IEEE Trans. Circuits Syst. Video Technol. 2019, 30, 2917–2931. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-unet: Unet-like pure transformer for medical image segmentation. In Proceedings of the Computer Vision–ECCV 2022 Workshops, Tel Aviv, Israel, 23–27 October 2022; pp. 205–218. [Google Scholar]
- Xiao, X.; Zhang, D.; Hu, G.; Jiang, Y.; Xia, S. CNN–MHSA: A Convolutional Neural Network and multi-head self-attention combined approach for detecting phishing websites. Neural Netw. 2020, 125, 303–312. [Google Scholar] [CrossRef]
- Sheng, C.; Wang, L.; Huang, Z.; Wang, T.; Guo, Y.; Hou, W.; Xu, L.; Wang, J.; Yan, X. Transformer-Based Deep Learning Network for Tooth Segmentation on Panoramic Radiographs. J. Syst. Sci. Complex. 2023, 36, 257–272. [Google Scholar] [CrossRef]
- Alex, K.; Ilya, S.; Geoffrey, E.H. Image net classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar]
- Ghiasi, G.; Lin, T.Y.; Le, Q.V. Drop Block: A regularization method for convolutional networks. In Proceedings of the 32nd International Conference on Neural Information Processing Systems, Montreal, QC, Canada, 2–8 December 2018; Curran Associates Inc.: Red Hook, NY, USA, 2018; pp. 10750–10760. [Google Scholar]
- Morid, M.A.; Borjali, A.; Del Fiol, G. A scoping review of transfer learning research on medical image analysis using ImageNet. Comput. Biol. Med. 2021, 128, 104115. [Google Scholar] [CrossRef] [PubMed]
- Hasan, A.S.M.M.; Sohel, F.; Diepeveen, D.; Laga, H.; Jones, M.G. A survey of deep learning techniques for weed detection from images. Comput. Electron. Agric. 2021, 184, 106067. [Google Scholar] [CrossRef]
- Touvron, H.; Cord, M.; Douze, M.; Massa, F.; Sablayrolles, A.; Jégou, H. Training data-efficient image transformers & distillation through attention. In Proceedings of the International Conference on Machine Learning, Virtual, 18–24 July 2021; PMLR: New York, NY, USA, 2021; pp. 10347–10357. [Google Scholar]
- Wang, Z.; Qu, S. Real-time semantic segmentation network based on attention mechanism and multi-scale pooling. Comput. Eng. 2023, 1–11. [Google Scholar] [CrossRef]
- Gao, J.J.; Zhang, X.; Guo, Y.; Liu, Y.K.; Guo, A.N.Q.; Shi, M.O.M.; Wang, P.; Yuan, Y. Research on the optimization method of pest image instance segmentation by incorporating Swin Transformer. J. Nanjing For. Univ. (Nat. Sci. Ed.) 2023, 47, 1–10. [Google Scholar]
- Garibaldi-Márquez, F.; Flores, G.; Mercado-Ravell, D.A.; Ramírez-Pedraza, A.; Valentín-Coronado, L.M. Weed Classification from Natural Corn Field-Multi-Plant Images Based on Shallow and Deep Learning. Sensors 2022, 22, 3021. [Google Scholar] [CrossRef]
- Picon, A.; San-Emeterio, M.G.; Bereciartua-Perez, A.; Klukas, C.; Eggers, T.; Navarra-Mestre, R. Deep learning-based segmentation of multiple species of weeds and corn crop using synthetic and real image datasets. Comput. Electron. Agric. 2022, 194, 106719. [Google Scholar] [CrossRef]



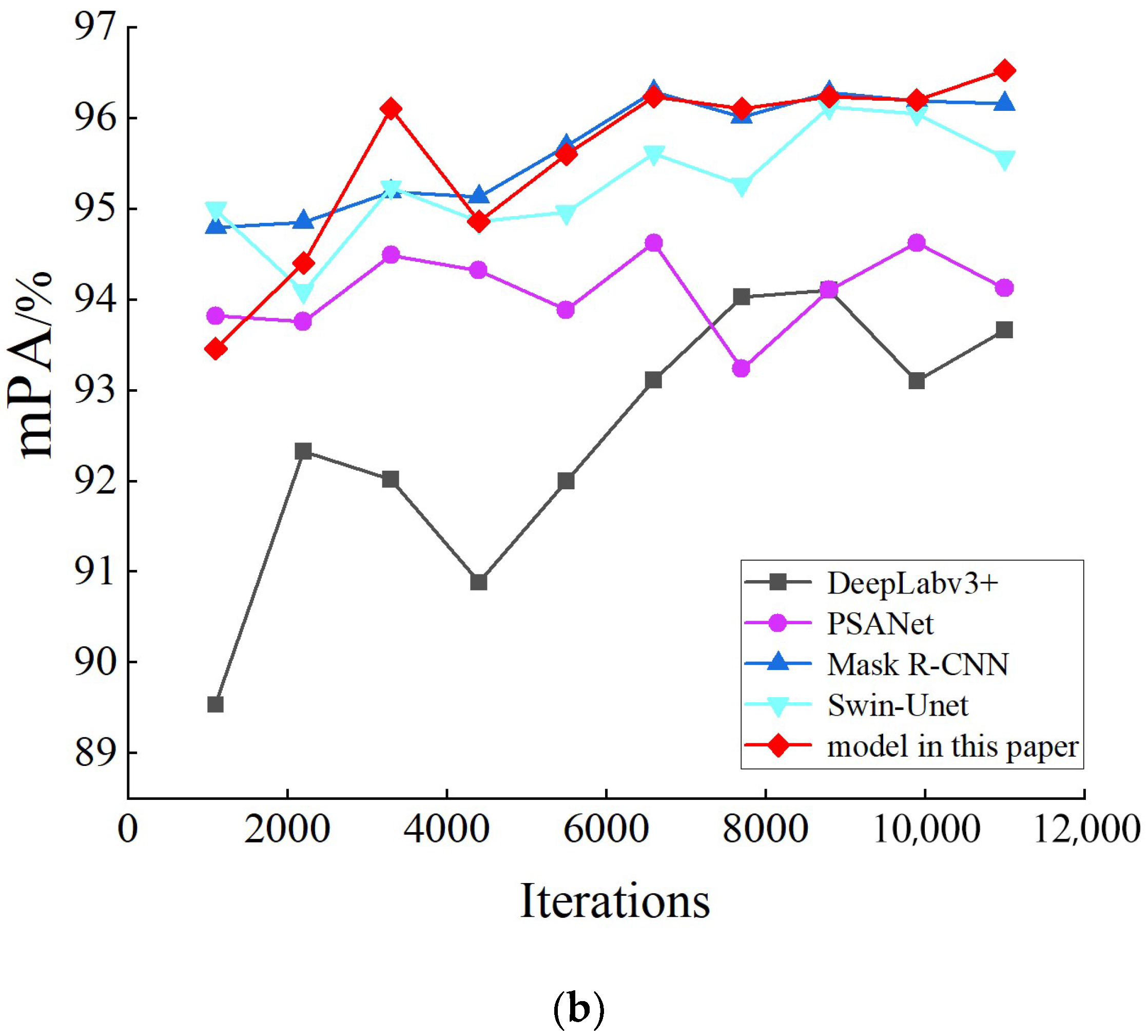

| Networks | mIoU (%) | mPA (%) | Segmentation Rate (FPS) |
|---|---|---|---|
| DeepLabv3+ | 90.48 | 92.47 | 14.9 |
| PSANet | 91.67 | 94.09 | 14.3 |
| Mask R-CNN | 91.97 | 95.06 | 15.3 |
| Swin-Unet | 92.03 | 95.27 | 15.0 |
| Model in this paper | 92.75 | 95.57 | 15.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Gong, J.; Zhang, Y.; Mostafa, K.; Yuan, G. Weed Identification in Maize Fields Based on Improved Swin-Unet. Agronomy 2023, 13, 1846. https://doi.org/10.3390/agronomy13071846
Zhang J, Gong J, Zhang Y, Mostafa K, Yuan G. Weed Identification in Maize Fields Based on Improved Swin-Unet. Agronomy. 2023; 13(7):1846. https://doi.org/10.3390/agronomy13071846
Chicago/Turabian StyleZhang, Jiaheng, Jinliang Gong, Yanfei Zhang, Kazi Mostafa, and Guangyao Yuan. 2023. "Weed Identification in Maize Fields Based on Improved Swin-Unet" Agronomy 13, no. 7: 1846. https://doi.org/10.3390/agronomy13071846
APA StyleZhang, J., Gong, J., Zhang, Y., Mostafa, K., & Yuan, G. (2023). Weed Identification in Maize Fields Based on Improved Swin-Unet. Agronomy, 13(7), 1846. https://doi.org/10.3390/agronomy13071846






