Development of Expressed Sequence Tag–Simple Sequence Repeat Markers Related to the Salt-Stress Response of Kenaf (Hibiscus cannabinus)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. RNA Extraction, Library Preparation, and Sequencing
2.3. Transcriptome-Based SSR and SNP Variation Analysis
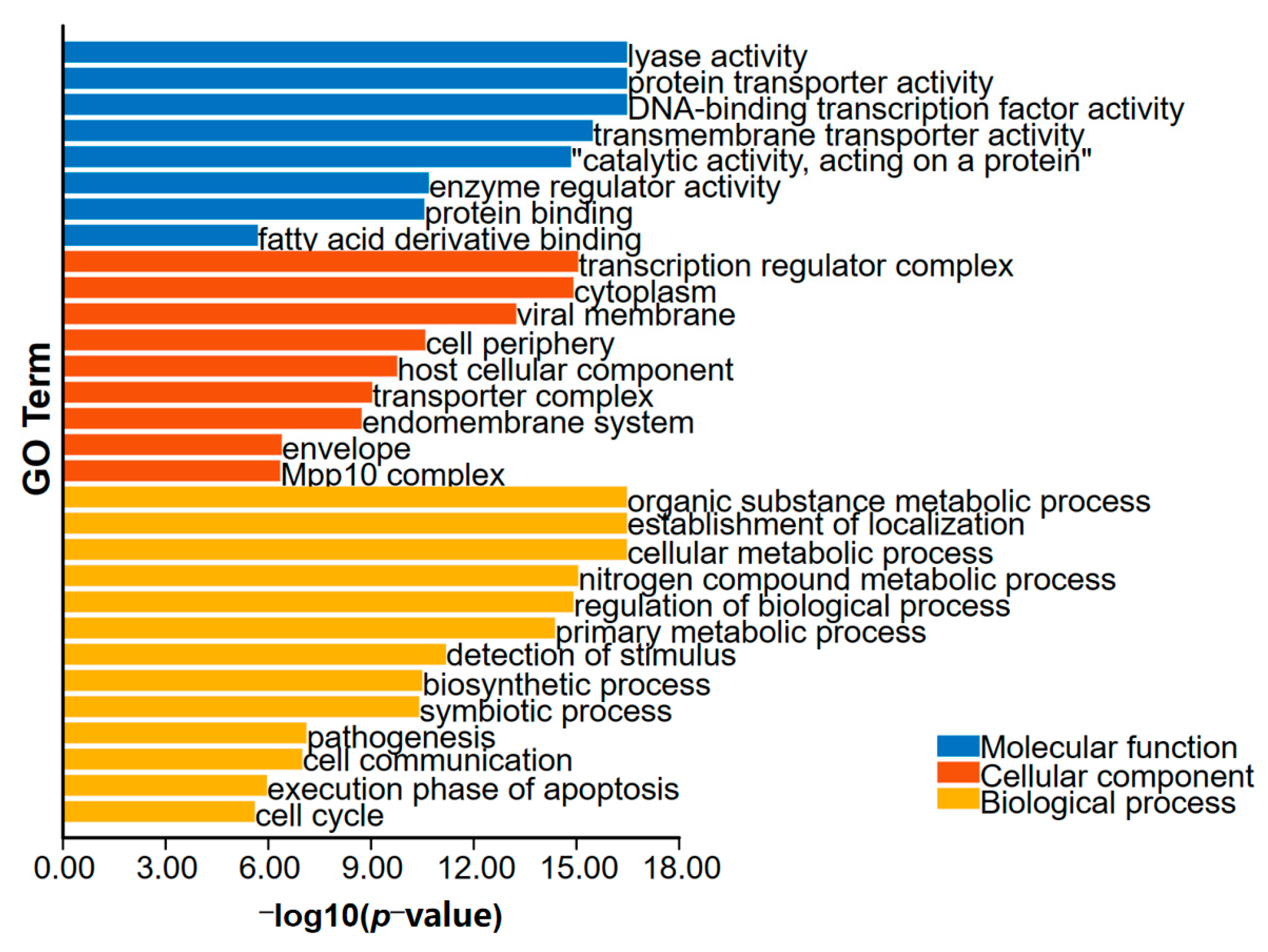
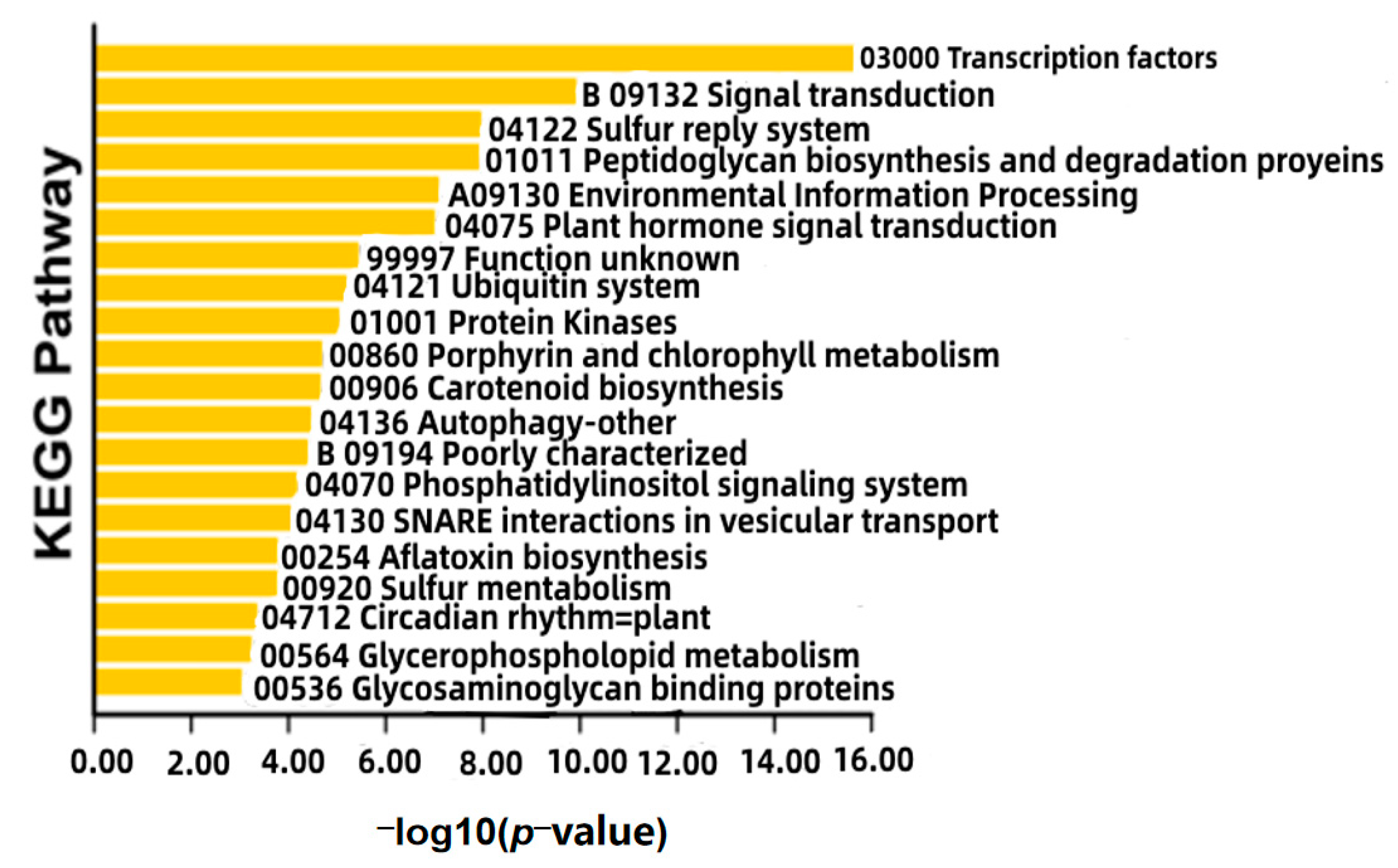
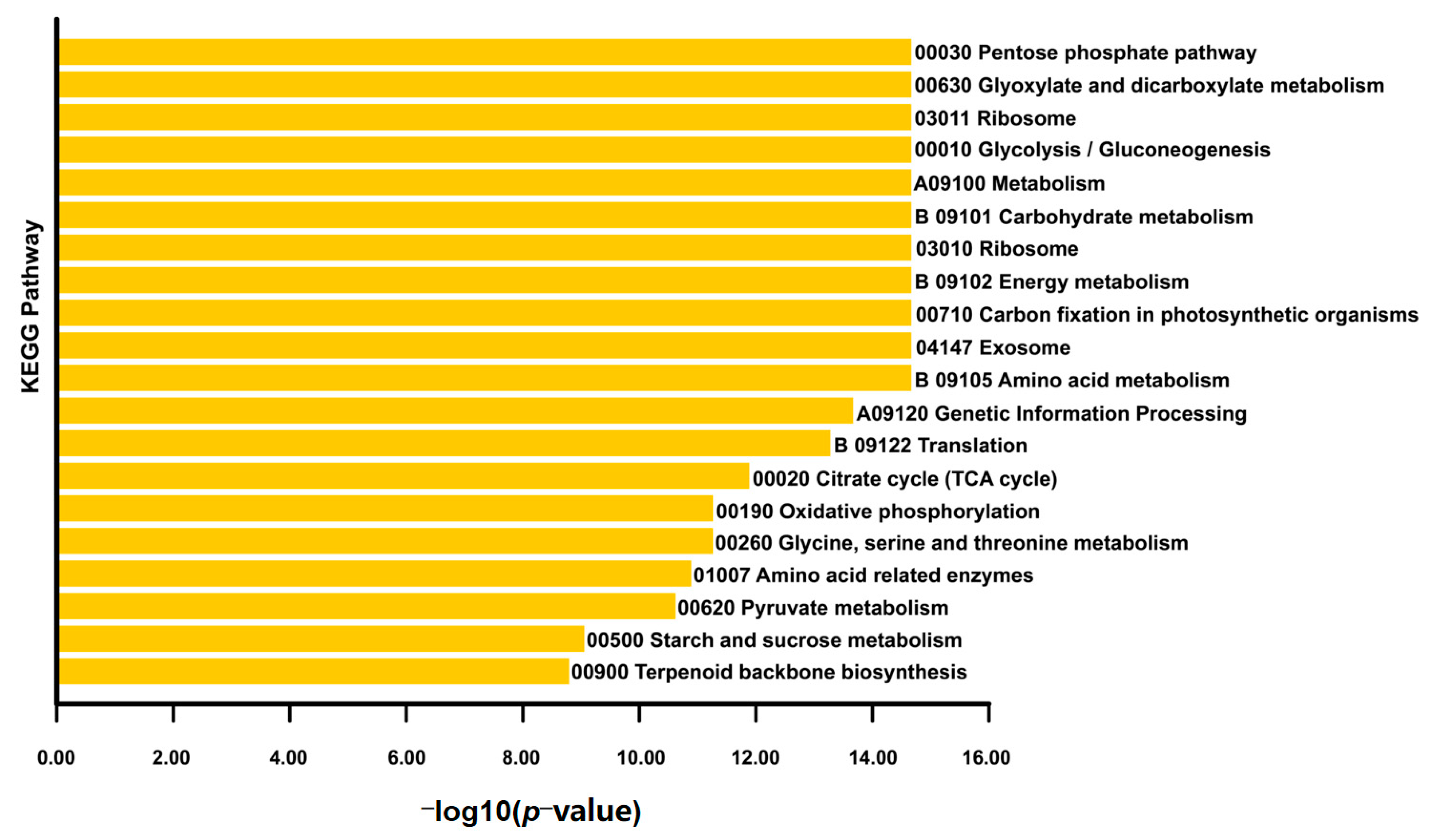
2.4. GO and KEGG Enrichment Analyses
2.5. Analysis of the Interaction Network for the Proteins Encoded by Differentially Expressed Genes
2.6. Genomic DNA Extraction
2.7. SSR Genotyping
2.8. Phylogenetic Analysis
2.9. Genetic Diversity Analysis
3. Results
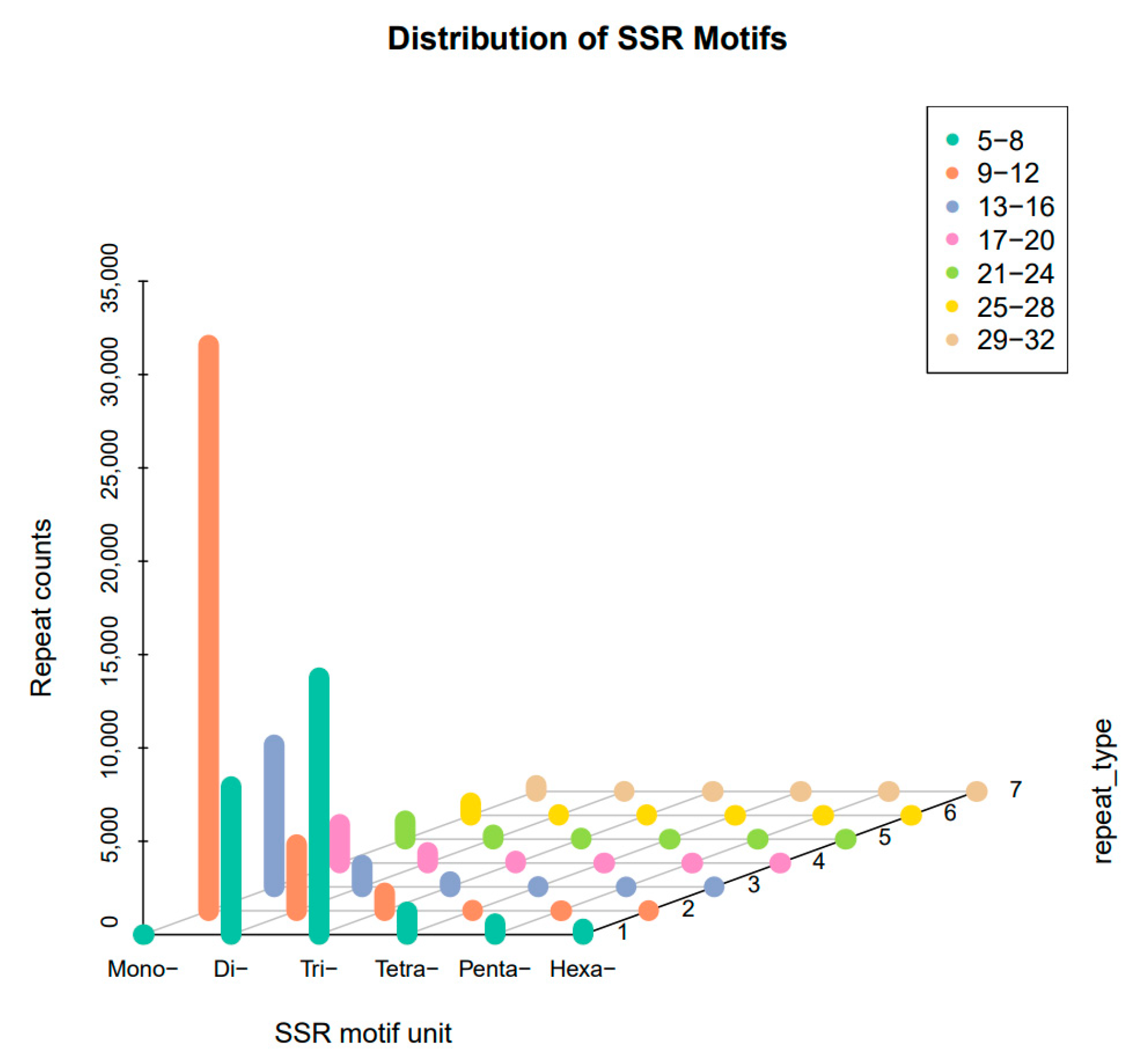
3.1. Analysis of Genomic SSR and SNP Characteristics
3.2. Interaction Network of Proteins Encoded by DEGs
3.3. EST-SSR Molecular Marker Verification
3.4. Genetic Diversity of Individual Loci
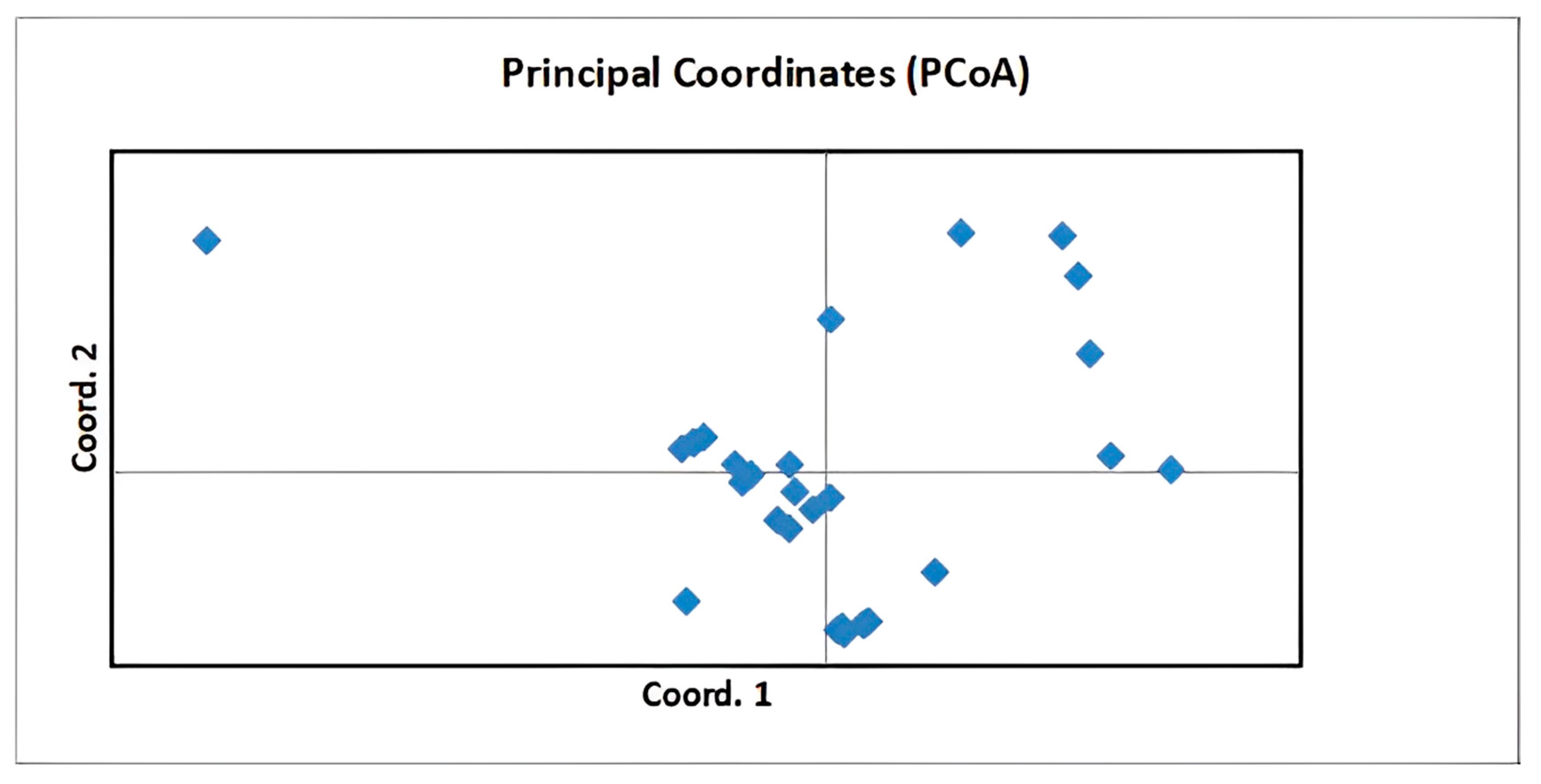
3.5. Genetic Structure Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ziska, L.H.; Bunce, J.A.; Shimono, H.; Gealy, D.R.; Baker, J.T.; Newton, P.C.; Reynolds, M.P.; Jagadish, K.S.; Zhu, C.; Howden, M.; et al. Food security and climate change: On the potential to adapt global crop production by active selection to rising atmospheric carbon dioxide. Proc. Biol. Sci. 2012, 279, 4097–4105. [Google Scholar] [CrossRef]
- Ludwig, M.; Wilmes, P.; Schrader, S. Measuring soil sustainability via soil resilience. Sci. Total Environ. 2018, 626, 1484–1493. [Google Scholar] [CrossRef] [PubMed]
- Savary, S.; Akter, S.; Almekinders, C.; Harris, J.; Korsten, L.; Rotter, R.; Waddington, S.; Watson, D. Mapping disruption and resilience mechanisms in food systems. Food Secur. 2020, 12, 695–717. [Google Scholar] [CrossRef]
- Chen, P.; Li, Z.; Luo, D.; Jia, R.; Lu, H.; Tang, M.; Hu, Y.; Yue, J.; Huang, Z. Comparative transcriptomic analysis reveals key genes and pathways in two different cadmium tolerance kenaf (Hibiscus cannabinus L.) cultivars. Chemosphere 2021, 263, 128211. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, A.; Wang, X.; Xu, J.; Zhang, G.; Su, J.; Qi, J.; Guan, C. Genetic diversity of kenaf (Hibiscus cannabinus) evaluated by inter-simple sequence repeat (ISSR). Biochem. Genet. 2013, 51, 800–810. [Google Scholar] [CrossRef]
- An, X.; Chen, J.; Jin, G.R. Transcriptome profiling of kenaf (Hibiscus cannabinus L.) under plumbic stress conditions implies the involvement of NAC transcription factors regulating reactive oxygen species-dependent programmed cell death. PeerJ 2020, 8, e8733. [Google Scholar] [CrossRef]
- An, X.; Chen, J.; Liu, T.; Li, W.; Luo, X.; Zou, L. Transcriptomic and Metabolic Profiling of Kenaf Stems under Salinity Stress. Plants 2022, 11, 1448. [Google Scholar] [CrossRef]
- An, X.; Luo, X.; Li, W.; Liu, T.; Zou, L. Development and Application of EST-SSR Markers Related to Lead Stress Responses in Kenaf Based on Transcriptome Sequencing Data. Sustainability 2023, 15, 1514. [Google Scholar] [CrossRef]
- An, X.; Jin, G.; Zhang, J.; Ma, G.; Jin, L.; Luo, X.; Chen, C.; Shi, X.; Zhou, J.; Wei, W.; et al. Research Progress on Tissue Culture and Genetic Transformation of Kenaf (Hibiscus cannabinus). Open Life Sci. 2017, 12, 465–472. [Google Scholar] [CrossRef]
- Kim, W.J.; Kim, D.S.; Kim, S.H.; Kim, J.B.; Goh, E.J.; Kang, S.Y. Analysis of genetic similarity detected by AFLP and PCoA among genotypes of kenaf (Hibiscus cannabinus L.). J. Crop Sci. Biotechnol. 2010, 13, 243–249. [Google Scholar] [CrossRef]
- Al-Mamun, M.; Rafii, M.Y.; Misran, A.b.; Berahim, Z.; Ahmad, Z.; Khan, M.M.H.; Oladosu, Y. Estimating Genetic Analysis Using Half Diallel Cross Underlying Kenaf (Hibiscus cannabinus L.) Fibre Yield in Tropical Climates. BioMed Res. Int. 2022, 2022, 1532987. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.K.; Varshney, R.K. The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphyica 2000, 113, 163–185. [Google Scholar] [CrossRef]
- Reddy, M.P.; Sarla, N.; Siddiq, E.A. Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica 2002, 128, 9–17. [Google Scholar] [CrossRef]
- Wen, M.; Wang, H.; Xia, Z.; Zou, M.; Lu, C.; Wang, W. Developmenrt of EST-SSR and genomic-SSR markers to assess genetic diversity in Jatropha Curcas L. BMC Res. Notes 2010, 3, 42. [Google Scholar] [CrossRef]
- Cheng, Z.; Lu, B.R.; Baldwin, B.S.; Sameshima, K.; Chen, J.K. Comparative studies of genetic diversity in kenaf (Hibiscus cannabinus L.) varieties based on analysis of agronomic and RAPD data. Hereditas 2002, 136, 231–239. [Google Scholar] [CrossRef]
- Cheng, Z.; Lu, B.R.; Sameshima, K.; Fu, D.X.; Chen, J.K. Identification and genetic relationships of kenaf (Hibiscus cannabinus L.) germplasm revealed by AFLP analysis. Genet. Resour. Crop Evol. 2004, 51, 393–401. [Google Scholar] [CrossRef]
- Satya, P.; Karan, M.; Chakraborty, K.; Biswas, C.; Karmakar, P.G. Comparative analysis of diversification and population structure of kenaf (Hibiscus cannabinus L.) and roselle (H. sabdariffa L.) using SSR and RGA (resistance gene analogue) markers. Plant Syst. Evol. 2013, 300, 1209–1218. [Google Scholar] [CrossRef]
- Li, H.; Li, D.; Chen, A.; Tang, H.; Li, J.; Huang, S. Characterization of the Kenaf (Hibiscus cannabinus) Global Transcriptome Using Illumina Paired-End Sequencing and Development of EST-SSR Markers. PLoS ONE 2016, 11, e0150548. [Google Scholar] [CrossRef]
- Kim, J.M.; Lyu, J.I.; Lee, M.-K.; Kim, D.-G.; Kim, J.-B.; Ha, B.-K.; Ahn, J.-W.; Kwon, S.-J. Cross-species transferability of EST-SSR markers derived from the transcriptome of kenaf (Hibiscus cannabinus L.) and their application to genus Hibiscus. Genet. Resour. Crop Evol. 2019, 66, 1543–1556. [Google Scholar] [CrossRef]
- Mehmood, F.; Shahzadi, I.; Waseem, S.; Mirza, B.; Ahmed, I.; Waheed, M.T. Chloroplast genome of Hibiscus rosa-sinensis (Malvaceae): Comparative analyses and identification of mutational hotspots. Genomics 2020, 1121, 581–591. [Google Scholar] [CrossRef]
- Zhang, L.M.; Wan, X.B.; Zhang, L.L.; XU, Y.; Lin, L.H.; Qi, J.M.; Zhang, L.W. Development of InDel Markers for Identification of a Single Mendelian Locus Controlling Leaf Shape in Kenaf (Hibiscus cannabinus). Trop. Plant Biol. 2019, 12, 78–84. [Google Scholar] [CrossRef]
- Wan, X.B.; Xu, J.T.; Li, D.X.; Lin, L.H.; Qi, J.M.; Zhang, L.W. Construction of DNA fingerprinting in Kenaf (Hibiscus cannabinus) using microsatellite markers. J. Plant Genet. Res. 2018, 19, 87–95. [Google Scholar] [CrossRef]
- Yuan, C.Y.; Zhang, C.; Wang, P.; Hu, S.; Chang, H.P.; Xiao, W.J.; Lu, X.T.; Jiang, S.B.; Ye, J.Z.; Guo, X.H. Genetic diversity analysis of okra (Abelmoschus esculentus L.) by inter-simple sequence repeat (ISSR) markers. Genet Mol. Res. 2014, 13, 3165–3175. [Google Scholar] [CrossRef]
- Wang, X.M.; Hou, X.Q.; Zhang, Y.Q.; Yang, R.; Feng, S.F.; Li, Y.; Ren, Y. Genetic diversity of the endemic and medicinally important plant Rheum officinale as revealed by Inter-Simpe Sequence Repeat (ISSR) Markers. Int. J. Mol. Sci. 2012, 13, 3900–3915. [Google Scholar] [CrossRef]
- Wan, X.B.; Li, D.X.; Xu, Y.; Xu, J.T.; Zhang, L.L.; Zhang, L.M.; Lin, L.H.; Qi, J.M.; Zhang, L.W. Development and polymorphism evaluation of EST-SSR markers in kenaf. Acta Agron. Sin. 2017, 43, 1170–1180. [Google Scholar] [CrossRef]
- Jeong, S.W.; Kwon, S.-J.; Ryu, J.; Kim, J.-B.; Ahn, J.-W.; Kim, S.H.; Jo, Y.D.; Choi, H.-I.; Im, S.B.; Kang, S.-Y. Development of EST-SSR markers through de novo RNA sequencing and application for biomass productivity in kenaf (Hibiscus cannabinus L.). Genes Genom. 2017, 39, 1139–1156. [Google Scholar] [CrossRef]
- Jin, G.R.; An, X.; Luo, X.H.; Chen, C.L.; Li, W.L.; Li, P.F.; Tian, D.Q.; Wang, B.; Wu, W.Q.; Zhu, G.L. The EST-SSR markers of the response gene of kenaf under drought stress. Mol Plant. Breeding 2018, 16, 4735–4742. [Google Scholar] [CrossRef]
- Yang, X.; Gao, K.; Chen, Z.; Yang, X.Y.; Rao, P.; Zhao, T.Y.; An, X.M. Development and Application of EST-SSR Markers in Koelreuteria paniculata Laxm. Using a Transcriptomic Approach. Biotechnology 2017, 16, 45–56. [Google Scholar] [CrossRef]
- Kim, J.M.; Lyu, J.I.; Kim, D.G.; Hung, N.N.; Ryu, J.; Kim, J.B.; Ahn, J.W.; Ha, B.K.; Kwon, S.J. Analysis of genetic diversity and relationships of Perilla frutescens using novel EST-SSR markers derived from transcriptome between wild-type and mutant Perilla. Mol. Biol. Rep. 2021, 48, 6387–6400. [Google Scholar] [CrossRef]
- Jia, Y.; Bai, J.Q.; Liu, M.L.; Jiang, Z.F.; Wu, Y.; Fang, M.F.; Li, Z.H. Transcriptome analysis ofthe endangered Notoptery giumincisum: Coldtolerance gene discovery and identification ofEST-SSR and SNPmarkers. Plant Divers. 2019, 41, 1–6. [Google Scholar] [CrossRef]
- Yildiz, M.; Kocak, M.; Baloch, F.S. Genetic bottlenecks in Turkish okra germplasm and utility of iPBS retrotransposon markers for genetic diversity assessment. Genet Mol. Res. 2015, 14, 10588–10602. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Takezaki, N. Estimation of genetic distances and phylogenetic trees from DNA analysis. In Proceedings of the 5th World Congress on Genetics Applied to Livestock Production, Guelph, ON, Canada, 7–12 August 1994; Volume 21, pp. 405–412. [Google Scholar]
- Yeh, F.C. Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg. J. Bot. 1997, 129, 157. [Google Scholar]
- Smouse, P.E.; Banks, S.C.; Peakall, R. Converting quadratic entropy to diversity: Both animals and alleles are diverse, but some are more diverse than others. PLoS ONE 2017, 12, e0185499. [Google Scholar] [CrossRef] [PubMed]
- Schafleitner, R.; Kumar, S.; Lin, C.Y.; Hegde, S.G.; Ebert, A. The okra (Abelmoschus esculentus) transcriptome as a source for gene sequence information and molecular markers for diversity analysis. Gene 2013, 517, 27–36. [Google Scholar] [CrossRef]
- Li, H.P.; Yao, Y.F.; Lian, D.M.; Lai, Z.F.; Hong, J.J. Transcriptome Sequencing and Analysis of Okra Fruit. Biotechnol. Bull. 2018, 34, 121–127. [Google Scholar] [CrossRef]
- Zhang, L.; Wan, X.; Xu, J.; Lin, L.; Qi, J. De novo assembly of kenaf (Hibiscus cannabinus) transcriptome using Illumina sequencing for gene discovery and marker identification. Mol. Breed. 2015, 35, 192. [Google Scholar] [CrossRef]
- Li, H.; Li, D.; Chen, A.; Tang, H.; Li, J.; Huang, S. RNA-seq for comparative transcript profiling of kenaf under salinity stress. J. Plant Res. 2017, 130, 365–372. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]


| Number | Variety Name | Origin |
|---|---|---|
| S31 | 85–245 | Zimbabwe |
| S32 | C2032 | Cuba |
| S33 | Burmese Kenaf | Myanmar |
| S34 | j-1-113 | USA |
| S35 | KG2006-014 | China |
| S36 | Whitten | USA |
| S37 | MSI-80 | USA |
| S38 | CPI-F8891 | China |
| S39 | PI-270116 | USA |
| S40 | FJ/026H | France |
| S41 | FJ/01FH | France |
| S42 | DY/069H | China |
| S43 | NY/061H | Nigeria |
| S44 | BL/012H | USA |
| S45 | DS/012H | Guatemala |
| S46 | SM/025H | El Salvador |
| S48 | 78-18RS10 | USA |
| S50 | GR2563 | USA |
| S52 | Masterfiber | Africa |
| S53 | MSI104gr | USA |
| S54 | MSI135 | USA |
| S55 | MSI136 | USA |
| S56 | MSI139 | USA |
| S57 | MSI77 | USA |
| S58 | MSI78 | USA |
| S59 | MSI180 | USA |
| S60 | Soudan Pre | Sudan |
| S61 | Indian Selection’98 | India |
| S62 | Zhe 1’96 | China |
| S64 | Zhejiang No. 2’96 | China |
| Type | No. |
|---|---|
| Total number of sequences examined | 175,216 |
| Total size of examined sequences (bp) | 268,192,545 |
| Total number of identified SSRs | 73,728 |
| Number of SSRs containing sequences | 53,444 |
| Number of sequences containing more than 1 SSR | 14,775 |
| Number of SSRs present in compound formation | 5717 |
| Type | Count | Frequency Per kb |
|---|---|---|
| Transition | ||
| C/T | 3367 | 0.01 |
| A/G | 3461 | 0.01 |
| Transversion | ||
| A/T | 1212 | 0 |
| A/C | 868 | 0 |
| T/G | 877 | 0 |
| C/G | 698 | 0 |
| Total | 10,483 | 0.04 |
| SNP Position in Codon | ||
| First | 2107 | |
| Second | 780 | |
| Third | 1931 |
| Primers | ID | SSR Type | SSR | Size | Forward Primer1 (5′-3′) | Tm (°C) | Size | Reverse Primer1 (5′-3′) | Tm (°C) | Size | Product Size (bp) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| KSSR59 | Cluster-12086.5227 | p3 | (CCA)5 | 15 | AAGCCGAAAAAGCCTCACCT | 60.179 | 20 | AGCTGGTGTTTCTTGGCTGT | 60.107 | 20 | 132 |
| KSSR74 | Cluster-12086.35062 | p3 | (TTG)5 | 15 | TGCCGCTGCTTTCTCCAATA | 60.036 | 20 | GCTTCATGCTTGTTTTGTGGA | 57.904 | 21 | 217 |
| KSSR91 | Cluster-12086.23640 | p3 | (TCT)5 | 15 | GACAGCAAGGTGATCCTCCC | 60.107 | 20 | ACGATGAAGACGACGAACCC | 60.109 | 20 | 230 |
| KSSR102 | Cluster-12086.10800 | p2 | (AT)8 | 16 | ACACTTTGACAACCGGAGCA | 60.107 | 20 | TGGAGAAACAGATTGACTTGGGA | 59.606 | 23 | 244 |
| KSSR118 | Cluster-12086.3729 | p2 | (CT)7 | 14 | GTCGGAAGTGGTGAATGGCT | 60.322 | 20 | ATAGGGAGGCTGATGGTGGT | 60.03 | 20 | 175 |
| KSSR70 | Cluster-12086.36583 | p3 | (TCT)5 | 15 | ACCTGATTGCCTCACTGCTC | 60.036 | 20 | CATCTTCAACGGCTGCCATG | 59.9 | 20 | 217 |
| KSSR79 | Cluster-12086.30425 | p3 | (GAG)5 | 15 | AAACCAGCAGACCTTTCAGT | 57.263 | 20 | GTTGGCAGAGTGAAGGGTGA | 59.891 | 20 | 229 |
| KSSR95 | Cluster-12086.9979 | p2 | (CT)6 | 12 | ACGTGAGTTCCATCAGCCAA | 59.604 | 20 | AGCGTGCACTTAAACGGGTA | 59.966 | 20 | 228 |
| KSSR111 | Cluster-12086.3242 | p4 | (ATAC)5 | 20 | AACTGGTGGTGCTCTGATGG | 59.963 | 20 | CCAACAACTATGCACTGGACG | 59.535 | 21 | 246 |
| Locus | N | Na | Ne | Genotype No | Major Allele Frquency | I | Ho | He | PIC | F |
|---|---|---|---|---|---|---|---|---|---|---|
| KSSR59 | 30 | 2.000 | 1.105 | 3 | 0.95 | 0.199 | 0.033 | 0.095 | 0.0904875 | 0.649 |
| KSSR74 | 29 | 2.000 | 1.991 | 2 | 0.534482759 | 0.691 | 0.931 | 0.498 | 0.373808112 | −0.871 |
| KSSR91 | 29 | 4.000 | 1.280 | 5 | 0.879310345 | 0.464 | 0.103 | 0.219 | 0.206366706 | 0.527 |
| KSSR102 | 20 | 6.000 | 3.175 | 8 | 0.5 | 1.417 | 0.350 | 0.685 | 0.649782813 | 0.489 |
| KSSR118 | 30 | 2.000 | 1.142 | 2 | 0.933333333 | 0.245 | 0.000 | 0.124 | 0.116701235 | 1.000 |
| KSSR70 | 30 | 3.000 | 1.106 | 3 | 0.95 | 0.230 | 0.067 | 0.096 | 0.093603549 | 0.306 |
| KSSR79 | 29 | 5.000 | 2.069 | 7 | 0.655172414 | 0.978 | 0.448 | 0.517 | 0.469219978 | 0.132 |
| KSSR95 | 24 | 3.000 | 1.135 | 3 | 0.9375 | 0.274 | 0.042 | 0.119 | 0.115107407 | 0.650 |
| KSSR111 | 28 | 4.000 | 1.115 | 4 | 0.946428571 | 0.268 | 0.107 | 0.103 | 0.101601953 | |
| Mean | 27.66666667 | 3.444 | 1.569 | 4.111111111 | 0.809580825 | 0.529 | 0.231 | 0.273 | 0.246297695 |
| S128 | S100 | S101 | S102 | S103 | S104 | S105 | S106 | S107 | S108 | S109 | S110 | S111 | S112 | S113 | S114 | S115 | S116 | S117 | S118 | S119 | S120 | S121 | S122 | S123 | S124 | S125 | S126 | S127 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S100 | ||||||||||||||||||||||||||||
| S101 | 0.5714 | |||||||||||||||||||||||||||
| S102 | 0.5385 | 0.4667 | ||||||||||||||||||||||||||
| S103 | 0.6154 | 0.6667 | 0.5833 | |||||||||||||||||||||||||
| S104 | 0.9 | 0.5 | 0.4615 | 0.6364 | ||||||||||||||||||||||||
| S105 | 0.3 | 0.1818 | 0.3333 | 0.375 | 0.375 | |||||||||||||||||||||||
| S106 | 0.6364 | 0.5833 | 0.5 | 0.7273 | 0.5455 | 0.3333 | ||||||||||||||||||||||
| S107 | 0.7273 | 0.6364 | 0.7 | 0.8182 | 0.7778 | 0.375 | 0.7 | |||||||||||||||||||||
| S108 | 0.6923 | 0.5714 | 0.5385 | 0.8182 | 0.7273 | 0.4 | 0.7 | 1 | ||||||||||||||||||||
| S109 | 0.6667 | 0.4667 | 0.6667 | 0.8 | 0.5833 | 0.2 | 0.7 | 1 | 0.6667 | |||||||||||||||||||
| S110 | 0.5714 | 0.4667 | 0.6667 | 0.8182 | 0.5833 | 0.2727 | 0.7 | 1 | 0.6923 | 1 | ||||||||||||||||||
| S111 | 0.6923 | 0.4667 | 0.5385 | 0.75 | 0.7273 | 0.3 | 0.6364 | 0.9 | 0.6923 | 0.8182 | 0.8333 | |||||||||||||||||
| S112 | 0.5714 | 0.5714 | 0.6667 | 0.9091 | 0.5833 | 0.3 | 0.8 | 0.9 | 0.6923 | 0.8182 | 0.8333 | 0.6923 | ||||||||||||||||
| S113 | 0.6667 | 0.5833 | 0.6364 | 0.75 | 0.7 | 0.375 | 0.6364 | 0.9 | 0.9 | 0.8889 | 0.9 | 0.8182 | 0.8182 | |||||||||||||||
| S114 | 0.5 | 0.5833 | 0.5 | 0.6923 | 0.7 | 0.3333 | 0.6364 | 0.6667 | 0.6667 | 0.7 | 0.6667 | 0.6154 | 0.75 | 0.6154 | ||||||||||||||
| S115 | 0.6154 | 0.5 | 0.5833 | 0.8182 | 0.6364 | 0.3 | 0.7 | 1 | 0.75 | 0.7273 | 0.75 | 0.75 | 0.75 | 0.9 | 0.6667 | |||||||||||||
| S116 | 0.75 | 0.6154 | 0.5833 | 0.8182 | 0.8 | 0.4444 | 0.7 | 1 | 0.9091 | 0.7273 | 0.75 | 0.75 | 0.75 | 0.9 | 0.6667 | 0.8182 | ||||||||||||
| S117 | 0.75 | 0.6154 | 0.5833 | 0.8182 | 0.8 | 0.4444 | 0.7 | 1 | 0.9091 | 0.7273 | 0.75 | 0.75 | 0.75 | 0.9 | 0.6667 | 0.8182 | 1 | |||||||||||
| S118 | 0.6667 | 0.7273 | 0.6364 | 0.9091 | 0.7 | 0.375 | 0.8 | 0.9 | 0.9 | 0.8889 | 0.9 | 0.8182 | 1 | 0.8182 | 0.75 | 0.9 | 0.9 | 0.9 | ||||||||||
| S119 | 0.7273 | 0.6364 | 0.7 | 0.8182 | 0.7778 | 0.375 | 0.7 | 1 | 1 | 1 | 1 | 0.9 | 0.9 | 0.9 | 0.6667 | 1 | 1 | 1 | 0.9 | |||||||||
| S120 | 0.75 | 0.6154 | 0.5833 | 0.8182 | 0.8 | 0.4444 | 0.7 | 1 | 0.9091 | 0.7273 | 0.75 | 0.75 | 0.75 | 0.9 | 0.6667 | 0.8182 | 1 | 1 | 0.9 | 1 | ||||||||
| S121 | 0.6429 | 0.7692 | 0.5 | 0.9091 | 0.6667 | 0.4 | 0.8 | 0.9 | 0.7692 | 0.6154 | 0.6429 | 0.6429 | 0.7692 | 0.8182 | 0.75 | 0.6923 | 0.8333 | 0.8333 | 1 | 0.9 | 0.8333 | |||||||
| S122 | 0.6154 | 0.4 | 0.4615 | 0.6667 | 0.8 | 0.3 | 0.5455 | 0.8 | 0.6154 | 0.7273 | 0.75 | 0.9091 | 0.6154 | 0.7273 | 0.6667 | 0.6667 | 0.6667 | 0.6667 | 0.7273 | 0.8 | 0.6667 | 0.5714 | ||||||
| S123 | 0.6154 | 0.6154 | 0.4615 | 0.8182 | 0.8 | 0.4444 | 0.7 | 0.8 | 0.75 | 0.5833 | 0.6154 | 0.6154 | 0.75 | 0.7273 | 0.8182 | 0.6667 | 0.8182 | 0.8182 | 0.9 | 0.8 | 0.8182 | 0.8333 | 0.6667 | |||||
| S124 | 0.5385 | 0.5833 | 0.5 | 0.75 | 0.5455 | 0.375 | 0.6364 | 0.7273 | 0.7273 | 0.7 | 0.7273 | 0.6667 | 0.8182 | 0.6667 | 0.6154 | 0.7273 | 0.7273 | 0.7273 | 0.8182 | 0.7273 | 0.7273 | 0.8182 | 0.5833 | 0.7273 | ||||
| S125 | 0.6923 | 0.5714 | 0.5385 | 0.75 | 0.7273 | 0.4444 | 0.6364 | 0.9 | 0.8333 | 0.6667 | 0.6923 | 0.6923 | 0.6923 | 0.8182 | 0.6154 | 0.75 | 0.9091 | 0.9091 | 0.8182 | 0.9 | 0.9091 | 0.7692 | 0.6154 | 0.75 | 0.8182 | |||
| S126 | 0.4375 | 0.3529 | 0.5 | 0.6154 | 0.4286 | 0.2727 | 0.5 | 0.7273 | 0.5333 | 0.6154 | 0.6429 | 0.5333 | 0.6429 | 0.6667 | 0.5 | 0.6923 | 0.5714 | 0.5714 | 0.6667 | 0.7273 | 0.5714 | 0.5 | 0.4667 | 0.4667 | 0.8182 | 0.6429 | ||
| S127 | 0.5 | 0.4545 | 0.3636 | 0.6 | 0.4 | 0.2 | 0.6 | 0.5556 | 0.5556 | 0.5556 | 0.5556 | 0.5 | 0.6667 | 0.5 | 0.5 | 0.5556 | 0.5556 | 0.5556 | 0.6667 | 0.5556 | 0.5556 | 0.6667 | 0.4 | 0.5556 | 0.875 | 0.6667 | 0.6667 | |
| S128 | 0.6429 | 0.6429 | 0.5 | 0.75 | 0.6667 | 0.4 | 0.6364 | 0.9 | 0.7692 | 0.6154 | 0.6429 | 0.6429 | 0.6429 | 0.8182 | 0.6154 | 0.6923 | 0.8333 | 0.8333 | 0.8182 | 0.9 | 0.8333 | 0.8462 | 0.5714 | 0.6923 | 0.6667 | 0.7692 | 0.5 | 0.5 |
| S129 | 0.5833 | 0.6364 | 0.5455 | 0.9 | 0.6 | 0.5 | 0.7778 | 0.8 | 0.8 | 0.7778 | 0.8 | 0.7273 | 0.9 | 0.8 | 0.6667 | 0.8 | 0.8 | 0.8 | 0.9 | 0.8 | 0.8 | 0.9 | 0.6364 | 0.8 | 0.7273 | 0.7273 | 0.5833 | 0.5556 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
An, X.; Liu, Q.; Ying, J.; Wei, J.; Dong, G.; Luo, X.; Li, W.; Liu, T.; Zhou, H.; Zou, L.; et al. Development of Expressed Sequence Tag–Simple Sequence Repeat Markers Related to the Salt-Stress Response of Kenaf (Hibiscus cannabinus). Agronomy 2023, 13, 1946. https://doi.org/10.3390/agronomy13071946
An X, Liu Q, Ying J, Wei J, Dong G, Luo X, Li W, Liu T, Zhou H, Zou L, et al. Development of Expressed Sequence Tag–Simple Sequence Repeat Markers Related to the Salt-Stress Response of Kenaf (Hibiscus cannabinus). Agronomy. 2023; 13(7):1946. https://doi.org/10.3390/agronomy13071946
Chicago/Turabian StyleAn, Xia, Qin Liu, Jinyao Ying, Jiqian Wei, Guoyun Dong, Xiahong Luo, Wenlue Li, Tingting Liu, Huaping Zhou, Lina Zou, and et al. 2023. "Development of Expressed Sequence Tag–Simple Sequence Repeat Markers Related to the Salt-Stress Response of Kenaf (Hibiscus cannabinus)" Agronomy 13, no. 7: 1946. https://doi.org/10.3390/agronomy13071946
APA StyleAn, X., Liu, Q., Ying, J., Wei, J., Dong, G., Luo, X., Li, W., Liu, T., Zhou, H., Zou, L., & Chen, C. (2023). Development of Expressed Sequence Tag–Simple Sequence Repeat Markers Related to the Salt-Stress Response of Kenaf (Hibiscus cannabinus). Agronomy, 13(7), 1946. https://doi.org/10.3390/agronomy13071946






