Over-Expression of ZmIAA29, an AUX/IAA Transcription Factor, Improved Maize Flowering Time
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Transgenic Identification
2.2. Phenotypic Identification
2.3. Vector Construction, Subcellular Localization, Yeast Two-Hybrid, and BiLUC
2.4. Combined Analysis of RNA-seq and DAP-seq
3. Results
3.1. Identification of ZmIAA29 Overexpression in Maize
3.2. Phenotype Characterization of ZmIAA29 Overexpression Maize
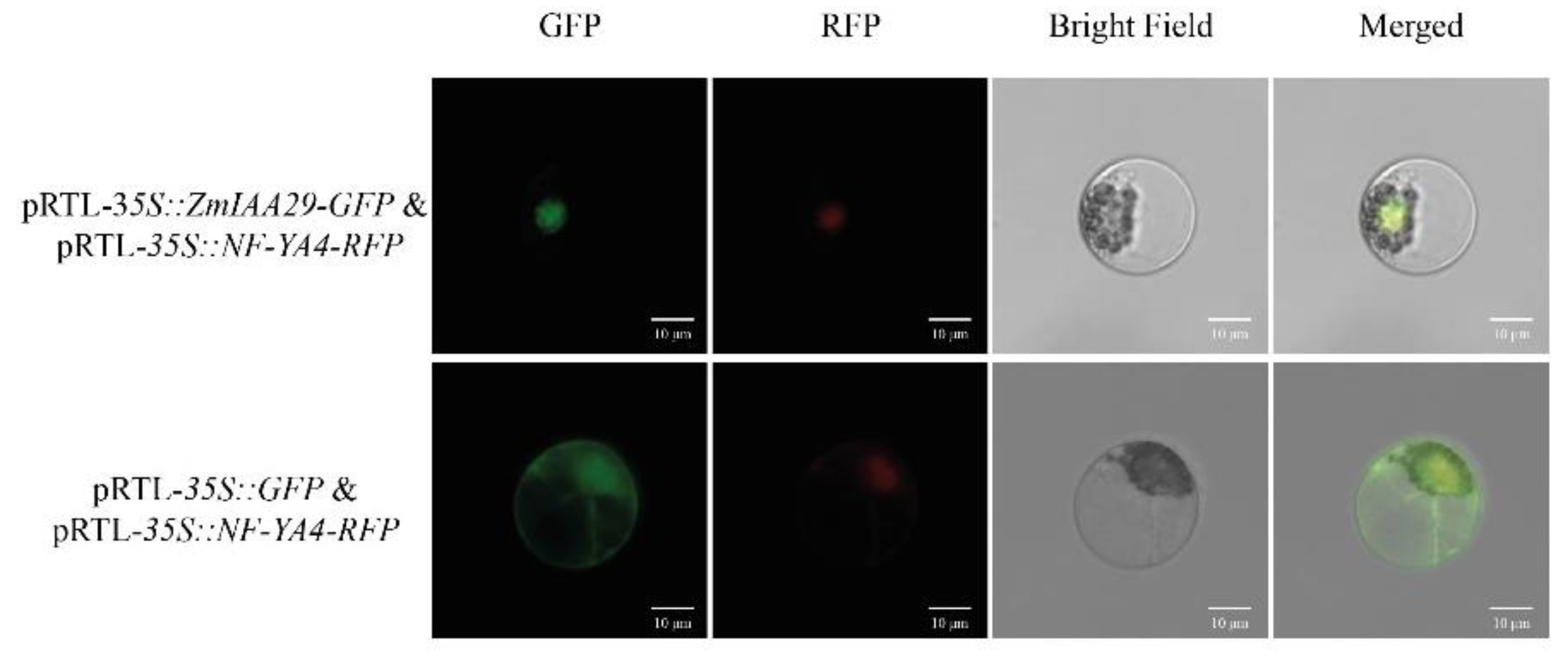
3.3. The Subcellular Localization of ZmIAA29
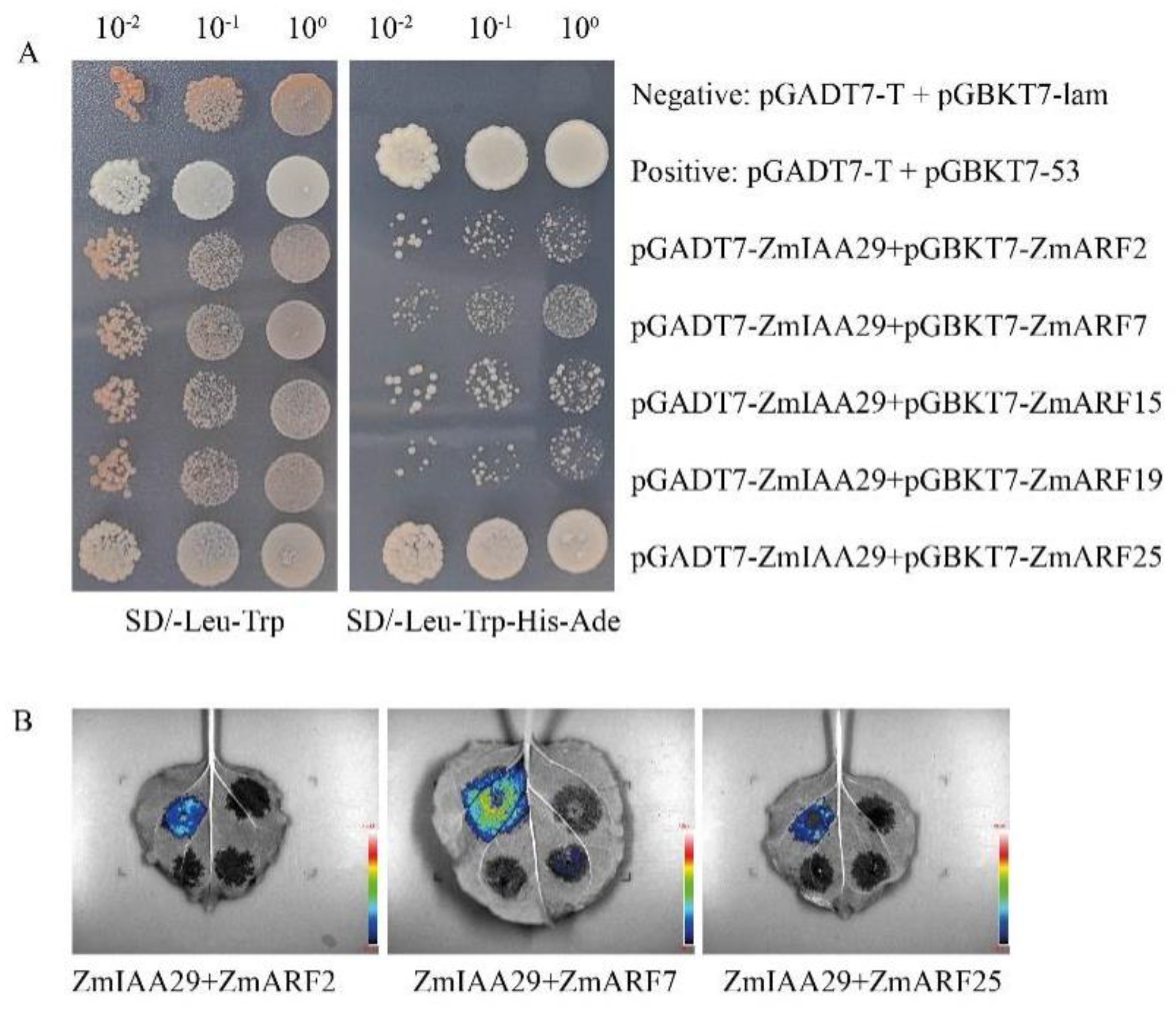
3.4. ZmIAA29 Interacts with ZmARF2, ZmARF7, and ZmARF25
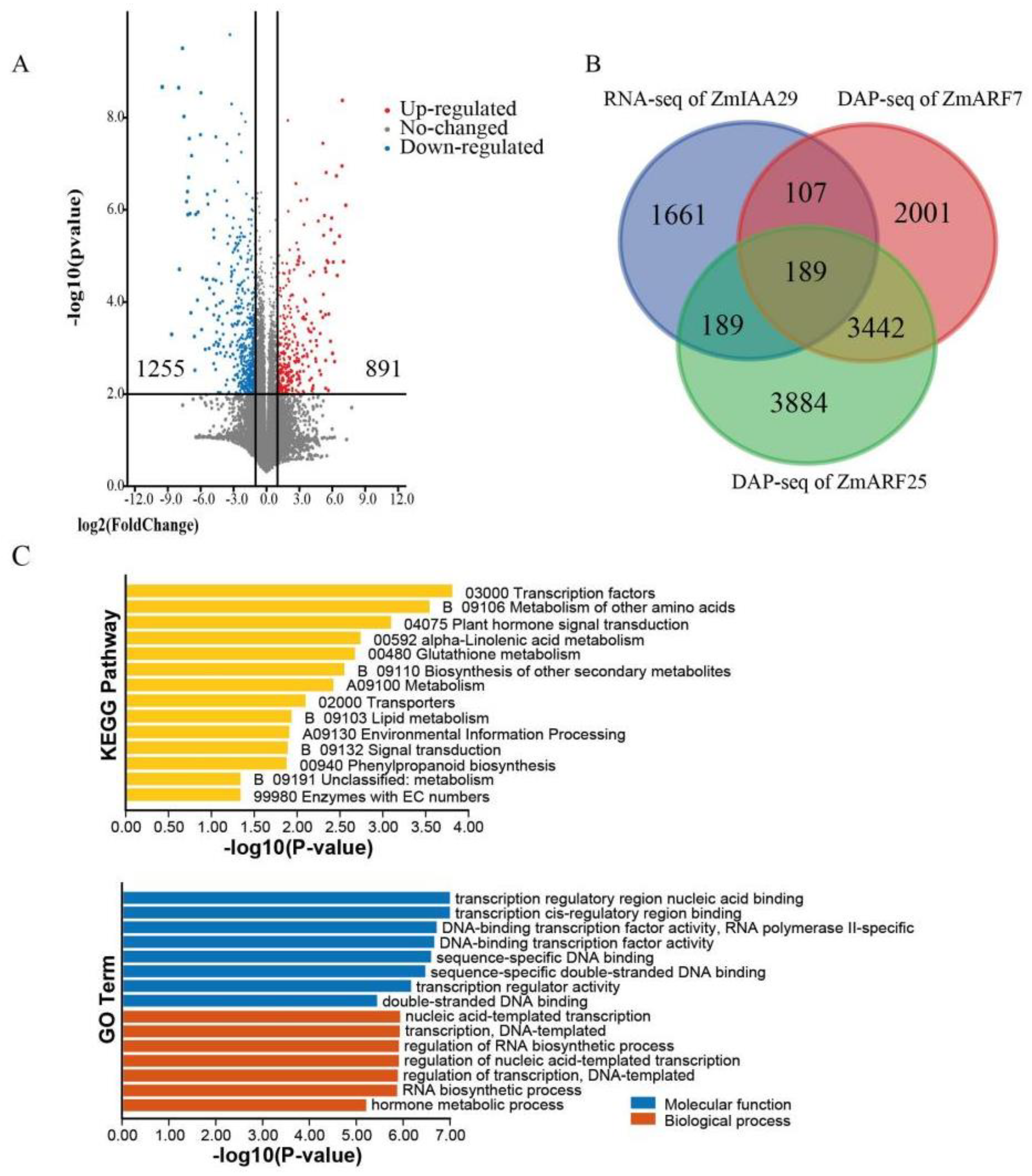
3.5. ZmIAA29 and ZmARFs Regulate Maize Growth through Plant Signal Transduction and Other Metabolic Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Godfray, H.C.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food security: The challenge of feeding 9 billion people. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ray, D.K.; Mueller, N.D.; West, P.C.; Foley, J.A. Yield Trends Are Insufficient to Double Global Crop Production by 2050. PLoS ONE 2013, 8, e66428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, H.; Zhang, X.; Chen, S.; Pei, D.; Liu, C. Effects of harvest and sowing time on the performance of the rotation of winter wheat–summer maize in the North China Plain. Ind. Crops Prod. 2007, 25, 239–247. [Google Scholar] [CrossRef]
- Wei, S.B.; Li, X.; Lu, Z.F.; Zhang, H.; Ye, X.Y.; Zhou, Y.J.; Li, J.; Yan, Y.Y.; Pei, H.C.; Duan, F.Y.; et al. A transcriptional regulator that boosts grain yields and shortens the growth duration of rice. Science 2022, 377, eabi8455. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Carrasco, V.P.; Hernandez-Garcia, J.; Mutte, S.K.; Weijers, D. The birth of a giant: Evolutionary insights into the origin of auxin responses in plants. EMBO J. 2023, 42, e113018. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Gallei, M.; Friml, J. Bending to auxin: Fast acid growth for tropisms. Trends Plant Sci. 2022, 27, 440–449. [Google Scholar] [CrossRef] [PubMed]
- Cancé, C.; Martin-Arevalillo, R.; Boubekeur, K.; Dumas, R. Auxin response factors are keys to the many auxin doors. New Phytol. 2022, 235, 402–419. [Google Scholar] [CrossRef]
- Figueiredo, M.R.A.; Strader, L.C. Intrinsic and extrinsic regulators of Aux/IAA protein degradation dynamics. Trends Biochem. Sci. 2022, 47, 865–874. [Google Scholar] [CrossRef]
- Galli, M.; Khakhar, A.; Lu, Z.; Chen, Z.; Sen, S.; Joshi, T.; Nemhauser, J.L.; Schmitz, R.J.; Gallavotti, A. The DNA binding landscape of the maize AUXIN RESPONSE FACTOR family. Nat. Commun. 2018, 9, 4526. [Google Scholar] [CrossRef] [Green Version]
- Luo, S.; Li, Q.; Liu, S.; Pinas, N.M.; Tian, H.; Wang, S. Constitutive Expression of OsIAA9 Affects Starch Granules Accumulation and Root Gravitropic Response in Arabidopsis. Front. Plant Sci. 2015, 6, 1156. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Peng, Z.; Liu, S.; He, Y.; Cheng, L.; Kong, F.; Wang, J.; Lu, G. Genome-wide analysis of Aux/IAA gene family in Solanaceae species using tomato as a model. Mol. Genet. Genet. Genet. Genom. MGG 2012, 287, 295–311. [Google Scholar] [CrossRef]
- Tiwari, S.B.; Hagen, G.; Guilfoyle, T.J. Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 2004, 16, 533–543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piya, S.; Shrestha, S.K.; Binder, B.; Stewart, C.N., Jr.; Hewezi, T. Protein-protein interaction and gene co-expression maps of ARFs and Aux/IAAs in Arabidopsis. Front. Plant Sci. 2014, 5, 744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woodward, A.W.; Bartel, B. A receptor for auxin. Plant Cell 2005, 17, 2425–2429. [Google Scholar] [CrossRef] [Green Version]
- Okushima, Y.; Overvoorde, P.J.; Arima, K.; Alonso, J.M.; Chan, A.; Chang, C.; Ecker, J.R.; Hughes, B.; Lui, A.; Nguyen, D.; et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 2005, 17, 444–463. [Google Scholar] [CrossRef] [Green Version]
- Hardtke, C.S.; Berleth, T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998, 17, 1405–1411. [Google Scholar] [CrossRef]
- Wang, D.; Pei, K.; Fu, Y.; Sun, Z.; Li, S.; Liu, H.; Tang, K.; Han, B.; Tao, Y. Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 2007, 394, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Pudake, R.N.; Guo, G.; Xing, G.; Hu, Z.; Zhang, Y.; Sun, Q.; Ni, Z. Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genom. 2011, 12, 178. [Google Scholar] [CrossRef] [Green Version]
- Nagpal, P.; Ellis, C.M.; Weber, H.; Ploense, S.E.; Barkawi, L.S.; Guilfoyle, T.J.; Hagen, G.; Alonso, J.M.; Cohen, J.D.; Farmer, E.E.; et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 2005, 132, 4107–4118. [Google Scholar] [CrossRef] [Green Version]
- Guseman, J.M.; Hellmuth, A.; Lanctot, A.; Feldman, T.P.; Moss, B.L.; Klavins, E.; Calderon Villalobos, L.I.; Nemhauser, J.L. Auxin-induced degradation dynamics set the pace for lateral root development. Development 2015, 142, 905–909. [Google Scholar] [CrossRef] [Green Version]
- Nagpal, P.; Walker, L.M.; Young, J.C.; Sonawala, A.; Timpte, C.; Estelle, M.; Reed, J.W. AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 2000, 123, 563–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tabata, R.; Ikezaki, M.; Fujibe, T.; Aida, M.; Tian, C.E.; Ueno, Y.; Yamamoto, K.T.; Machida, Y.; Nakamura, K.; Ishiguro, S. Arabidopsis auxin response factor6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant Cell Physiol. 2010, 51, 164–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Yan, D.W.; Yuan, T.T.; Gao, X.; Lu, Y.T. A gain-of-function mutation in IAA8 alters Arabidopsis floral organ development by change of jasmonic acid level. Plant Mol. Biol. 2013, 82, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.H.; Wan, J.; Liang, R.Q.; Zhang, X.H.; Qiu, X.Q.; Meng, S.J.; Xu, N.K.; Lin, Y.; Dang, K.T.; Wang, Q.Y.; et al. Candidate Gene Association Analysis of Maize Transcription Factors in Flowering Time. Sci. Agric. Sin. 2022, 55, 12–25. [Google Scholar] [CrossRef]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Liu, W.; Wang, H.; Du, Q.; Fu, Z.; Li, W.X.; Tang, J. ZmEHD1 Is Required for Kernel Development and Vegetative Growth through Regulating Auxin Homeostasis. Plant Physiol. 2020, 182, 1467–1480. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Xue, Y.; Li, B.; Lin, Y.; Li, H.; Guo, Z.; Li, W.; Fu, Z.; Ding, D.; Tang, J. The chimeric gene atp6c confers cytoplasmic male sterility in maize by impairing the assembly of the mitochondrial ATP synthase complex. Mol. Plant 2022, 15, 872–886. [Google Scholar] [CrossRef]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef]
- Wan, J.; Wang, Q.; Zhao, J.; Zhang, X.; Guo, Z.; Hu, D.; Meng, S.; Lin, Y.; Qiu, X.; Mu, L.; et al. Gene expression variation explains maize seed germination heterosis. BMC Plant Biol. 2022, 22, 301. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, R.H.; Estelle, M. Diversity and specificity: Auxin perception and signaling through the TIR1/AFB pathway. Curr. Opin. Plant Biol. 2014, 21, 51–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ori, N. Dissecting the Biological Functions of ARF and Aux/IAA Genes. Plant Cell 2019, 31, 1210–1211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knapp, W.R.; Knapp, J.S. Response of Winter Wheat to Date of Planting and Fall Fertilization 1. Agron. J. 1978, 70, 1048–1053. [Google Scholar] [CrossRef]
- Winter, S.R.; Musick, J.T. Wheat Planting Date Effects on Soil Water Extraction and Grain Yield. Agron. J. 1993, 85, 912–916. [Google Scholar] [CrossRef]
- Wang, F.; Li, S.; Kong, F.; Lin, X.; Lu, S. Altered regulation of flowering expands growth ranges and maximizes yields in major crops. Front. Plant Sci. 2023, 14, 1094411. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Chen, L.; Zhang, X.; Yang, N.; Guo, J.; Wang, M.; Ji, S.; Zhao, X.; Yin, P.; Cai, L.; et al. Convergent selection of a WD40 protein that enhances grain yield in maize and rice. Science 2022, 375, eabg7985. [Google Scholar] [CrossRef]
- Guo, L.; Wang, X.; Zhao, M.; Huang, C.; Li, C.; Li, D.; Yang, C.J.; York, A.M.; Xue, W.; Xu, G.; et al. Stepwise cis-Regulatory Changes in ZCN8 Contribute to Maize Flowering-Time Adaptation. Curr. Biol. 2018, 28, 3005–3015. [Google Scholar] [CrossRef] [Green Version]
- Stephenson, E.; Estrada, S.; Meng, X.; Ourada, J.; Muszynski, M.G.; Habben, J.E.; Danilevskaya, O.N. Over-expression of the photoperiod response regulator ZmCCT10 modifies plant architecture, flowering time and inflorescence morphology in maize. PLoS ONE 2019, 14, e0203728. [Google Scholar] [CrossRef] [Green Version]
- Salvi, S.; Tuberosa, R.; Chiapparino, E.; Maccaferri, M.; Veillet, S.; van Beuningen, L.; Isaac, P.; Edwards, K.; Phillips, R.L. Toward positional cloning of Vgt1, a QTL controlling the transition from the vegetative to the reproductive phase in maize. Plant Mol. Biol. 2002, 48, 601–613. [Google Scholar] [CrossRef]
- Parent, B.; Leclere, M.; Lacube, S.; Semenov, M.A.; Welcker, C.; Martre, P.; Tardieu, F. Maize yields over Europe may increase in spite of climate change, with an appropriate use of the genetic variability of flowering time. Proc. Natl. Acad. Sci. USA 2018, 115, 10642–10647. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Jia, H.; Li, M.; Huang, Y.; Chen, W.; Yin, P.; Yang, Z.; Chen, Q.; Tian, F.; Zhang, Z.; et al. Divergent selection of KNR6 maximizes grain production by balancing the flowering-time adaptation and ear size in maize. Plant Biotechnol. J. 2023, 21, 1311. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Huang, Y.; Qi, P.; Lian, G.; Hu, X.; Han, N.; Wang, J.; Zhu, M.; Qian, Q.; Bian, H. Functional analysis of auxin receptor OsTIR1/OsAFB family members in rice grain yield, tillering, plant height, root system, germination, and auxinic herbicide resistance. New Phytol. 2021, 229, 2676–2692. [Google Scholar] [CrossRef] [PubMed]
- Nam, H.; Han, S.; Lee, S.; Nam, H.; Lim, H.; Lee, G.; Cho, H.S.; Dang, T.V.T.; Choi, S.; Lee, M.M.; et al. CPR5-mediated nucleo-cytoplasmic localization of IAA12 and IAA19 controls lateral root development during abiotic stress. Proc. Natl. Acad. Sci. USA 2023, 120, e2209781120. [Google Scholar] [CrossRef]
- Hamann, T.; Benkova, E.; Baurle, I.; Kientz, M.; Jurgens, G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002, 16, 1610–1615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, X.; Li, Y.; Guan, Y.; Wang, S.; Luo, H.; Li, X.; Li, H.; Zhang, Z. Auxin-induced AUXIN RESPONSE FACTOR4 activates APETALA1 and FRUITFULL to promote flowering in woodland strawberry. Hortic. Res. 2021, 8, 115. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhang, K.; Guo, L.; Liu, X.; Zhang, Z. AUXIN RESPONSE FACTOR3 plays distinct role during early flower development. Plant Signal. Behav. 2018, 13, e1467690. [Google Scholar] [CrossRef] [Green Version]
- Mena, M.; Mandel, M.A.; Lerner, D.R.; Yanofsky, M.F.; Schmidt, R.J. A characterization of the MADS-box gene family in maize. Plant J. 1995, 8, 845–854. [Google Scholar] [CrossRef]
- Liu, Y.; Jia, Z.; Li, X.; Wang, Z.; Chen, F.; Mi, G.; Forde, B.; Takahashi, H.; Yuan, L. Involvement of a truncated MADS-box transcription factor ZmTMM1 in root nitrate foraging. J. Exp. Bot. 2020, 71, 4547–4561. [Google Scholar] [CrossRef] [Green Version]
- Su, P.; Sui, C.; Wang, S.; Liu, X.; Zhang, G.; Sun, H.; Wan, K.; Yan, J.; Guo, S. Genome-wide evolutionary analysis of AUX/IAA gene family in wheat identifies a novel gene TaIAA15-1A regulating flowering time by interacting with ARF. Int. J. Biol. Macromol. 2023, 227, 285–296. [Google Scholar] [CrossRef]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 1999, 96, 5844–5849. [Google Scholar] [CrossRef]
- Shi, Q.; Kong, F.; Zhang, H.; Jiang, Y.; Heng, S.; Liang, R.; Ma, L.; Liu, J.; Lu, X.; Li, P.; et al. Molecular mechanisms governing shade responses in maize. Biochem. Biophys. Res. Commun. 2019, 516, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Wang, F.; Kong, J.; Xu, Q.; Li, T.; Chen, L.; Chen, H.; Jiang, H.; Li, C.; Cheng, B. Functional analysis of ZmMADS1a reveals its role in regulating starch biosynthesis in maize endosperm. Sci. Rep. 2019, 9, 3253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adamczyk, B.J.; Fernandez, D.E. MADS domain heterodimers are required for pollen maturation and tube growth in Arabidopsis. Plant Physiol. 2009, 149, 1713–1723. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.; Liu, Q.; Wang, X.; Huang, C.; Xu, G.; Hey, S.; Lin, H.Y.; Li, C.; Xu, D.; Wu, L.; et al. ZmMADS69 functions as a flowering activator through the ZmRap2.7-ZCN8 regulatory module and contributes to maize flowering time adaptation. New Phytol. 2019, 221, 2335–2347. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Li, X.; Sun, T.P. Four class A AUXIN RESPONSE FACTORs promote tomato fruit growth despite suppressing fruit set. Nat Plants 2023, 9, 706–719. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, C.; Dang, K.; Xie, Q.; Sahito, J.H.; Yuan, B.; Wan, J.; Qiu, X.; Zhao, J.; Lin, Y.; Meng, S.; et al. Over-Expression of ZmIAA29, an AUX/IAA Transcription Factor, Improved Maize Flowering Time. Agronomy 2023, 13, 2028. https://doi.org/10.3390/agronomy13082028
Ma C, Dang K, Xie Q, Sahito JH, Yuan B, Wan J, Qiu X, Zhao J, Lin Y, Meng S, et al. Over-Expression of ZmIAA29, an AUX/IAA Transcription Factor, Improved Maize Flowering Time. Agronomy. 2023; 13(8):2028. https://doi.org/10.3390/agronomy13082028
Chicago/Turabian StyleMa, Chenhui, Kuntai Dang, Qiankun Xie, Javed Hussain Sahito, Baiyu Yuan, Jiong Wan, Xiaoqian Qiu, Jiawen Zhao, Yanan Lin, Shujun Meng, and et al. 2023. "Over-Expression of ZmIAA29, an AUX/IAA Transcription Factor, Improved Maize Flowering Time" Agronomy 13, no. 8: 2028. https://doi.org/10.3390/agronomy13082028






