Association Mapping of Quantitative Trait Loci for Agronomic Traits in a Winter Wheat Collection Grown in Kazakhstan
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Field Experiments
2.2. Phenotyping of the Collection
2.3. Genotyping of the Collection
2.4. Analysis of Linkage Disequilibrium and Population Structure
2.5. Genome-Wide Association Study
2.6. Candidate Gene Analysis
3. Results
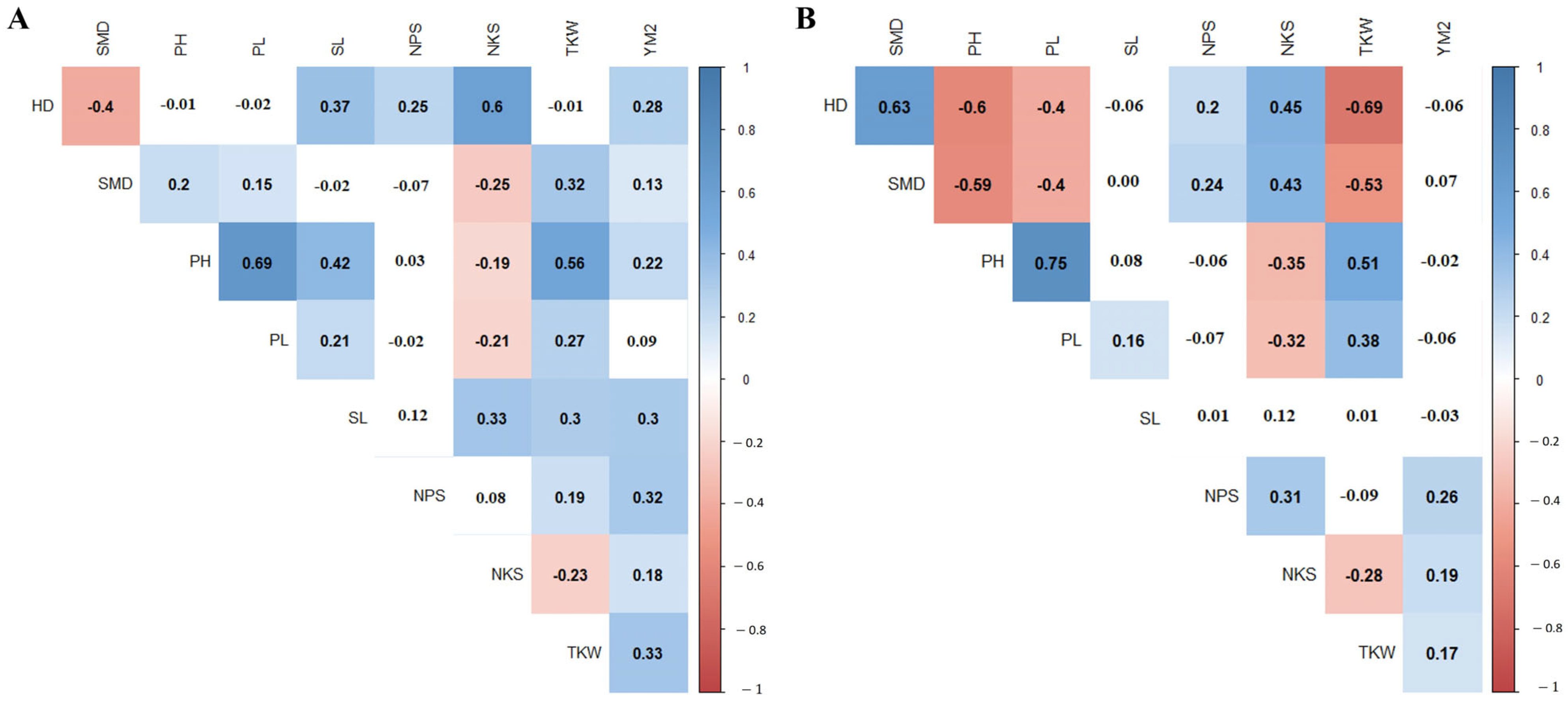
3.1. Descriptive Statistics of Phenotypic Traits
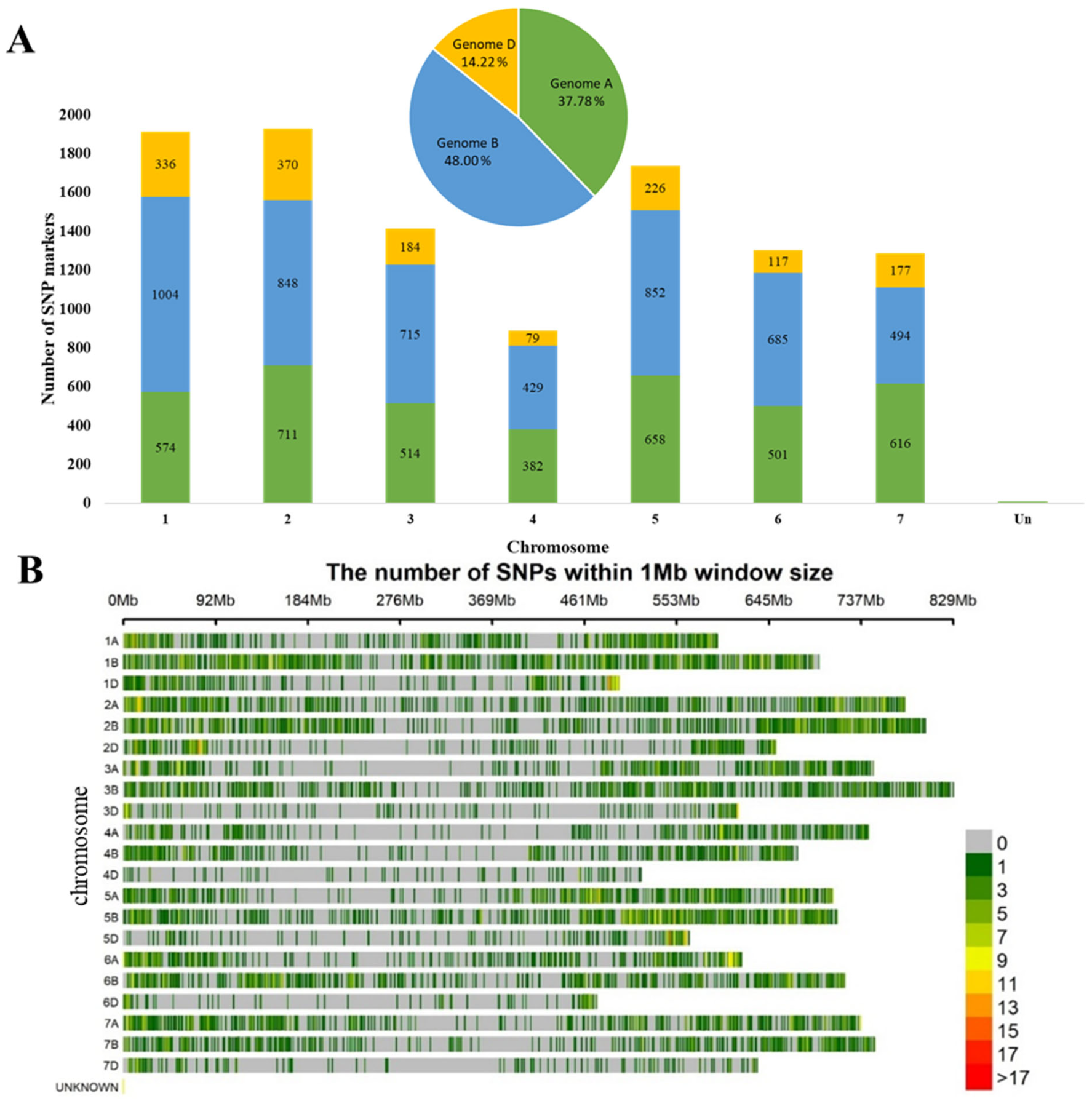
3.2. SNP Genotyping and Population Structure in the Studied Winter Wheat Collection
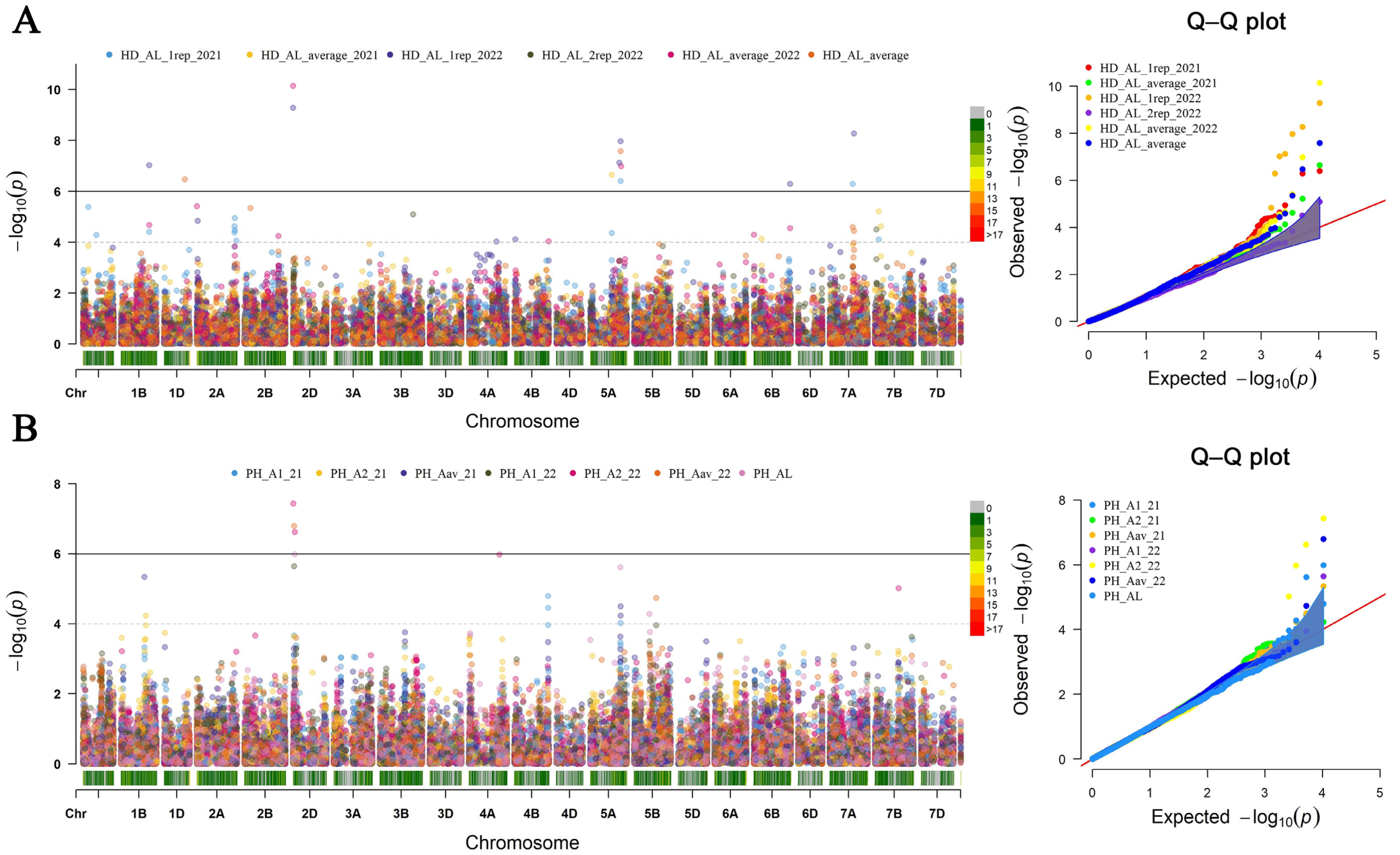
3.3. Identification of Marker–Trait Associations for Agronomic Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations. Available online: https://www.fao.org/faostat/en/#home (accessed on 25 February 2023).
- Shiferaw, B.; Smale, M.; Braun, H.J.; Duveiller, E.; Reynolds, M.; Muricho, G. Crops that feed the World 10. Past successes and future challenges to the role played by wheat in global food security. Food Secur. 2013, 5, 291–317. [Google Scholar] [CrossRef] [Green Version]
- Amalova, A.; Yermekbayev, K.; Griffiths, S.; Winfield, M.O.; Morgounov, A.; Abugalieva, S.; Turuspekov, Y. Population Structure of Modern Winter Wheat Accessions from Central Asia. Plants 2023, 12, 2233. [Google Scholar] [CrossRef]
- United States Department of Agriculture. Available online: https://www.usda.gov/ (accessed on 25 February 2023).
- Eltaher, S.; Baenziger, P.S.; Belamkar, V.; Emara, H.A.; Nower, A.A.; Salem, K.F.; Alqudah, A.M.; Sallam, A. GWAS revealed effect of genotype× environment interactions for grain yield of Nebraska winter wheat. BMC Genom. 2021, 22, 2. [Google Scholar] [CrossRef]
- Weigt, D.; Kiel, A.; Siatkowski, I.; Zyprych-Walczak, J.; Tomkowiak, A.; Kwiatek, M. Comparison of the Androgenic Response of Spring and Winter Wheat (Triticum aestivum L.). Plants 2019, 9, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Liu, Y. The conversion of spring wheat into winter wheat and vice versa: False claimor Lamarckian inheritance? J. Biosci. 2010, 35, 321–325. [Google Scholar] [CrossRef]
- Huang, M.; Mheni, N.; Brown-Guedira, G.; McKendry, A.; Griffey, C.; Van Sanford, D.; Costa, J.; Sneller, C. Genetic analysis of heading date in winter and spring wheat. Euphytica 2018, 214, 128. [Google Scholar] [CrossRef]
- Halder, J.; Gill, H.S.; Zhang, J.; Altameemi, R.; Olson, E.; Turnipseed, B.; Sehgal, S.K. Genome-wide association analysis of spike and kernel traits in the U.S. hard winter wheat. Plant Genome 2023, 16, e20300. [Google Scholar] [CrossRef]
- Collard, B.C.; Jahufer, M.Z.Z.; Brouwer, J.B.; Pang, E.C.K. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica 2005, 142, 169–196. [Google Scholar] [CrossRef]
- Xu, Y.; Li, P.; Yang, Z.; Xu, C. Genetic mapping of quantitative trait loci in crops. Crop J. 2017, 5, 175–184. [Google Scholar] [CrossRef] [Green Version]
- Pang, Y.; Liu, C.; Wang, D.; Amand, P.S.; Bernardo, A.; Li, W.; Li, L.; Wang, L.; Yuan, X.; Dong, L.; et al. High-resolution genome-wide association study identifies genomic regions and candidate genes for important agronomic traits in wheat. Mol. Plant 2020, 13, 1311–1327. [Google Scholar] [CrossRef]
- Sukumaran, S.; Dreisigacker, S.; Lopes, M.; Chavez, P.; Reynolds, M.P. Genome-wide association study for grain yield and related traits in an elite spring wheat population grown in temperate irrigated environments. Theor. Appl. Genet. 2015, 128, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Zanke, C.D.; Ling, J.; Plieske, J.; Kollers, S.; Ebmeyer, E.; Korzun, V.; Argillier, O.; Stiewe, G.; Hinze, M.; Neumann, F.; et al. Analysis of main effect QTL for thousand grain weight in European winter wheat (Triticum aestivum L.) by genome-wide association mapping. Front. Plant Sci. 2015, 6, 644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turuspekov, Y.; Baibulatova, A.; Yermekbayev, K.; Tokhetova, L.; Chudinov, V.; Sereda, G.; Ganal, M.; Griffiths, S.; Abugalieva, S. GWAS for plant growth stages and yield components in spring wheat (Triticum aestivum L.) harvested in three regions of Kazakhstan. BMC Plant Biol. 2017, 17, 190. [Google Scholar] [CrossRef] [Green Version]
- Mir, R.R.; Kumar, S.; Shafi, S. Genetic Dissection for Yield and Yield-Related Traits in Bread Wheat (Triticum aestivum L.). In Physiological, Molecular, and Genetic Perspectives of Wheat Improvement; Springer: Cham, Switzerland, 2021; pp. 209–227. [Google Scholar] [CrossRef]
- Anuarbek, S.; Abugalieva, S.; Pecchioni, N.; Laidò, G.; Maccaferri, M.; Tuberosa, R.; Turuspekov, Y. Quantitative trait loci for agronomic traits in tetraploid wheat for enhancing grain yield in Kazakhstan environments. PLoS ONE. 2020, 15, e0234863. [Google Scholar] [CrossRef] [PubMed]
- Amalova, A.; Abugalieva, S.; Babkenov, A.; Babkenova, S.; Turuspekov, Y. Genome-wide association study of yield components in spring wheat collection harvested under two water regimes in Northern Kazakhstan. PeerJ 2021, 9, e11857. [Google Scholar] [CrossRef] [PubMed]
- Reif, J.C.; Maurer, H.P.; Korzun, V.; Ebmeyer, E.; Miedaner, T.; Würschum, T. Mapping QTLs with main and epistatic effects underlying grain yield and heading time in soft winter wheat. Theor. Appl. Genet. 2011, 123, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Muqaddasi, Q.H.; Brassac, J.; Ebmeyer, E.; Kollers, S.; Korzun, V.; Argillier, O.; Stiewe, G.; Röder, M.S. Prospects of GWAS and predictive breeding for European winter wheat’s grain protein content, grain starch content, and grain hardness. Sci. Rep. 2020, 10, 12541. [Google Scholar] [CrossRef] [PubMed]
- Duggan, B.L.; Domitruk, D.R.; Fowler, D.B. Yield component variation in winter wheat grown under drought stress. Can. J. Plant Sci. 2000, 80, 739–745. [Google Scholar] [CrossRef] [Green Version]
- Sallam, A.; Eltaher, S.; Alqudah, A.M.; Belamkar, V.; Baenziger, P.S. Combined GWAS and QTL mapping revealed candidate genes and SNP network controlling recovery and tolerance traits associated with drought tolerance in seedling winter wheat. Genomics 2022, 114, 110358. [Google Scholar] [CrossRef]
- Lin, M.; Zhang, D.; Liu, S.; Zhang, G.; Yu, J.; Fritz, A.K.; Bai, G. Genome-wide association analysis on pre-harvest sprouting resistance and grain color in US winter wheat. BMC Genom. 2016, 17, 794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, L.X.; Morgounov, A.; Wanyera, R.; Keser, M.; Singh, S.K.; Sorrells, M. Identification of Ug99 stem rust resistance loci in winter wheat germplasm using genome-wide association analysis. Theor. Appl. Genet. 2012, 125, 749–758. [Google Scholar] [CrossRef]
- Tessmann, E.W.; Dong, Y.; Van Sanford, D.A. GWAS for Fusarium head blight traits in a soft red winter wheat mapping panel. Crop Sci. 2019, 59, 1823–1837. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.K.; Jiang, X.L.; Peng, T.; Shi, C.L.; Han, S.X.; Tian, B. Mapping quantitative trait loci with additive effects and additive × additive epistatic interactions for biomass yield, grain yield, and straw yield using a doubled haploid population of wheat (Triticum aestivum L.). Genet. Mol. Res. 2014, 13, 1412–1424. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Zhang, H.; Deng, Z.; Wu, R.; Li, D.; Wang, M.; Tian, J. Genome-wide association study for kernel weight-related traits using SNPs in a Chinese winter wheat population. Euphytica 2016, 212, 173–185. [Google Scholar] [CrossRef]
- El-Feki, W.M.; Byrne, P.F.; Reid, S.D.; Haley, S.D. Mapping quantitative trait loci for agronomic traits in winter wheat under different soil moisture levels. Agronomy 2018, 8, 133. [Google Scholar] [CrossRef] [Green Version]
- Ward, B.P.; Brown-Guedira, G.; Kolb, F.L.; Van Sanford, D.A.; Tyagi, P.; Sneller, C.H.; Griffey, C.A. Genome-wide association studies for yield-related traits in soft red winter wheat grown in Virginia. PLoS ONE 2019, 14, e0208217. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Guan, P.; Xin, M.; Wang, Y.; Chen, X.; Zhao, A.; Liu, M.; Li, H.; Zhang, M.; Lu, L.; et al. Genome-wide association study identifies QTL for thousand grain weight in winter wheat under normal-and late-sown stressed environments. Theor. Appl. Genet. 2021, 134, 143–157. [Google Scholar] [CrossRef]
- Chidzanga, C.; Mullan, D.; Roy, S.; Baumann, U.; Garcia, M. Nested association mapping-based GWAS for grain yield and related traits in wheat grown under diverse Australian environments. Theor. Appl. Genet 2022, 135, 4437–4456. [Google Scholar] [CrossRef]
- Acevedo, E.; Silva, P.; Silva, H. Wheat growth and physiology. Bread Wheat Improv. Prod. 2002, 30, 39–70. [Google Scholar]
- Hag, D.A.E.; Dalia, A.A. Effect of seeding rates on yield and yield components of two bread wheat cultivars. J. Agric. Res. 2016, 42, 71–81. [Google Scholar]
- Dospekhov, B. Methods of Field Experience; Kolos: Moscow, Russia, 1985. [Google Scholar]
- ADAPTAWHEAT 2012. 7th Framework Programme of the European Union. Available online: https://cordis.europa.eu/project/id/289842 (accessed on 12 February 2022).
- Allen, A.M.; Barker, G.L.; Berry, S.T.; Coghill, J.A.; Gwilliam, R.; Kirby, S.; Robinson, P.; Brenchley, R.C.; D’Amore, R.; McKenzie, N.; et al. Transcript-specific single-nucleotide polymorphism discovery and linkage analysis in hexaploid bread wheat (Triticum aestivum L.). Plant Biotechnol. J. 2011, 9, 1086–1099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkinson, P.A.; Winfield, M.O.; Barker, G.L.; Allen, A.M.; Burridge, A.; Coghill, J.A.; Edwards, K.J. CerealsDB 2.0: An integrated resource for plant breeders and scientists. BMC Bioinform. 2012, 13, 219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pritchard, J.K.; Stephens, M.; Donnelly., P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Earl, D.A.; VonHoldt, B.M. STRUCTURE HARVESTER version 0.6.94: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [Green Version]
- RStudio Team. RStudio: Integrated Development for R. MA: RStudio 2023.03.1 version; RStudio Inc.: Boston, MA, USA, 2015. [Google Scholar]
- Genievskaya, Y.; Turuspekov, Y.; Rsaliyev, A.; Abugalieva, S. Genome-wide association mapping for resistance to leaf, stem, and yellow rusts of common wheat under field conditions of South Kazakhstan. PeerJ 2020, 8, e9820. [Google Scholar] [CrossRef] [PubMed]
- Genome Associated Prediction Integrated Tool. Available online: http://www.maizegenetics.net/GAPIT (accessed on 15 January 2023).
- Yin, L.; Zhang, H.; Tang, Z.; Xu, J.; Yin, D.; Zhang, Z.; Yuan, X.; Zhu, M.; Zhao, S.; Li, X.; et al. rMVP: A memory-efficient. visualization-enhanced. and parallel-accelerated tool for genome-wide association study. Genom. Proteom. Bioinform. 2021, 19, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Ensembl Plants. Available online: https://plants.ensembl.org/Triticum_aestivum/Info/Index (accessed on 15 February 2023).
- Mandea, V.; Mustățea, P.; Marinciu, C.M.; Şerban, G.; Meluca, C.; Păunescu, G.; Dragomic, C.; Bunta, G.; Filiche, E.; Voinea, L.; et al. Yield components compensation in winter wheat (Triticum aestivum L.) is cultivar dependent. Rom. Agric. Res. 2019, 36, 27–33. [Google Scholar] [CrossRef]
- Würschum, T.; Leiser, W.L.; Langer, S.M.; Tucker, M.R.; Longin, C.F.H. Phenotypic and genetic analysis of spike and kernel characteristics in wheat reveals long-term genetic trends of grain yield components. Theor. Appl. Genet. 2018, 131, 2071–2084. [Google Scholar] [CrossRef]
- Mecha, B.; Alamerew, S.; Assefa, A.; Dutamo, D.; Assefa, E. Correlation and path coefficient studies of yield and yield associated traits in bread wheat (Triticum aestivum L.) genotypes. Adv Plants Agric Res. 2017, 6, 128–136. [Google Scholar]
- Mackay, T.F.; Stone, E.A.; Ayroles, J.F. The genetics of quantitative traits: Challenges and prospects. Nat. Rev. Genet. 2009, 10, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wang, X.; Zhang, G.; Jiang, P.; Chen, W.; Hao, Y.; Ma, X.; Xu, S.; Jia, J.; Kong, L.; et al. QTL mapping for yield-related traits in wheat based on four RIL populations. Theor. Appl. Genet. 2020, 133, 917–933. [Google Scholar] [CrossRef] [PubMed]
- Tahmasebi, S.; Heidari, B.; Pakniyat, H.; McIntyre, C.L. Mapping QTLs associated with agronomic and physiological traits under terminal drought and heat stress conditions in wheat (Triticum aestivum L.). Genome 2017, 60, 26–45. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Hao, C.; Zhang, Y.; Cheng, J.; Zhang, Z.; Liu, J.; Yi, X.; Cheng, X.; Sun, D.; Xu, Y.; et al. A combined association mapping and linkage analysis of kernel number per spike in common wheat (Triticum aestivum L.). Front. Plant Sci. 2017, 8, 1412. [Google Scholar] [CrossRef] [Green Version]
- Hu, P.; Zheng, Q.; Luo, Q.; Teng, W.; Li, H.; Li, B.; Li, Z. Genome-wide association study of yield and related traits in common wheat under salt-stress conditions. BMC Plant Biol. 2021, 21, 27. [Google Scholar] [CrossRef]
- Khan, H.; Krishnappa, G.; Kumar, S.; Mishra, C.N.; Krishna, H.; Devate, N.B.; Rathan, N.D.; Parkash, O.; Yadav, S.S.; Srivastava, P.; et al. Genome-wide association study for grain yield and component traits in bread wheat (Triticum aestivum L.). Front. Genet. 2022, 13, 982589. [Google Scholar] [CrossRef]
- Amalova, A.; Abugalieva, S.; Chudinov, V.; Sereda, G.; Tokhetova, L.; Abdikhalyk, A.; Turuspekov, Y. QTL mapping of agronomic traits in wheat using the UK Avalon× Cadenza reference mapping population grown in Kazakhstan. PeerJ 2021, 9, e10733. [Google Scholar] [CrossRef]
- Amalova, A.; Yermekbayev, K.; Griffiths, S.; Abugalieva, S.; Babkenov, A.; Fedorenko, E.; Abugalieva, A.; Turuspekov, Y. Identification of quantitative trait loci of agronomic traits in bread wheat using a Pamyati Azieva× Paragon mapping population harvested in three regions of Kazakhstan. PeerJ 2022, 10, e14324. [Google Scholar] [CrossRef]
- Abou-Elwafa, S.F.; Shehzad, T. Genetic diversity, GWAS and prediction for drought and terminal heat stress tolerance in bread wheat (Triticum aestivum L.). Genet. Resour. Crop Evol. 2020, 68, 711–728. [Google Scholar] [CrossRef]
- International Wheat Genome Sequencing Consortium (IWGSC); Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Singh, N.K. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar]
- Xu, X.; Zhang, L.; Zhao, W.; Fu, L.; Han, Y.; Wang, K.; Yan, L.; Li, Y.; Zhang, X.H.; Min, D.H. Genome-wide analysis of the serine carboxypeptidase-like protein family in Triticum aestivum reveals TaSCPL184-6D is involved in abiotic stress response. BMC Genom. 2021, 22, 1–19. [Google Scholar] [CrossRef]
- Wang, L.; Shen, R.; Chen, L.T.; Liu, Y.G. Characterization of a novel DUF1618 gene family in rice. J. Integr. Plant Biol. 2014, 56, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Tomasik, P.; Horton, D. Enzymatic conversions of starch. Adv. Carbohydr. Chem. Biochem. 2012, 68, 59–436. [Google Scholar] [PubMed]
- Nozoye, T.; Nagasaka, S.; Kobayashi, T.; Takahashi, M.; Sato, Y.; Sato, Y.; Uozumi, N.; Nishizawa, N.K. Phytosiderophore efflux transporters are crucial for iron acquisition in graminaceous plants. J. Biol. Chem. 2011, 286, 5446–5454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, K.; Li, Y. Functional genomics of the protein kinase superfamily from wheat. Mol. Breed. 2019, 39, 141. [Google Scholar] [CrossRef]
- Ma, W.; Cui, S.; Lu, Z.; Yan, X.; Cai, L.; Lu, Y.; Cai, K.; Zhou, H.; Ma, R.; Zhou, S.; et al. YTH Domain Proteins Play an Essential Role in Rice Growth and Stress Response. Plants 2022, 11, 2206. [Google Scholar] [CrossRef]
- Guo, J.; Sun, B.; He, H.; Zhang, Y.; Tian, H.; Wang, B. Current understanding of bHLH transcription factors in plant abiotic stress tolerance. Int. J. Mol. Sci. 2021, 22, 4921. [Google Scholar] [CrossRef]
- Han, G.; Qiao, Z.; Li, Y.; Yang, Z.; Wang, C.; Zhang, Y.; Liu, L.; Wang, B. RING zinc finger proteins in plant abiotic stress tolerance. Front. Plant Sci. 2022, 13, 1055. [Google Scholar] [CrossRef]
- Kumar, R.; Mishra, R.K.; Mishra, V.; Qidwai, A.; Pandey, A.; Shukla, S.K.; Parndey, M.; Dikshit, A. Detoxification and tolerance of heavy metals in plants. In Plant Metal Interaction; Elsevier: Amsterdam, The Netherlands, 2016; pp. 335–359. [Google Scholar]
- Gao, J.; Wallis, J.G.; Jewell, J.B.; Browse, J. Trimethylguanosine synthase1 (TGS1) is essential for chilling tolerance. Plant Physiol. 2017, 174, 1713–1727. [Google Scholar] [CrossRef] [Green Version]
- Kunihiro, A.; Yamashino, T.; Nakamichi, N.; Niwa, Y.; Nakanishi, H.; Mizuno, T. Phytochrome-interacting factor 4 and 5 (PIF4 and PIF5) activate the homeobox ATHB2 and auxin-inducible IAA29 genes in the coincidence mechanism underlying photoperiodic control of plant growth of Arabidopsis thaliana. Plant Cell Physiol. 2011, 52, 1315–1329. [Google Scholar] [CrossRef] [Green Version]

| Site/Region | KRIAPG (Almaty Region) | KBS (Turkestan Region) | ||
|---|---|---|---|---|
| Latitude/Longitude | 43°21′/76°53′ | 41°46′/69°45′ | ||
| Soil type | Light chestnut (humus 2.0–2.5%) | Light serozem (humus 1.1%) | ||
| Conditions | Rainfed | Rainfed | ||
| Year | 2020–2021 | 2021–2022 | 2020–2021 | 2021–2022 |
| Annual rainfall, mm | 464.7 | 568.9 | 279.4 | 421.0 |
| Mean temperature, °C | 10.5 | 12.2 | 17.5 | 11.7 |
| Max temperature, °C | 26.9 | 26.5 | 31.6 | 23.3 |
| Min temperature, °C | 1.8 | 1.1 | 2.7 | 4.0 |
| Traits | Site | Check Cultivar | Min | Max | Mean ± SE |
|---|---|---|---|---|---|
| HD, days | KRIAPG | 102.75 ± 31.75 | 44.00 | 143.00 | 102.37 ± 1.29 |
| KBS | 92.00 ± 12.00 | 82.50 | 115.00 | 102.98 ± 0.38 | |
| SMD, days | KRIAPG | 36.50 ± 5.50 | 24.50 | 44.00 | 35.35 ± 0.18 |
| KBS | 36.00 ± 6.50 | 26.50 | 52.00 | 39.46 ± 0.31 | |
| PH, cm | KRIAPG | 71.92 ± 20.75 | 35.00 | 132.00 | 70.58 ± 0.81 |
| KBS | 42.00 ± 0.00 | 26.40 | 58.25 | 41.95 ± 0.45 | |
| PL, cm | KRIAPG | 30.83 ± 8.83 | 13.42 | 61.33 | 25.47 ± 0.41 |
| KBS | 16.75 ± 0.25 | 6.90 | 31.55 | 17.38 ± 0.30 | |
| SL, cm | KRIAPG | 8.52 ± 0.19 | 3.17 | 12.65 | 9.52 ± 0.08 |
| KBS | 7.20 ± 1.00 | 5.40 | 28.85 | 8.29 ± 0.14 | |
| NPS, pcs | KRIAPG | 3.17 ± 0.83 | 1.33 | 6.17 | 3.27 ± 0.06 |
| KBS | 2.50 ± 0.50 | 3.00 | 8.00 | 4.57 ± 0.06 | |
| NKS, pcs | KRIAPG | 50.49 ± 4.86 | 23.17 | 70.88 | 47.04 ± 0.48 |
| KBS | 18.50 ± 1.50 | 25.00 | 70.00 | 42.60 ± 0.50 | |
| TKW, g | KRIAPG | 40.66 ± 6.61 | 19.85 | 58.80 | 35.25 ± 0.43 |
| KBS | 30.27 ± 4.15 | 13.49 | 33.42 | 20.44 ± 0.29 | |
| YM2, g/m2 | KRIAPG | 354.80 ± 176.09 | 22.95 | 997.28 | 393.48 ± 7.88 |
| KBS | 36.72 ± 1.85 | 15.05 | 266.98 | 81.33 ± 2.16 |
| Traits | Total QTLs | Stable QTLs | KRIAPG (Almaty Region) | KBS (Shymkent Region) | Both Regions |
|---|---|---|---|---|---|
| HD, days | 187 | 37 | 22 | 8 | 7 |
| SMD, days | 96 | 8 | 3 | 5 | NA |
| PH, cm | 171 | 68 | 11 | 52 | 5 |
| PL, cm | 105 | 8 | 3 | 4 | 1 |
| SL, cm | 76 | 9 | 3 | 6 | NA |
| NPS, pcs | 52 | 4 | 1 | 3 | NA |
| NKS, pcs | 123 | 25 | 10 | 12 | 3 |
| TKW, g | 55 | 1 | 1 | NA | NA |
| YM2, g/m2 | 86 | 13 | 5 | 4 | 4 |
| Total | 951 | 173 | 59 | 94 | 19 |
| Traits | SNP | Chromosome | Physical Position (Mb) | p-Value | Effect | PVE (%) | Conditions |
|---|---|---|---|---|---|---|---|
| HD | AX-95186349 | 1A | 102,166,440 | 4.19 × 10−6 | −3.59 | 6.00 | AL2021 |
| HD | AX-94958010 | 1B | 548,536,119 | 9.61 × 10−8 | 10.98 | 12.88 | AL2021, 2022, average; SH 2021, average |
| HD | AX-94687276 | 2D | 5,326,043 | 7.24 × 10−11 | −19.50 | 39.08 | AL2022, average; |
| HD | AX-94440472 | 3B | 507,287,720 | 5.08 × 10−7 | 5.12 | 64.32 | AL2021, SH2022 |
| HD | AX-94567204 | 3D | 496,732,990 | 4.15 × 10−9 | −30.26 | 95.13 | SH2022 |
| HD | AX-94720837 | 5A | 416,225,444 | 2.27 × 10−7 | −3.67 | 29.92 | AL2021 |
| HD | AX-94999352 | 5A | 563,498,900 | 7.60 × 10−8 | −8.64 | 3.24 | AL2022 |
| HD | AX-94464997 | 5A | 591,156,115 | 1.08 × 10−8 | 9.18 | 9.47 | AL2021,2022, average |
| HD | AX-94675648 | 6B | 704,187,628 | 5.18 × 10−7 | −10.74 | 5.39 | AL2022 |
| HD | AX-94994788 | 7A | 446,323,817 | 5.18 × 10−7 | −5.80 | 40.95 | AL2021, average |
| HD | AX-95074391 | 7A | 468,461,397 | 5.39 × 10−9 | 17.34 | 18.86 | AL2022, average |
| PH | AX-94442698 | 1B | 457,863,915 | 4.61 × 10−6 | 2.88 | 15.74 | AL2021, SH2021 |
| PH | AX-94384624 | 2D | 10,323,263 | 3.72 × 10−8 | −13.42 | 35.21 | AL2021, 2022, average |
| PH | AX-94711247 | 3A | 687,675,656 | 2.43 × 10−6 | 5.56 | 48.06 | SH2022, average |
| PH | AX-94517571 | 5A | 585,434,191 | 2.44 × 10−6 | −4.61 | 10.65 | AL2021, average |
| PL | AX-94599879 | 2D | 32,190,116 | 2.72 × 10−7 | 4.13 | 43.37 | AL2022, average |
| PL | AX-95633357 | 5D | 46,980,690 | 3.77 × 10−12 | −31.38 | 46.21 | SH2021 |
| NKS | AX-94594842 | 2A | 36,131,037 | 4.67 × 10−7 | −3.34 | 18.44 | SH2022, average |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amalova, A.; Yessimbekova, M.; Ortaev, A.; Rsaliyev, S.; Griffiths, S.; Burakhoja, A.; Turuspekov, Y.; Abugalieva, S. Association Mapping of Quantitative Trait Loci for Agronomic Traits in a Winter Wheat Collection Grown in Kazakhstan. Agronomy 2023, 13, 2054. https://doi.org/10.3390/agronomy13082054
Amalova A, Yessimbekova M, Ortaev A, Rsaliyev S, Griffiths S, Burakhoja A, Turuspekov Y, Abugalieva S. Association Mapping of Quantitative Trait Loci for Agronomic Traits in a Winter Wheat Collection Grown in Kazakhstan. Agronomy. 2023; 13(8):2054. https://doi.org/10.3390/agronomy13082054
Chicago/Turabian StyleAmalova, Akerke, Minura Yessimbekova, Anarbai Ortaev, Shynbolat Rsaliyev, Simon Griffiths, Aigerym Burakhoja, Yerlan Turuspekov, and Saule Abugalieva. 2023. "Association Mapping of Quantitative Trait Loci for Agronomic Traits in a Winter Wheat Collection Grown in Kazakhstan" Agronomy 13, no. 8: 2054. https://doi.org/10.3390/agronomy13082054
APA StyleAmalova, A., Yessimbekova, M., Ortaev, A., Rsaliyev, S., Griffiths, S., Burakhoja, A., Turuspekov, Y., & Abugalieva, S. (2023). Association Mapping of Quantitative Trait Loci for Agronomic Traits in a Winter Wheat Collection Grown in Kazakhstan. Agronomy, 13(8), 2054. https://doi.org/10.3390/agronomy13082054







