Hyperspectral Estimation of Chlorophyll Content in Apple Tree Leaf Based on Feature Band Selection and the CatBoost Model
Abstract
:1. Introduction
2. Materials and Methods
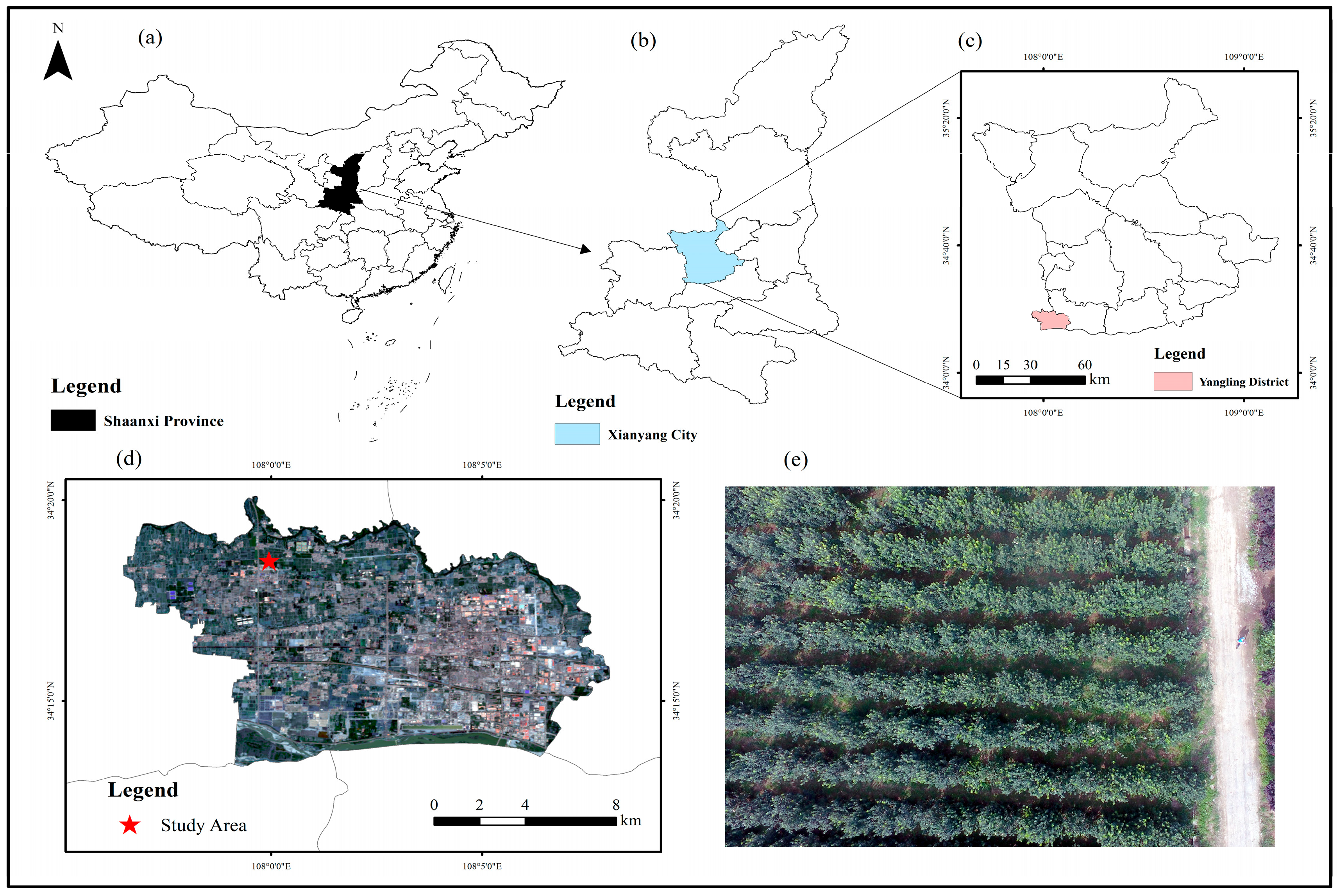
2.1. Study Area
2.2. Data Measurement
2.3. Hyperspectral Data Acquisition and Preprocessing
2.3.1. Hyperspectral Data Acquisition
2.3.2. Hyperspectral Data Preprocessing
2.4. Feature Band Selection Method
2.4.1. Competitive Adaptive Reweighted Sampling Algorithm
2.4.2. Random Frog Algorithm
- (1)
- Enter an initial band variable subset F0, which includes K random bands during initialization, and set the number of iterations N.
- (2)
- Select a candidate band variable subset F* based on F0, including K* bands. Establish a PLS model for F0, and calculate and rank the absolute regression coefficients of each band in descending order. If K* = K, then F* = F0; if K* < K, generate K* bands form a candidate band variable subset F*; if K* > K, the first Q bands form a candidate subset F*.
- (3)
- Select F* to replace the initial band variable subset F0, iterate N times, and complete the calculation.
- (4)
- Calculate the probability value of each band being selected after N iterations. The magnitude of this probability value is used as the criterion for whether the variable is selected. The higher the probability value, the more likely it is that the selected band is prioritized.
2.4.3. Elastic Net Algorithm
2.4.4. Improved Feature Band Selection Algorithm
2.5. Estimation Algorithm and Model Evaluation
2.5.1. Estimation Algorithm
2.5.2. Model Evaluation
3. Results
3.1. Original Spectral Characteristics of Apple Tree Leaves
3.2. Correlation Analysis between Different Spectral Transformations and LCC
3.3. Feature Band Selection
3.3.1. Feature Band Selection Based on the CARS Algorithm
3.3.2. Feature Band Selection Based on the RF Algorithm
3.3.3. Selection of Feature Bands Using the EN Algorithm
3.4. Estimation Results of LCC Based on a Single Band Selection Algorithm and Three Models
3.5. CatBoost Estimation Results of LCC Based on Improved Band Selection Algorithm and Grid Search Optimization
3.5.1. Band Selection Based on Improved Feature Selection Algorithm
3.5.2. CatBoost Estimation Results Based on Grid Search Parameter Optimization
4. Discussion
4.1. Selected Optimized Spectral Transformation Method
4.2. Advantages of Combining Dimensionality-Reduction Algorithms
4.3. Competitiveness of the CatBoost Algorithm for Performing Hyperspectral Estimation
4.4. Challenges and Future Research
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LCC | Leaf chlorophyll content |
| OR | Original spectrum |
| CR | Continuum removal |
| MSC | Multiplicative scatter correction |
| SD | Second derivative |
| CARS | Competitive adaptive reweighted sampling |
| EDF | Exponential decay function |
| RMSECV | Root mean squared error of cross validation |
| RF | Random frog |
| EN | Elastic net |
| PLSR | Partial least squares regression |
| RFR | Random forest regression |
| R2 | Determination coefficient |
| RMSE | Root mean square error |
| RPD | Relative prediction deviation |
| HRS | Hyperspectral remote sensing |
| CA | Correlation analysis |
| ANNs | Artificial neural networks |
| SG | Savitzky–Golay |
| CV | Cross validation |
| LASSO | Least absolute shrinkage and selection operator |
| MSE | Mean square error |
| GBDT | Gradient boosting decision tree |
| UAV | Unmanned aerial vehicle |
References
- Croft, H.; Chen, J.; Wang, R.; Mo, G.; Luo, S.; Luo, X.; He, L.; Gonsamo, A.; Arabian, J.; Zhang, Y.; et al. The global distribution of leaf chlorophyll content. Remote Sens. Environ. 2020, 236, 15. [Google Scholar] [CrossRef]
- Feng, W.; Yao, X.; Tian, Y.; Cao, W.; Zhu, Y. Monitoring leaf pigment status with hyperspectral remote sensing in wheat. Aust. J. Agric. Res. 2008, 59, 748–760. [Google Scholar] [CrossRef]
- Zhu, W.; Sun, Z.; Yang, T.; Li, J.; Peng, J.; Zhu, K.; Li, S.; Gong, H.; Lyu, Y.; Li, B.; et al. Estimating leaf chlorophyll content of crops via optimal unmanned aerial vehicle hyperspectral data at multi-scales. Comput. Electron. Agric. 2020, 178, 16. [Google Scholar] [CrossRef]
- Amirruddin, A.D.; Muharam, F.M.; Ismail, M.H.; Ismail, M.F.; Tan, N.P.; Karam, D.S. Hyperspectral remote sensing for assessment of chlorophyll sufficiency levels in mature oil palm (Elaeis guineensis) based on frond numbers: Analysis of decision tree and random forest. Comput. Electron. Agric. 2020, 169, 105221. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Gritz, Y.; Merzlyak, M.N. Relationships between leaf chlorophyll content and spectral reflectance and algorithms for non-destructive chlorophyll assessment in higher plant leaves. J. Plant Physiol. 2003, 160, 271–282. [Google Scholar] [CrossRef]
- Li, C.; Zhu, X.; Wei, Y.; Cao, S.; Guo, X.; Yu, X.; Chang, C. Estimating apple tree canopy chlorophyll content based on Sentinel-2A remote sensing imaging. Sci. Rep. 2018, 8, 10. [Google Scholar] [CrossRef] [Green Version]
- Shah, S.H.; Angel, Y.; Houborg, R.; Ali, S.; McCabe, M.F. A Random Forest Machine Learning Approach for the Retrieval of Leaf Chlorophyll Content in Wheat. Remote Sens. 2019, 11, 920. [Google Scholar] [CrossRef] [Green Version]
- Ali, A.; Imran, M.M. Evaluating the potential of red edge position (REP) of hyperspectral remote sensing data for real time estimation of LAI & chlorophyll content of kinnow mandarin (Citrus reticulata) fruit orchards. Sci. Hortic. 2020, 267, 109326. [Google Scholar] [CrossRef]
- Zhang, L.; Han, W.; Niu, Y.; Chavez, J.; Shao, G.; Zhang, H. Evaluating the sensitivity of water stressed maize chlorophyll and structure based on UAV derived vegetation indices. Comput. Electron. Agric. 2021, 185, 106174. [Google Scholar] [CrossRef]
- Zhang, Y.; Xia, C.; Zhang, X.; Cheng, X.; Feng, G.; Wang, Y.; Gao, Q. Estimating the maize biomass by crop height and narrowband vegetation indices derived from UAV-based hyperspectral images. Ecol. Indic. 2021, 129, 107985. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhao, C.; Yang, H.; Yang, G.; Han, L.; Li, Z.; Feng, H.; Xu, B.; Wu, J.; Lei, L. Estimation of maize above-ground biomass based on stem-leaf separation strategy integrated with LiDAR and optical remote sensing data. PeerJ 2019, 7, e7593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marang, I.J.; Filippi, P.; Weaver, T.B.; Evans, B.J.; Whelan, B.M.; Bishop, T.F.A.; Murad, M.O.F.; Al-Shammari, D.; Roth, G. Machine Learning Optimised Hyperspectral Remote Sensing Retrieves Cotton Nitrogen Status. Remote Sens. 2021, 13, 1428. [Google Scholar] [CrossRef]
- Yang, M.; Hassan, M.; Xu, K.; Zheng, C.; Rasheed, A.; Zhang, Y.; Jin, X.; Xia, X.; Xiao, Y.; He, Z. Assessment of Water and Nitrogen Use Efficiencies Through UAV-Based Multispectral Phenotyping in Winter Wheat. Front. Plant Sci. 2020, 11, 927. [Google Scholar] [CrossRef]
- Liang, L.; Geng, D.; Yan, J.; Qiu, S.; Di, L.; Wang, S.; Xu, L.; Wang, L.; Kang, J.; Li, L. Estimating Crop LAI Using Spectral Feature Extraction and the Hybrid Inversion Method. Remote Sens. 2020, 12, 3534. [Google Scholar] [CrossRef]
- Zhao, D.; Zhen, J.; Zhang, Y.; Miao, J.; Shen, Z.; Jiang, X.; Wang, J.; Jiang, J.; Tang, Y.; Wu, G. Mapping mangrove leaf area index (LAI) by combining remote sensing images with PROSAIL-D and XGBoost methods. Remote Sens. Ecol. Conserv. 2022, 9, 370–389. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N. Remote estimation of chlorophyll content in higher plant leaves. Int. J. Remote Sens. 1997, 18, 2691–2697. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Vina, A.; Ciganda, V.; Rundquist, D.C.; Arkebauer, T.J. Remote estimation of canopy chlorophyll content in crops. Geophys. Res. Lett. 2005, 32, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Lin, D.; Li, G.; Zhu, Y.; Liu, H.; Li, L.T.; Fahad, S.; Zhang, X.; Wei, C.; Jiao, Q. Predicting copper content in chicory leaves using hyperspectral data with continuous wavelet transforms and partial least squares. Comput. Electron. Agric. 2021, 187, 11. [Google Scholar] [CrossRef]
- Shi, T.; Chen, Y.; Liu, Y.; Wu, G. Visible and near-infrared reflectance spectroscopy-An alternative for monitoring soil contamination by heavy metals. J. Hazard. Mater. 2014, 265, 166–176. [Google Scholar] [CrossRef]
- Xiao, D.; Huang, J.; Li, J.; Fu, Y.; Li, Z. Inversion study of cadmium content in soil based on reflection spectroscopy and MSC-ELM model. Spectroc. Acta Part A-Molec. Biomol. Spectr. 2022, 283, 15. [Google Scholar] [CrossRef]
- Fu, Y.; Yang, G.; Li, Z.; Li, H.; Li, Z.; Xu, X.; Song, X.; Zhang, Y.; Duan, D.; Zhao, C.; et al. Progress of hyperspectral data processing and modelling for cereal crop nitrogen monitoring. Comput. Electron. Agric. 2020, 172, 14. [Google Scholar] [CrossRef]
- Cui, Y.; Meng, F.; Fu, P.; Yang, X.; Zhang, Y.; Liu, P. Application of hyperspectral analysis of chlorophyll a concentration inversion in Nansi Lake. Ecol. Inform. 2021, 64, 11. [Google Scholar] [CrossRef]
- Wu, M.; Lin, N.; Li, G.; Liu, H.; Li, D. Hyperspectral estimation of petroleum hydrocarbon content in soil using ensemble learning method and LASSO feature extraction. Environ. Pollut. Bioavail. 2022, 34, 308–320. [Google Scholar] [CrossRef]
- Zhang, J.; Fu, P.; Meng, F.; Yang, X.; Xu, J.; Cui, Y. Estimation algorithm for chlorophyll-a concentrations in water from hyperspectral images based on feature derivation and ensemble learning. Ecol. Inform. 2022, 71, 101783. [Google Scholar] [CrossRef]
- Feilhauer, H.; Asner, G.P.; Martin, R.E. Multi-method ensemble selection of spectral bands related to leaf biochemistry. Remote Sens. Environ. 2015, 164, 57–65. [Google Scholar] [CrossRef]
- Jiang, H.; Xu, W.; Ding, Y.; Chen, Q. Quantitative analysis of yeast fermentation process using Raman spectroscopy: Comparison of CARS and VCPA for variable selection. Spectroc. Acta Part A-Molec. Biomol. Spectr. 2020, 228, 8. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Gitelson, A.A. Remote estimation of gross primary productivity in soybean and maize based on total crop chlorophyll content. Remote Sens. Environ. 2012, 117, 440–448. [Google Scholar] [CrossRef]
- Luo, L.; Chang, Q.; Wang, Q.; Huang, Y. Identification and Severity Monitoring of Maize Dwarf Mosaic Virus Infection Based on Hyperspectral Measurements. Remote Sens. 2021, 13, 4560. [Google Scholar] [CrossRef]
- Wang, T.; Gao, M.; Cao, C.; You, J.; Zhang, X.; Shen, L. Winter wheat chlorophyll content retrieval based on machine learning using in situ hyperspectral data. Comput. Electron. Agric. 2022, 193, 17. [Google Scholar] [CrossRef]
- Wen, P.; Shi, Z.; Li, A.; Ning, F.; Zhang, Y.; Wang, R.; Li, J. Estimation of the vertically integrated leaf nitrogen content in maize using canopy hyperspectral red edge parameters. Precis. Agric. 2021, 22, 984–1005. [Google Scholar] [CrossRef]
- Sun, H.; Liu, N.; Wu, L.; Zheng, T.; Li, M.; Wu, J. Visualization of water content distribution in potato leaves based on hyperspectral image. Spectrosc. Spectr. Anal. 2019, 39, 910–916. [Google Scholar] [CrossRef]
- Gao, D.; Li, M.; Zhang, J.; Song, D.; Sun, H.; Qiao, L.; Zhao, R. Improvement of chlorophyll content estimation on maize leaf by vein removal in hyperspectral image. Comput. Electron. Agric. 2021, 184, 9. [Google Scholar] [CrossRef]
- Fan, S.; Huang, W.; Guo, Z.; Zhang, B.; Zhao, C. Prediction of Soluble Solids Content and Firmness of Pears Using Hyperspectral Reflectance Imaging. Food Anal. Method. 2015, 8, 1936–1946. [Google Scholar] [CrossRef]
- Zhang, J.; Cheng, T.; Guo, W.; Xu, X.; Qiao, H.; Xie, Y.; Ma, X. Leaf area index estimation model for UAV image hyperspectral data based on wavelength variable selection and machine learning methods. Plant Methods 2021, 17, 49. [Google Scholar] [CrossRef]
- Wang, K.; Qi, Y.; Guo, W.; Zhang, J.; Chang, Q. Retrieval and Mapping of Soil Organic Carbon Using Sentinel-2A Spectral Images from Bare Cropland in Autumn. Remote Sens. 2021, 13, 1072. [Google Scholar] [CrossRef]
- Yang, X.; Yang, R.; Ye, Y.; Yuan, Z.; Wang, D.; Hua, K. Winter wheat SPAD estimation from UAV hyperspectral data using cluster-regression methods. Int. J. Appl. Earth Obs. Geoinf. 2021, 105, 11. [Google Scholar] [CrossRef]
- Ta, N.; Chang, Q.; Zhang, Y. Estimation of Apple Tree Leaf Chlorophyll Content Based on Machine Learning Methods. Remote Sens. 2021, 13, 3902. [Google Scholar] [CrossRef]
- Maimaitijiang, M.; Sagan, V.; Sidike, P.; Hartling, S.; Esposito, F.; Fritschi, F.B. Soybean yield prediction from UAV using multimodal data fusion and deep learning. Remote Sens. Environ. 2020, 237, 20. [Google Scholar] [CrossRef]
- Zhu, Y.; Yang, G.; Yang, H.; Zhao, F.; Han, S.; Chen, R.; Zhang, C.; Yang, X.; Liu, M.; Cheng, J.; et al. Estimation of Apple Flowering Frost Loss for Fruit Yield Based on Gridded Meteorological and Remote Sensing Data in Luochuan, Shaanxi Province, China. Remote Sens. 2021, 13, 1630. [Google Scholar] [CrossRef]
- Jay, S.; Gorretta, N.; Morel, J.; Maupas, F.; Bendoula, R.; Rabatel, G.; Dutartre, D.; Comar, A.; Baret, F. Estimating leaf chlorophyll content in sugar beet canopies using millimeter- to centimeter-scale reflectance imagery. Remote Sens. Environ. 2017, 198, 173–186. [Google Scholar] [CrossRef]
- Lin, M.J.; Hsu, B.D. Photosynthetic plasticity of Phalaenopsis in response to different light environments. J. Plant Physiol. 2004, 161, 1259–1268. [Google Scholar] [CrossRef]
- Sui, X.; Mao, S.; Wang, L.; Zhang, B.; Zhang, Z. Effect of Low Light on the Characteristics of Photosynthesis and Chlorophyll a Fluorescence During Leaf Development of Sweet Pepper. J. Integr. Agric. 2012, 11, 1633–1643. [Google Scholar] [CrossRef]
- Cerovic, Z.G.; Masdoumier, G.; Ben Ghozlen, N.; Latouche, G. A new optical leaf-clip meter for simultaneous non-destructive assessment of leaf chlorophyll and epidermal flavonoids. Physiol. Plantarum 2012, 146, 251–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, B.; Feng, Y.; Ma, C.; Zhang, J.; Song, X.; Wang, M.; Sheng, D.; Feng, W.; Jiao, N. Suitability of different multivariate analysis methods for monitoring leaf N accumulation in winter wheat using in situ hyperspectral data. Comput. Electron. Agric. 2022, 198, 8. [Google Scholar] [CrossRef]
- Silalahi, D.D.; Midi, H.; Arasan, J.; Mustafa, M.S.; Caliman, J.P. Robust generalized multiplicative scatter correction algorithm on pretreatment of near infrared spectral data. Vib. Spectrosc. 2018, 97, 55–65. [Google Scholar] [CrossRef]
- Jiang, H.; Zhang, H.; Chen, Q.; Mei, C.; Liu, G. Identification of solid state fermentation degree with FT-NIR spectroscopy: Comparison of wavelength variable selection methods of CARS and SCARS. Spectroc. Acta Part A-Molec. Biomol. Spectr. 2015, 149, 1–7. [Google Scholar] [CrossRef]
- Wang, H.; Yang, G.; Zhang, Y.; Bao, Y.; He, Y. Detection of fungal disease on tomato leaves with competitive adaptive reweighted sampling and correlation analysis methods. Spectrosc. Spectr. Anal. 2017, 37, 2115–2119. [Google Scholar] [CrossRef]
- Xu, S.; Xu, X.; Blacker, C.; Gaulton, R.; Zhu, Q.; Yang, M.; Yang, G.; Zhang, J.; Yang, Y.; Yang, M.; et al. Estimation of Leaf Nitrogen Content in Rice Using Vegetation Indices and Feature Variable Optimization with Information Fusion of Multiple-Sensor Images from UAV. Remote Sens. 2023, 15, 854. [Google Scholar] [CrossRef]
- Sun, J.; Yang, W.; Zhang, M.; Feng, M.; Xiao, L.; Ding, G. Estimation of water content in corn leaves using hyperspectral data based on fractional order Savitzky-Golay derivation coupled with wavelength selection. Comput. Electron. Agric. 2021, 182, 105989. [Google Scholar] [CrossRef]
- Yun, Y.; Li, H.; Wood, L.R.E.; Fan, W.; Wang, J.; Cao, D.; Xu, Q.; Liang, Y. An efficient method of wavelength interval selection based on random frog for multivariate spectral calibration. Spectroc. Acta Part A-Molec. Biomol. Spectr. 2013, 111, 31–36. [Google Scholar] [CrossRef]
- Ren, G.; Wang, Y.; Ning, J.; Zhang, Z. Highly identification of keemun black tea rank based on cognitive spectroscopy: Near infrared spectroscopy combined with feature variable selection. Spectroc. Acta Part A-Molec. Biomol. Spectr. 2020, 230, 118079. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xu, Q.; Liang, Y. Random frog: An efficient reversible jump Markov Chain Monte Carlo-like approach for variable selection with applications to gene selection and disease classification. Anal. Chim. Acta 2012, 740, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Hastie, T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B-Stat. Methodol. 2005, 67, 301–320. [Google Scholar] [CrossRef] [Green Version]
- Tibshirani, R. Regression shrinkage and selection via the Lasso. J. R. Stat. Soc. Ser. B-Methodol. 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Brewick, P.T.; Masri, S.F.; Carboni, B.; Lacarbonara, W. Enabling reduced-order data-driven nonlinear identification and modeling through naive elastic net regularization. Int. J. Non-Linear Mech. 2017, 94, 46–58. [Google Scholar] [CrossRef]
- Chen, W.; Liu, X.; He, X.; Min, S.; Zhang, L. Near-infrared spectrum quantitative analysis model based on principal components selected by elastic net. Spectrosc. Spectr. Anal. 2010, 30, 2932–2935. [Google Scholar] [CrossRef]
- Satpathi, A.; Setiya, P.; Das, B.; Nain, A.S.; Jha, P.K.; Singh, S.; Singh, S. Comparative Analysis of Statistical and Machine Learning Techniques for Rice Yield Forecasting for Chhattisgarh, India. Sustainability 2023, 15, 2786. [Google Scholar] [CrossRef]
- Cao, C.; Wang, T.; Gao, M.; Li, Y.; Li, D.; Zhang, H. Hyperspectral inversion of nitrogen content in maize leaves based on different dimensionality reduction algorithms. Comput. Electron. Agric. 2021, 190, 14. [Google Scholar] [CrossRef]
- Yang, J.; Guo, Z.; Huang, Y.; Gao, H.; Jin, K.; Wu, X.; Yang, J. Early classification and detection of melon graft healing state based on hyperspectral imaging. Spectrosc. Spectr. Anal. 2022, 42, 2218–2224. [Google Scholar] [CrossRef]
- Cheng, J.; Chen, Z. Wavelength selection of near-infrared spectra based on improved SiPLS-random frog algorithm. Spectrosc. Spectr. Anal. 2020, 40, 3451–3456. [Google Scholar] [CrossRef]
- Sudu, B.; Rong, G.; Guga, S.; Li, K.; Zhi, F.; Guo, Y.; Zhang, J.; Bao, Y. Retrieving SPAD Values of Summer Maize Using UAV Hyperspectral Data Based on Multiple Machine Learning Algorithm. Remote Sens. 2022, 14, 5407. [Google Scholar] [CrossRef]
- Wu, L.; Huang, G.; Fan, J.; Zhang, F.; Wang, X.; Zeng, W. Potential of kernel-based nonlinear extension of Arps decline model and gradient boosting with categorical features support for predicting daily global solar radiation in humid regions. Energy Conv. Manag. 2019, 183, 280–295. [Google Scholar] [CrossRef]
- Kohavi, R.; Li, C.H. Oblivious Decision Trees Graphs and Top down Pruning. In Proceedings of the 14th International Joint Conference on Artificial Intelligence, Montreal, QC, Canada, 20–25 August 1995; Volume 2, pp. 1071–1077. [Google Scholar]
- Pham, T.D.; Yokoya, N.; Xia, J.; Ha, N.T.; Le, N.N.; Nguyen, T.T.T.; Dao, T.H.; Vu, T.T.P.; Pham, T.D.; Takeuchi, W. Comparison of Machine Learning Methods for Estimating Mangrove Above-Ground Biomass Using Multiple Source Remote Sensing Data in the Red River Delta Biosphere Reserve, Vietnam. Remote Sens. 2020, 12, 1334. [Google Scholar] [CrossRef] [Green Version]
- Wold, S.; Sjostrom, M.; Eriksson, L. PLS-regression: A basic tool of chemometrics. Chemometr. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Xie, J.; Pan, Q.; Li, F.; Tang, Y.; Hou, S.; Xu, C. Simultaneous detection of trace adulterants in food based on multi-molecular infrared (MM-IR) spectroscopy. Talanta 2021, 222, 7. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Mutanga, O.; Adam, E.; Cho, M.A. High density biomass estimation for wetland vegetation using WorldView-2 imagery and random forest regression algorithm. Int. J. Appl. Earth Obs. Geoinf. 2012, 18, 399–406. [Google Scholar] [CrossRef]
- Ge, Y.; Bai, G.; Stoerger, V.; Schnable, J.C. Temporal dynamics of maize plant growth, water use, and leaf water content using automated high throughput RGB and hyperspectral imaging. Comput. Electron. Agric. 2016, 127, 625–632. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Sun, C.; Luo, L.; He, Y. Determination of tea polyphenols content by infrared spectroscopy coupled with iPLS and random frog techniques. Comput. Electron. Agric. 2015, 112, 28–35. [Google Scholar] [CrossRef]
- Liu, N.; Xing, Z.; Zhao, R.; Qiao, L.; Li, M.; Liu, G.; Sun, H. Analysis of Chlorophyll Concentration in Potato Crop by Coupling Continuous Wavelet Transform and Spectral Variable Optimization. Remote Sens. 2020, 12, 2826. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y.; Du, L.; Liu, X.; Shi, S.; Chen, B. Improving the Selection of Vegetation Index Characteristic Wavelengths by Using the PROSPECT Model for Leaf Water Content Estimation. Remote Sens. 2021, 13, 821. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, Q.; Yi, X.; Ma, L.; Zhang, L.; Huang, C.; Zhang, Z.; Lv, X. Estimation of Cotton Leaf Area Index (LAI) Based on Spectral Transformation and Vegetation Index. Remote Sens. 2022, 14, 136. [Google Scholar] [CrossRef]
- Upreti, D.; Huang, W.J.; Kong, W.P.; Pascucci, S.; Pignatti, S.; Zhou, X.; Ye, H.; Casa, R. A Comparison of Hybrid Machine Learning Algorithms for the Retrieval of Wheat Biophysical Variables from Sentinel-2. Remote Sens. 2019, 11, 481. [Google Scholar] [CrossRef] [Green Version]
- Han, Z.; Deng, L. Application driven key wavelengths mining method for aflatoxin detection using hyperspectral data. Comput. Electron. Agric. 2018, 153, 248–255. [Google Scholar] [CrossRef]
- Yu, J.; Zhangzhong, L.; Lan, R.; Zhang, X.; Xu, L.; Li, J. Ensemble Learning Simulation Method for Hydraulic Characteristic Parameters of Emitters Driven by Limited Data. Agronomy 2023, 13, 986. [Google Scholar] [CrossRef]
- Niu, D.; Diao, L.; Zang, Z.; Che, H.; Zhang, T.; Chen, X. A Machine-Learning Approach Combining Wavelet Packet Denoising with Catboost for Weather Forecasting. Atmosphere 2021, 12, 1618. [Google Scholar] [CrossRef]
- Hancock, J.T.; Khoshgoftaar, T.M. CatBoost for big data: An interdisciplinary review. J. Big Data 2020, 7, 94. [Google Scholar] [CrossRef]













| Sample Sets | No. of Samples | Max. | Min. | Mean | Standard Deviation |
|---|---|---|---|---|---|
| All Samples | 160 | 51.44 | 15.00 | 33.93 | 10.10 |
| Modeling set | 112 | 51.44 | 15.00 | 33.86 | 10.57 |
| Validation set | 48 | 48.05 | 18.34 | 34.10 | 9.01 |
| Spectral Transformation | Feature Band Selection/nm | Number |
|---|---|---|
| OR | 402, 417, 418, 419, 420, 437, 438, 458, 463, 532, 659, 732, 836, 914, 926, 936, 953, 956, 963, 969, 970, 980, 987 | 23 |
| CR | 425, 435, 558, 594, 652, 696, 709, 710, 711, 712, 728, 729, 730, 731, 732, 743, 744, 745, 968, 969, 970 | 21 |
| MSC | 418, 419, 556, 603, 660, 849, 925, 926, 953, 961, 962, 980, 987 | 13 |
| SD | 416, 420, 426, 433, 439, 444, 448, 457, 458, 459, 463, 486, 488, 501, 503, 506, 542, 545, 554, 575, 577, 611, 644, 655, 671, 705, 708, 710, 718, 767, 770, 775, 779, 833, 852, 854, 882, 887, 890, 897, 898, 903, 906, 908, 915, 927, 934, 947, 951, 955, 964, 968 | 52 |
| Spectral Transformation | Feature Band Selection/nm | Number |
|---|---|---|
| OR | 447, 899, 914, 920, 926, 951, 974, 988 | 8 |
| CR | 588, 592, 654, 655, 710, 728, 744, 745, 755, 968, 969 | 11 |
| MSC | 898, 914, 921, 926, 941, 987 | 6 |
| SD | 444, 575, 611, 625, 763, 767, 770, 779, 808, 816, 823, 908 | 12 |
| Different Alpha Values | Spectrum Transform | Value | Number |
|---|---|---|---|
| Optimal value | OR | 0.55 | 43 |
| CR | 0.60 | 38 | |
| MSC | 0.10 | 133 | |
| SD | 0.95 | 16 | |
| Fixed value | OR | 0.10 | 123 |
| CR | 0.10 | 87 | |
| MSC | 0.10 | 133 | |
| SD | 0.10 | 116 |
| Selection Method | Spectrum Transform | Sensitive Band Selection/nm | Number |
|---|---|---|---|
| EN-CARS | OR | 401, 402, 404, 423, 424, 437, 447, 535, 663, 670, 705, 706, 710, 711, 713, 727, 729, 778, 955, 983 | 20 |
| CR | 522, 530, 531, 536, 735, 745, 748, 756, 926, 956 | 10 | |
| MSC | 641, 642, 650, 684, 686, 705, 712, 713, 727, 728, 729, 779, 785, 801, 839, 847, 848, 849, 963 | 19 | |
| SD | 444, 556, 575, 577, 705, 710, 718, 734, 753, 756, 779, 816, 905, 908, 909, 957 | 16 | |
| EN-RF | OR | 401, 402, 403, 404, 423, 425, 428, 433, 434, 437, 438, 439, 447, 662, 663, 664, 672, 705, 706, 711, 712, 713, 714, 726, 727, 728, 729, 955, 963, 983 | 30 |
| CR | 530, 531, 535, 536, 537, 744, 745, 756, 757, 771, 866, 867, 926 | 13 | |
| MSC | 642, 644, 705, 728, 783, 808, 847, 963 | 8 | |
| SD | 444, 552, 560, 575, 590, 705, 706, 710, 712, 717, 718, 734, 756, 779, 816 | 15 |
| Selection Method | Spectrum Transform | Default CatBoost | CatBoost Based on Grid Search Optimization | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Optimal Parameter | Optimized CatBoost | |||||||||
| R2 | RMSE | RPD | Iterations | Learning Rate | Depth | R2 | RMSE | RPD | ||
| EN-RF | OR | 0.823 | 3.754 | 2.401 | 400 | 0.010 | 9 | 0.832 | 3.650 | 2.469 |
| CR | 0.814 | 4.100 | 2.198 | 200 | 0.030 | 10 | 0.840 | 3.565 | 2.528 | |
| MSC | 0.868 | 3.288 | 2.740 | 300 | 0.013 | 9 | 0.900 | 2.814 | 3.202 | |
| SD | 0.871 | 3.575 | 2.521 | 100 | 0.029 | 9 | 0.892 | 2.936 | 3.069 | |
| EN-CARS | OR | 0.837 | 3.824 | 2.356 | 100 | 0.100 | 11 | 0.846 | 3.505 | 2.571 |
| CR | 0.828 | 3.976 | 2.266 | 200 | 0.051 | 9 | 0.856 | 3.379 | 2.666 | |
| MSC | 0.873 | 3.483 | 2.587 | 200 | 0.015 | 9 | 0.885 | 3.027 | 2.977 | |
| SD | 0.909 | 2.623 | 3.435 | 100 | 0.079 | 10 | 0.923 | 2.472 | 3.646 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Chang, Q.; Chen, Y.; Liu, Y.; Jiang, D.; Zhang, Z. Hyperspectral Estimation of Chlorophyll Content in Apple Tree Leaf Based on Feature Band Selection and the CatBoost Model. Agronomy 2023, 13, 2075. https://doi.org/10.3390/agronomy13082075
Zhang Y, Chang Q, Chen Y, Liu Y, Jiang D, Zhang Z. Hyperspectral Estimation of Chlorophyll Content in Apple Tree Leaf Based on Feature Band Selection and the CatBoost Model. Agronomy. 2023; 13(8):2075. https://doi.org/10.3390/agronomy13082075
Chicago/Turabian StyleZhang, Yu, Qingrui Chang, Yi Chen, Yanfu Liu, Danyao Jiang, and Zijuan Zhang. 2023. "Hyperspectral Estimation of Chlorophyll Content in Apple Tree Leaf Based on Feature Band Selection and the CatBoost Model" Agronomy 13, no. 8: 2075. https://doi.org/10.3390/agronomy13082075
APA StyleZhang, Y., Chang, Q., Chen, Y., Liu, Y., Jiang, D., & Zhang, Z. (2023). Hyperspectral Estimation of Chlorophyll Content in Apple Tree Leaf Based on Feature Band Selection and the CatBoost Model. Agronomy, 13(8), 2075. https://doi.org/10.3390/agronomy13082075






