The Effects of Localized Plant–Soil–Microbe Interactions on Soil Nitrogen Cycle in Maize Rhizosphere Soil under Long-Term Fertilizers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Determination Method
2.2.1. Method for Determination of Soil Physical and Chemical Properties, Enzyme Activity, and Root Exudates
2.2.2. Metagenomics Sequencing, Contig Assembly, and Annotation
2.3. Data Analysis
3. Results
3.1. Effects of Long-Term Fertilization on the Soil N Cycle
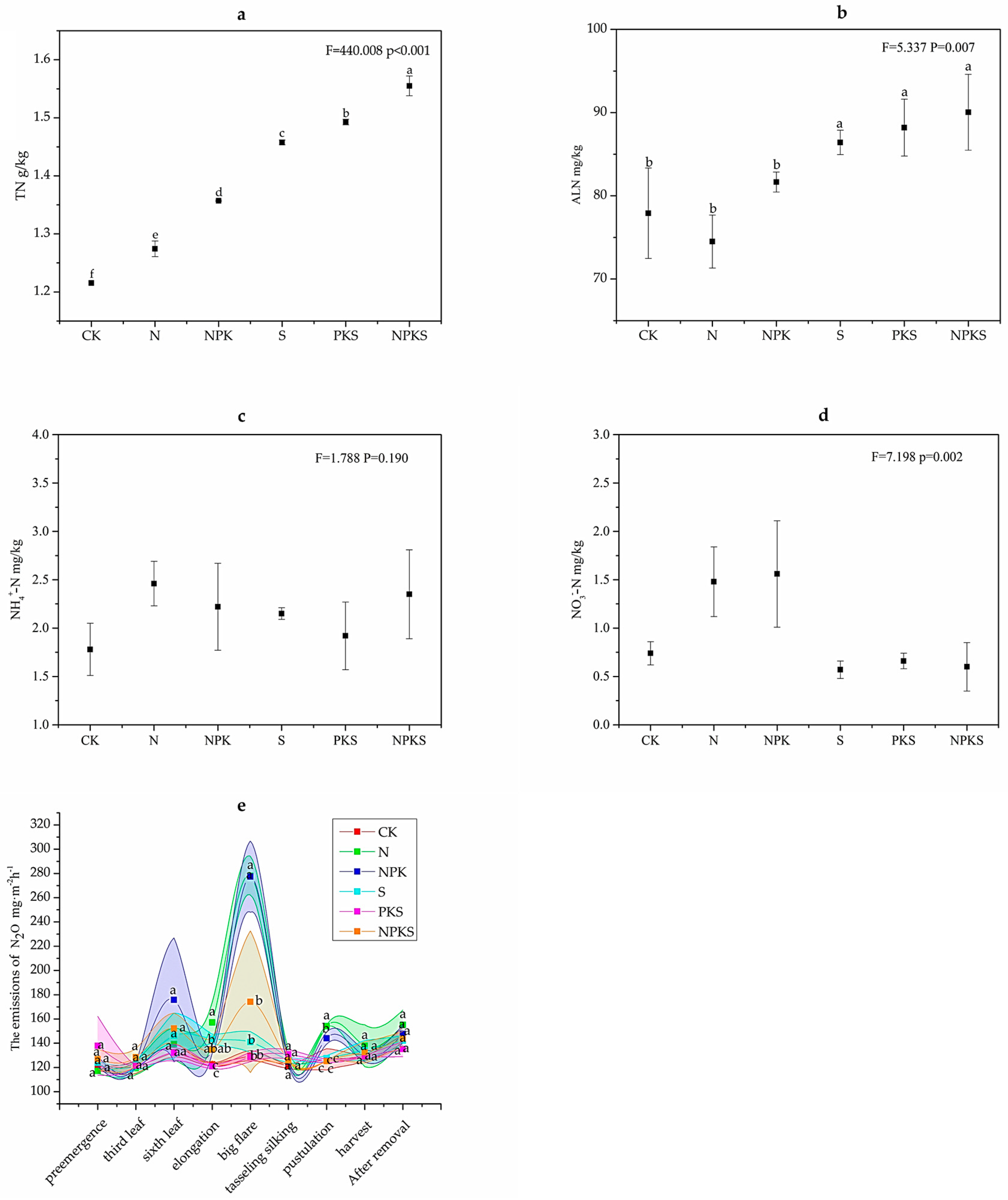
3.1.1. Effects of Long-Term Fertilization on Soil Nitrogen Content and N2O Emissions
3.1.2. Effects of Long-Term Fertilization on Enzyme Activities Related to the N Cycle in Maize Rhizosphere Soil
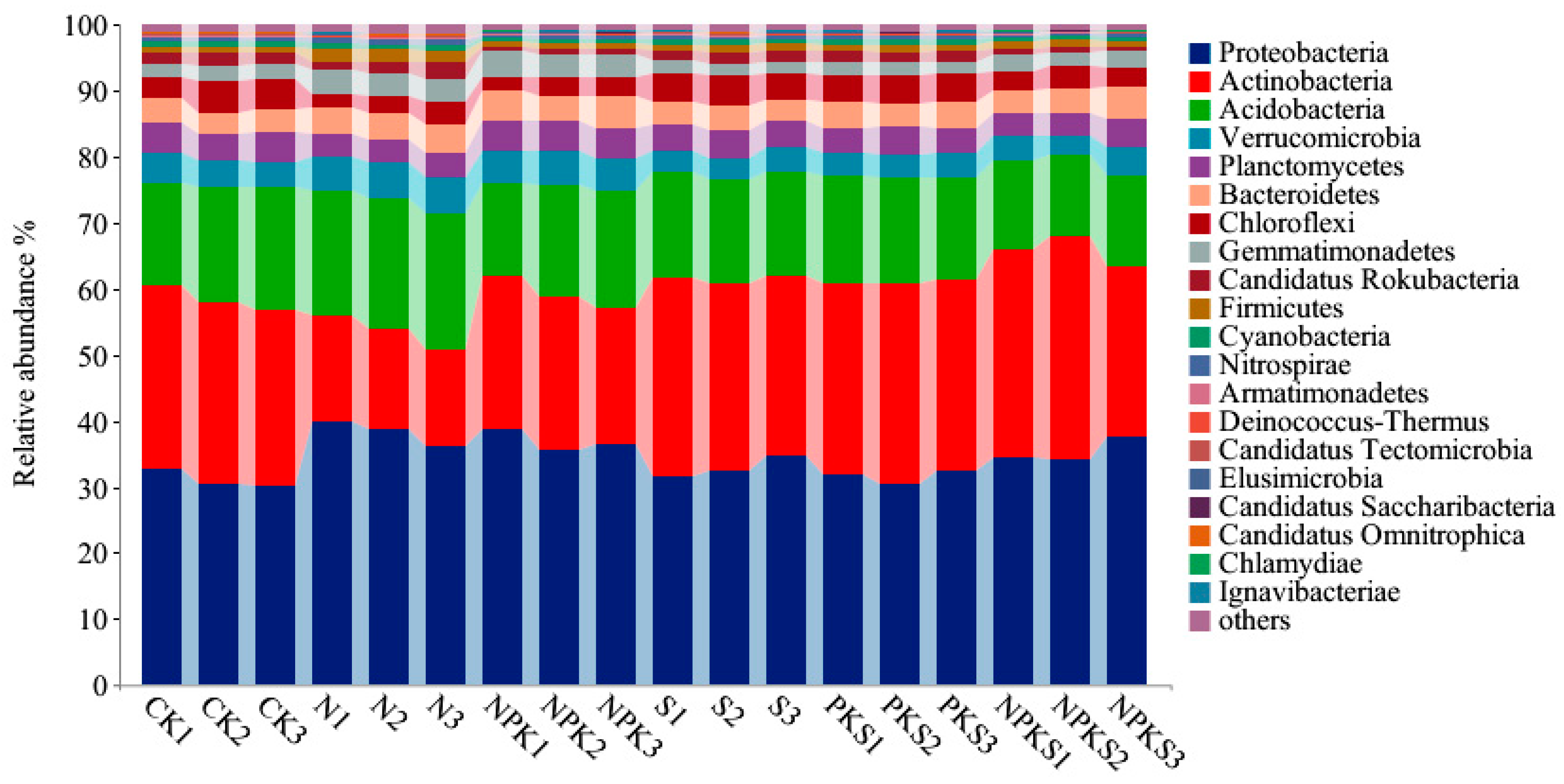
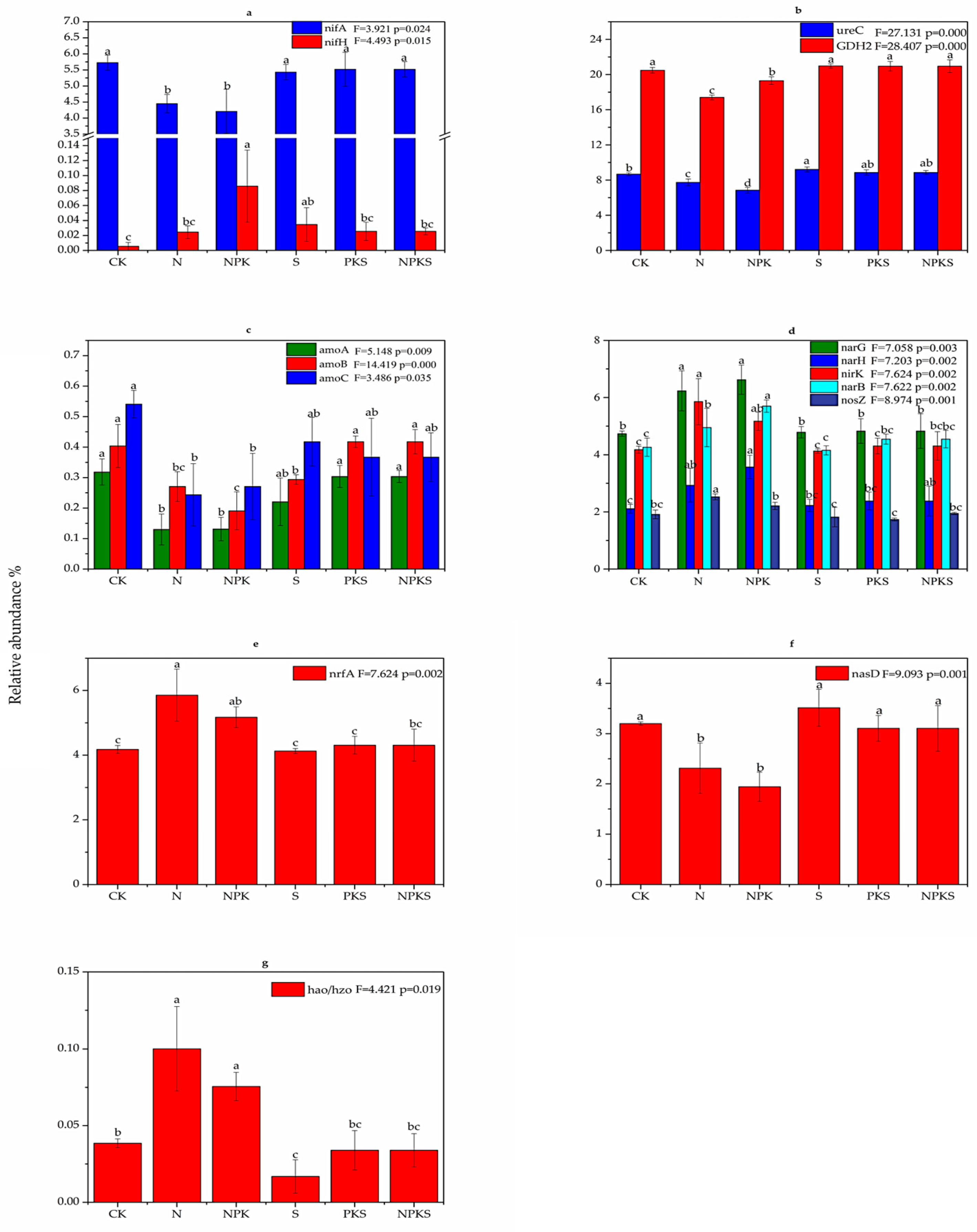
3.1.3. Effect of Long-Term Fertilization on N Cycle Functional Genes in Rhizosphere Soil of Maize
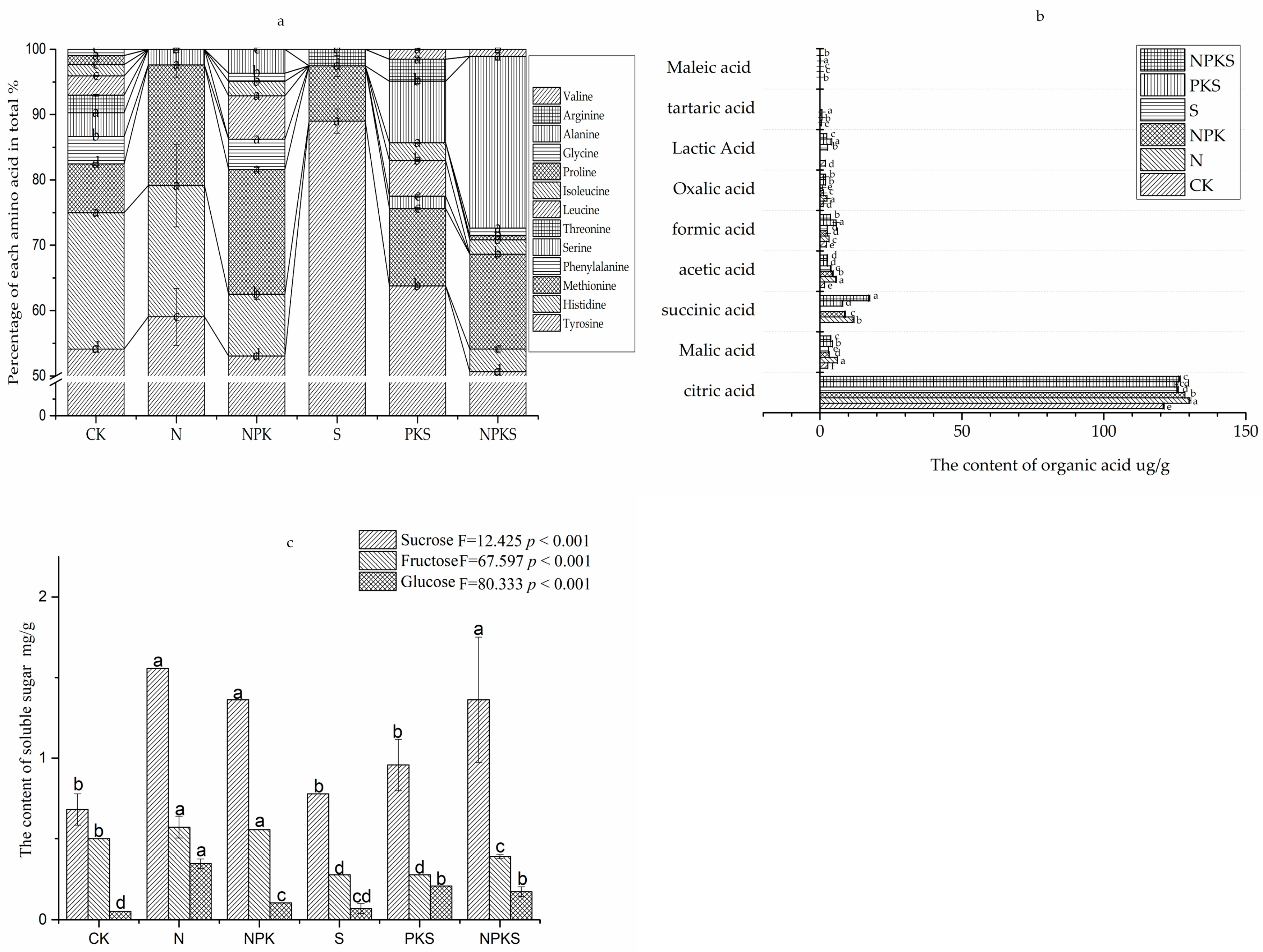
3.2. Effect of Long-Term Fertilization on Root Exudates of Maize
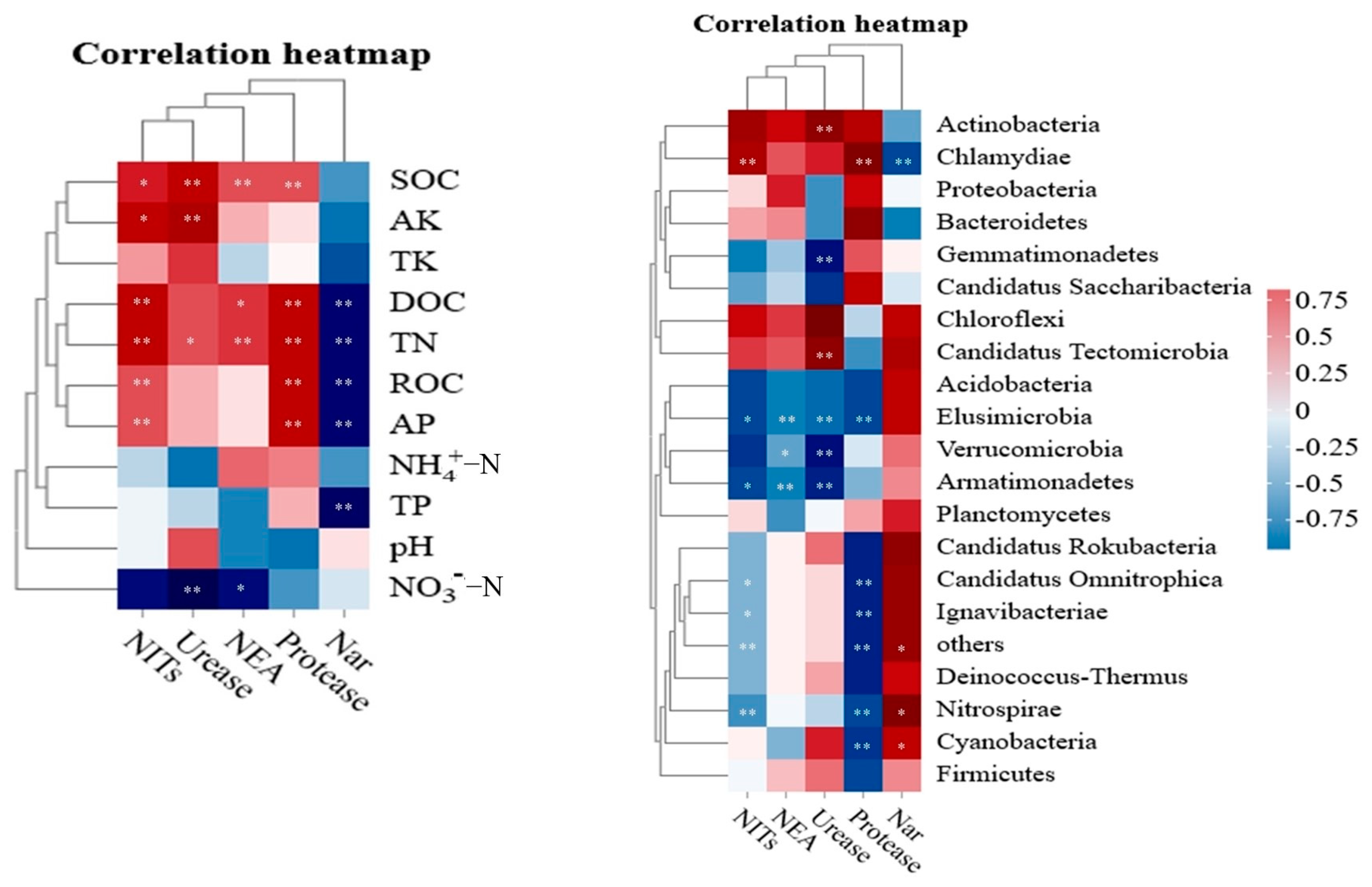
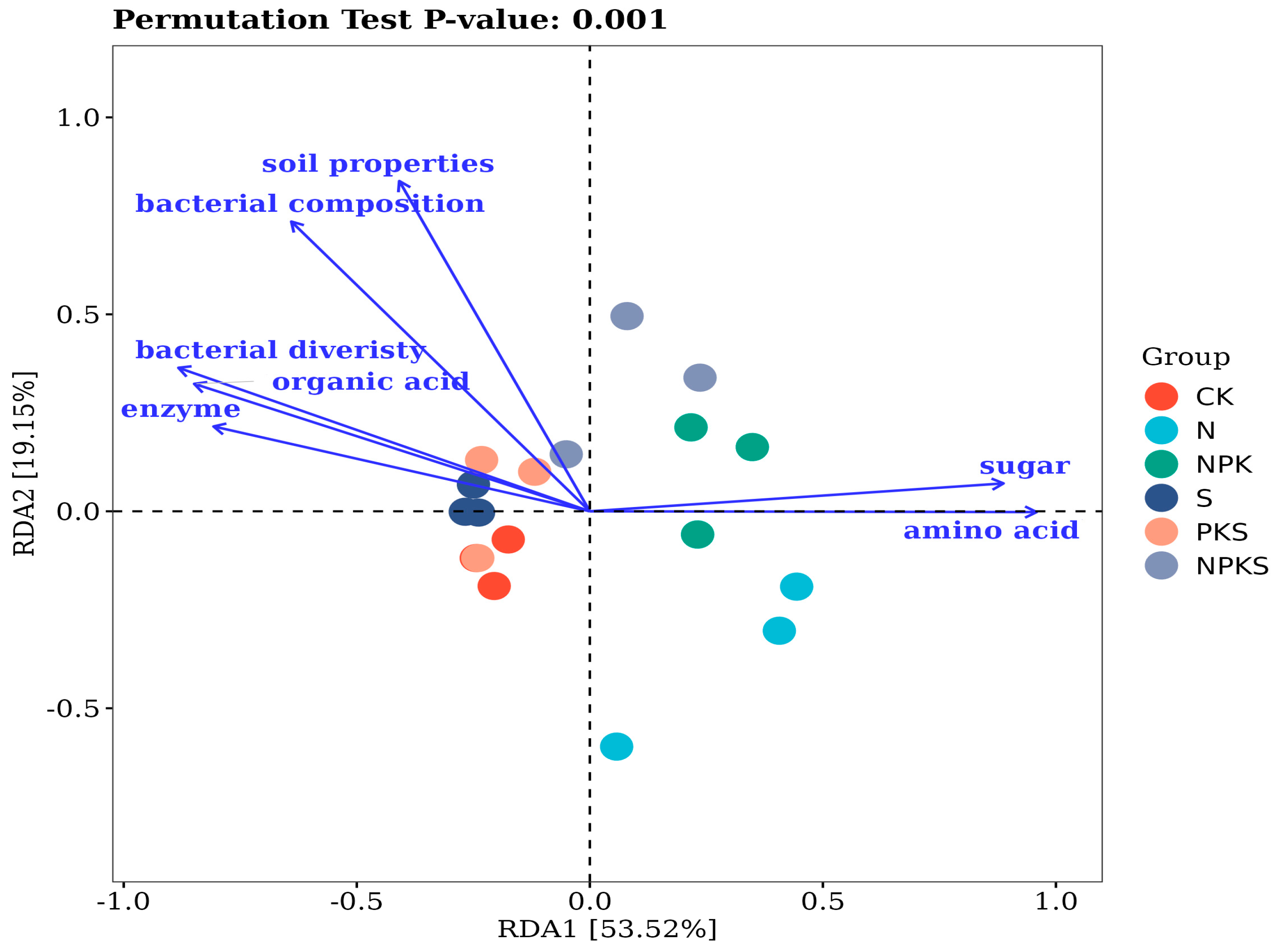
3.3. Effect of Plant–Soil–Microorganism Interaction on the N Cycle
4. Discussion
4.1. Effects of Long-Term Fertilization on the Soil N Cycle
4.2. Role of Root Exudates in the N Cycle of Maize
4.3. Role of Plant–Soil–Microorganism Systems in N Cycle
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| pH | SOC g/kg | DOC mg/kg | ROC g/kg | MBC mg/kg | TP g/kg | AP mg/kg | AK mg/kg | TK g/kg | |
|---|---|---|---|---|---|---|---|---|---|
| CK | 7.48 ± 0.06 a | 18.77 ± 0.42 c | 90.99 ± 6.20 c | 3.92 ± 0.06 d | 17.87 ± 0.06 a | 6.20 ± 0.10 c | 3.07 ± 0.38 c | 211.69 ± 4.01 d | 5.08 ± 0.15 a |
| N | 6.37 ± 0.16 c | 16.50 ± 0.27 d | 95.54 ± 9.65 c | 3.65 ± 0.19 d | 16.47 ± 0.10 b | 6.00 ± 0.08 cd | 2.42 ± 0.41 c | 156.37 ± 2.50 f | 4.93 ± 0.46 a |
| NPK | 6.77 ± 0.14 bc | 18.68 ± 0.00 c | 177.43 ± 5.79 b | 4.95 ± 0.09 c | 14.40 ± 0.16 c | 9.92 ± 0.33 b | 19.93 ± 2.14 b | 187.70 ± 4.24 e | 5.03 ± 0.59 a |
| S | 6.97 ± 0.52 b | 19.84 ± 0.31 b | 181.54 ± 8.27 b | 4.89 ± 0.06 c | 14.32 ± 0.10 c | 10.19 ± 0.29 b | 23.40 ± 2.19 a | 249.68 ± 3.07 c | 5.15 ± 0.09 a |
| PKS | 7.11 ± 0.10 ab | 19.46 ± 0.20 b | 195.62 ± 15.09 b | 5.47 ± 0.14 b | 12.48 ± 0.20 d | 5.29 ± 0.97 d | 4.20 ± 1.22 c | 303.23 ± 8.91 b | 5.05 ± 0.88 a |
| NPKS | 7.17 ± 0.13 ab | 21.28 ± 0.23 a | 241.12 ± 17.55 a | 6.22 ± 0.11 a | 11.58 ± 0.15 e | 11.27 ± 0.22 a | 23.15 ± 0.80 a | 325.39 ± 5.79 a | 5.20 ± 0.05 a |
| NH4+–N/TN % | NO3−–N/TN % | ATN/TN % | |
|---|---|---|---|
| CK | 0.15 | 0.06 | 57.20 |
| N | 0.19 | 0.12 | 56.57 |
| NPK | 0.16 | 0.11 | 55.43 |
| S | 0.15 | 0.04 | 56.11 |
| PKS | 0.13 | 0.04 | 55.80 |
| NPKS | 0.15 | 0.04 | 57.20 |
| Composition | |||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| ROC | 0.97 | 0.19 | −0.06 | 0 | |
| DOC | 0.93 | 0.24 | −0.11 | 0.07 | |
| TN | 0.92 | 0.17 | −0.18 | 0.2 | |
| SOC | 0.89 | −0.16 | 0.01 | 0.2 | |
| NITs | 0.89 | −0.06 | |||
| Protease | 0.88 | −0.35 | |||
| Phenylalanine | 0.81 | 0.08 | −0.09 | 0.5 | −0.28 |
| Threonine | 0.8 | −0.32 | 0.49 | −0.03 | −0.08 |
| Histidine | 0.77 | −0.41 | −0.26 | −0.1 | 0.35 |
| Proline | 0.77 | −0.03 | 0.48 | −0.38 | −0.17 |
| Arginine | −0.73 | −0.08 | 0.55 | 0.36 | 0.05 |
| Citric acid | −0.89 | −0.33 | |||
| Oxalic acid | 0.85 | 0.4 | |||
| Sucrose | 0.9 | ||||
| Elusimicrobia | 0.96 | 0.06 | 0.06 | ||
| Nitrospirae | 0.93 | 0.22 | −0.21 | ||
| Firmicutes | 0.89 | 0.13 | −0.39 | ||
| Simpson | 0.91 | ||||
| Bacteria | Soil | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Simpson | Elusimicrobia | Nitrospirae | Firmicutes | NITs | Protease | SOC | DOC | ROC | TN | ||
| Plant | Phenylalanine | 0.231 | 0.327 | 0.038 | −0.438 | −0.231 | −0.162 | −0.416 | −0.365 | −0.210 | −0.533 * |
| Threonine | 0.430 | 0.281 | 0.380 | 0.011 | −0.551 * | −0.637 ** | −0.218 | −0.564 * | −0.389 | −0.639 ** | |
| Histidine | −0.278 | 0.705 ** | 0.621 ** | 0.171 | −0.768 ** | −0.534 * | −0.696 ** | −0.683 ** | −0.654 ** | −0.778 ** | |
| Proline | 0.181 | −0.128 | 0.014 | −0.168 | −0.396 | −0.164 | 0.231 | −0.076 | 0.074 | −0.161 | |
| Arginine | 0.458 | −0.354 | −0.189 | 0.1156 | 0.499 * | 0.118 | 0.400 | 0.360 | 0.306 | 0.429 | |
| Sucrose | −0.695 ** | 0.168 | 0.017 | 0.193 | −0.115 | 0.346 | −0.228 | 0.175 | 0.058 | 0.204 | |
| Citric acid | 0.684 ** | 0.141 | 0.218 | −0.23 | −0.131 | −0.539 * | −0.207 | −0.536 * | −0.404 | −0.581 * | |
| Oxalic acid | −0.552 * | 0.346 | 0.332 | 0.626 ** | −0.357 | −0.119 | −0.252 | −0.034 | −0.079 | 0.047 | |
| Soil | DOC | 0.018 | −0.740 ** | −0.715 ** | −0.294 | 0.620 ** | 0.877 ** | ||||
| ROC | 0.172 | −0.763 ** | −0.749 ** | −0.421 | 0.593 ** | 0.801 ** | |||||
| TN | −0.012 | −0.754 ** | −0.717 ** | −0.250 | 0.655 ** | 0.832 ** | |||||
| SOC | 0.306 | −0.879 ** | −0.646 ** | −0.308 | 0.547 * | 0.634 ** | |||||
| r2 | Pr (>r) | |
|---|---|---|
| SOC | 0.744 | 0.001 |
| Methionine | 0.820 | 0.001 |
| Lactic Acid | 0.737 | 0.001 |
| Sucrose | 0.744 | 0.001 |
| Urease | 0.706 | 0.001 |
| Shannon | 0.922 | 0.001 |
| Actinobacteria | 0.899 | 0.001 |
| RDA1 | RDA2 | r2 | Pr (>r) | |
|---|---|---|---|---|
| bacterial composition | −0.638 | 0.770 | 0.900 | 0.001 |
| bacterial diversity | −0.914 | 0.406 | 0.841 | 0.001 |
| amino acids | 0.100 | −0.012 | 0.836 | 0.001 |
| soil properties | −0.425 | 0.905 | 0.832 | 0.001 |
| organic acids | −0.925 | 0.380 | 0.760 | 0.001 |
| sugar | 0.997 | 0.074 | 0.719 | 0.001 |
| enzymes | −0.960 | 0.279 | 0.640 | 0.001 |
References
- Shah, T.; Lateef, S.; Noor, M.A. Carbon and Nitrogen Cycling in Agroecosystems: An Overview. In Carbon Nitrogen Cycling in Soil; Springer: Singapore, 2020; pp. 1–15. [Google Scholar]
- Xu, G.; Wang, Z.; Gao, M.; Tian, D.; Huang, R.; Liu, J.; Li, J. Effects of straw and biochar return in soil on soil aggregate and carbon sequestration. Environ. Sci. 2018, 39, 355–362. [Google Scholar]
- Liu, H.; Wu, M.; Gao, H.; Gao, J.; Wang, S. Application of (15) N tracing and bioinformatics for estimating microbial-mediated nitrogen cycle processes in oil-contaminated soils. Environ. Res. 2023, 217, 114799. [Google Scholar]
- Govednik, A.; Potočnik, Ž.; Eler, K.; Mihelič, R.; Suhadolc, M. Combined effects of long-term tillage and fertilisation regimes on soil organic carbon, microbial biomass, and abundance of the total microbial communities and N-functional guilds. Appl. Soil Ecol. 2023, 188, 104876. [Google Scholar] [CrossRef]
- Sandén, T.; Spiegel, H.; Stüger, H.P.; Schlatter, N.; Haslmayr, H.P.; Zavattaro, L.; Grignani, C.; Bechini, L.; D′Hose, T.; Molendijk, L.; et al. European long-term field experiments: Knowledge gained about alternative management practices. Soil Use Manag. 2018, 34, 167–176. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Liu, H.; Ning, Y.; Xu, C.; Zhang, H.; Lu, X.; Wang, J.; Xu, X.; Feng, Y.; Zhang, Y. Reduced nitrogen fertilization under flooded conditions cut down soil N2O and CO2 efflux: An incubation experiment. J. Environ. Manag. 2022, 324, 116335. [Google Scholar]
- Bento, C.B.; Brandani, C.B.; Filoso, S.; Martinelli, L.A.; Carmo, J.B.d. Effects of extensive-to-intensive pasture conversion on soil nitrogen availability and CO2 and N2O fluxes in a Brazilian oxisol. Agric. Ecosyst. Environ. 2021, 321, 107633. [Google Scholar]
- Dămătîrcă, C.; Moretti, B.; Bertora, C.; Ferrarini, A.; Lerda, C.; Mania, I.; Celi, L.; Gorra, R.; Zavattaro, L. Residue incorporation and organic fertilisation improve carbon and nitrogen turnover and stabilisation in maize monocropping. Agric. Ecosyst. Environ. 2023, 342, 108255. [Google Scholar]
- Ma, L.; Gao, W.; Luan, H.; Tang, J.; Li, M.; Huang, S. Effects of partial substitution of chemical fertilizer with manure and/or straw on the abundance of functional genes related to soil N-cycling. J. Plant Nutr. Fertil. 2021, 27, 1767–1778. [Google Scholar]
- Wang, D.; Yin, W.; Li, H.; Chen, L.; Zhao, P.; Long, G. Effects of intercropping and nitrogen application on soil microbial metabolic functional diversity in maize cropping soil. Chin. J. Appl. Ecol. 2022, 33, 793–800. [Google Scholar]
- Ye, L.F.; Liu, H.Y.; Dan Deng, H.; Zheng, Y.P.; Han, Y.W.; Gao, X.T.; Abbott, L.K.; Zhao, C.M.; Li, J.H. Effects of decadal nitrogen and phosphorus fertilization on microbial taxonomic and functional attributes associated with soil organic carbon decomposition and concentration in an alpine meadow. Ecol. Indic. 2023, 146, 109790. [Google Scholar]
- Leadbeater, D.R.; Oates, N.C.; Bennett, J.P.; Li, Y.; Dowle, A.A.; Taylor, J.D.; Alponti, J.S.; Setchfield, A.T.; Alessi, A.M.; Helgason, T. Mechanistic strategies of microbial communities regulating lignocellulose deconstruction in a UK salt marsh. Microbiome 2021, 9, 48. [Google Scholar] [CrossRef]
- Kuypers, M.M.M.; Marchant, H.K.; Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 2018, 16, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, Q.; Cui, Q.; Ni, S.-Q. Nitrogen recovery through fermentative dissimilatory nitrate reduction to ammonium (DNRA): Carbon source comparison and metabolic pathway. Chem. Eng. J. 2022, 441, 135938. [Google Scholar]
- Chun, C.; Wu, Z.; Huang, Q.; Han, C.; Zhong, W. Effect of organic matter promotion on nitrogen-cycling genes and functional microorganisms in acidic red soil. Environ. Sci. 2020, 41, 2468–2475. [Google Scholar]
- An, X.; Jiang, S.; Xie, C.; Xu, Y.; Xu, C.; Shen, Q. Effects of reducing chemical fertilizers combined with organic fertilizers on soil microbial community in litchi orchards. Chin. J. Appl. Ecol. 2022, 33, 1099–1108. [Google Scholar]
- Tao, L. The Soil Microbial Activity and Microbial Functional Diversity of Cotton Field in Northern Xinjiang Response to Organic Fertilizer Partial Substitution for Chemical Fertilizer. Master’s Thesis, Shihezi University, Shihezi, China, 2013. [Google Scholar]
- Zhang, S. Effects of Chemical Fertilizers Partialy Substituted bu Organic Fertilizers on Patato Dry Matter Accumulation and Distribution and Soil Biological Characters. Ph.D. Thesis, Gansu Agricultural University, Lanzhou, China, 2014. [Google Scholar]
- Castellano-Hinojosa, A.; Strauss, S.L.; González-López, J.; Bedmar, E.J. Changes in the diversity and predicted functional composition of the bulk and rhizosphere soil bacterial microbiomes of tomato and common bean after inorganic N-fertilization. Rhizosphere 2021, 18, 100362. [Google Scholar] [CrossRef]
- Van Der Heijden, M.G.A.; Richard, D.B.; Van Straalen, N.M. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef]
- Cui, J.; Zhu, R.; Wang, X.; Xu, X.; Ai, C.; He, P.; Liang, G.; Zhou, W.; Zhu, P. Effect of high soil C/N ratio and nitrogen limitation caused by the long-term combined organic-inorganic fertilization on the soil microbial community structure and its dominated SOC decomposition. J. Environ. Manag. 2022, 303, 114155. [Google Scholar] [CrossRef]
- Enebe, M.C.; Babalola, O.O. Functional diversity of bacterial communities in the rhizosphere of maize grown on a soil under organic and inorganic fertilization. Sci. Afr. 2022, 16, e01212. [Google Scholar]
- Liu, J.; Jin, L.; Shan, Y.; Burgess, K.S.; Ge, X. Elevation explains variation in soil microbial diversity and community composition under experimental warming and fertilization treatments in mountain meadows. Appl. Soil Ecol. 2022, 171, 104311. [Google Scholar] [CrossRef]
- Das, P.P.; Singh, K.R.; Nagpure, G.; Mansoori, A.; Singh, R.P.; Ghazi, I.A.; Kumar, A.; Singh, J. Plant-soil-microbes: A tripartite interaction for nutrient acquisition and better plant growth for sustainable agricultural practices. Environ. Res. 2022, 214, 113821. [Google Scholar]
- Fadiji, A.E.; Yadav, A.N.; Santoyo, G.; Babalola, O.O. Understanding the plant-microbe interactions in environments exposed to abiotic stresses: An overview. Microbiol. Res. 2023, 271, 127368. [Google Scholar]
- Pascale, A.; Proietti, S.; Pantelides, I.S.; Stringlis, I.A. Modulation of the Root Microbiome by Plant Molecules: The Basis for Targeted Disease Suppression and Plant Growth Promotion. Front. Plant Sci. 2019, 10, 1741. [Google Scholar] [CrossRef]
- Li, W.; Li, Y.; Lv, J.; He, X.; Wang, J.; Teng, D.; Jiang, L.; Wang, H.; Lv, G. Rhizosphere effect alters the soil microbiome composition and C, N transformation in an arid ecosystem. Appl. Soil Ecol. 2022, 170, 104296. [Google Scholar] [CrossRef]
- de Vries, F.T.; Griffiths, R.I.; Knight, C.G.; Nicilitch, O.; Williams, A. Harnessing rhizosphere microbiomes for dought-resilient crop production. Science 2020, 368, 270–274. [Google Scholar] [PubMed]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [PubMed]
- Jacoby, R.; Peukert, M.; Succurro, A.; Koprivova, A.; Kopriva, S. The Role of Soil Microorganisms in Plant Mineral Nutrition-Current Knowledge and Future Directions. Front. Plant Sci. 2017, 8, 1617. [Google Scholar]
- Wu, L.; Lin, X.; Lin, W. Advances and perspective in research on plant-soil-microbe interactions mediated by root exudates. Chin. J. Plant Ecol. 2014, 38, 298–310. [Google Scholar]
- Ahmad, E.; Sharma, P.K.; Khan, M.S. Roles of Root Exudates in Different Processes in the Nitrogen Cycle in the Rhizosphere. Soil Nitrogen Ecol. 2021, 62, 179–200. [Google Scholar]
- Wang, J.; Liao, L.; Wang, G.; Liu, H.; Wu, Y.; Liu, G.; Zhang, C. N-induced root exudates mediate the rhizosphere fungal assembly and affect species coexistence. Sci. Total Environ. 2022, 804, 150148. [Google Scholar]
- Xu, Y. Effects of Simulated Root Secretions on the Abundance of Nitrification and Denitrification Genes. Master’s Thesis, Anhui Jianzhu University, Hefei, China, 2018. [Google Scholar]
- Yan, C.; Huang, J.; Li, Z.; Peng, C.; Cao, C. Effect of plant and its exudated on urease activity, nirtification and denitrification in wetland soil. Ecol. Environ. Sci. 2017, 26, 303–308. [Google Scholar]
- Bian, X.; Zhao, W.; Yue, Z.; Wang, H.; Jiao, H.; Sui, H. Research process of soil enzymes on carbon and nitrogen cycle in agricultural ecosystem. Chin. Agric. Sci. Bull. 2016, 32, 171–178. [Google Scholar]
- Li, Y.; Wang, C.; Chang, H.; Zhang, Y.; Liu, S.; He, W. Metagenomics reveals the effect of long-term fertilization on carbon cycle in the maize rhizosphere. Front. Microbiol. 2023, 14, 1170214. [Google Scholar]
- Huang, R.; Lan, T.; Song, X.; Li, J.; Ling, J.; Deng, O.; Wang, C.; Gao, X.; Li, Q.; Tang, X.; et al. Soil labile organic carbon impacts C:N:P stoichiometry in urban park green spaces depending on vegetation types and time after planting. Appl. Soil Ecol. 2021, 163, 103926. [Google Scholar]
- Xu, C.; Lin, X.; Xia, B. Response of root exudates of maize seedlings (Zea mays L. ) to pyrene contamination. Acta Ecol. Sin. 2010, 30, 3280–3288. [Google Scholar]
- Li, Y.; Wang, C.; Wang, T.; Liu, Y.; Jia, S.; Gao, Y.; Liu, S. Effects of Different Fertilizer Treatments on Rhizosphere Soil Microbiome Composition and Functions. Land 2020, 9, 329. [Google Scholar]
- Yuan, X.; Niu, D.; Weber-Grullon, L.; Fu, H. Nitrogen deposition enhances plant-microbe interactions in a semiarid grassland: The role of soil physicochemical properties. Geoderma 2020, 373, 114446. [Google Scholar] [CrossRef]
- Li, Q.; Jia, W.; Zhang, Q.; Cheng, X. Localized plant-soil-microbe interactions regulate spatial variations of soil oxidase activities within afforested systems in a subtropical area. Geoderma 2022, 406, 115499. [Google Scholar]
- Ziadi, N.; Angers, D.A.; Gagnon, B.; Lalande, R.; Morel, C.; Rochette, P.; Chantigny, M.H. Long-term tillage and synthetic fertilization affect soil functioning and crop yields in a corn–soybean rotation in eastern Canada. Can. J. Soil Sci. 2014, 94, 365–376. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Liu, X.; Hao, L.; Xie, H.; Zhang, G.; Chen, Z.; Zhang, Y. Effects of straw returning in conjunction with different nitrogen fertilizer dosages on corn yield and soil properties. Chin. J. Ecol. 2020, 39, 507–516. [Google Scholar]
- Sun, R.; Wang, D.; Lin, J.; Liu, Q.; Yang, L. Effects of long term integrated fertilization with organic manure and chemical fertilizers on soil nutrients in Taihu lake region. Soils 2009, 41, 384–388. [Google Scholar]
- Chu, H.; Hosen, Y.; Yagi, K. NO, N2O, CH4 and CO2 fluxes in winter barley field of Japanese Andisol as affected by N fertilizer management. Soil Biol. Biochem. 2007, 39, 330–339. [Google Scholar] [CrossRef]
- Bei, S.; Zhang, Y.; Liu, J.; Mou, Y.; Lun, X. Greenhouse gas emission under the treatments of fertilization and wheat straw returning during the maize growing seasons. Environ. Chem. 2012, 31, 407–414. [Google Scholar]
- Jian, S.; Li, J.; Chen, J.; Wang, G.; Mayes, M.A.; Dzantor, K.E.; Hui, D.; Luo, Y. Soil extracellular enzyme activities, soil carbon and nitrogen storage under nitrogen fertilization: A meta-analysis. Soil Biol. Biochem. 2016, 101, 32–43. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Fu, X.; Ghimire, R.; Sainju, U.M.; Jia, Y.; Zhao, F. Responses of soil bacterial community and enzyme activity to organic matter components under long-term fertilization on the Loess Plateau of China. Appl. Soil Ecol. 2021, 166, 103992. [Google Scholar] [CrossRef]
- Miao, F.; Li, Y.; Cui, S.; Jagadamma, S.; Yang, G.; Zhang, Q. Soil extracellular enzyme activities under long-term fertilization management in the croplands of China: A meta-analysis. Nutr. Cycl. Agroecosyst. 2019, 114, 125–138. [Google Scholar]
- Liu, H.; Lin, Y.H.; Zhang, Y.S.; Tan, X.X.; Wang, X.H. Effects of long-term fertilization on biodiversity and enzyme activity in grey desert soil. Acta Ecol. Sin. 2008, 28, 3898–3904. [Google Scholar]
- Xue, D.; Yao, H.; He, Z.; Huang, C. Relationshios between red soil enzyme activity and fertility. Chin. J. Appl. Ecol. 2005, 16, 1455–1458. [Google Scholar]
- Shao, X.; Zheng, J. Soil Organic Carbon, Black Carbon, and Enzyme Activity Under Long-Term Fertilization. J. Integr. Agric. 2014, 13, 517–524. [Google Scholar]
- Chang, N.; Zhai, Z.; Wang, L.; Deng, J. Impacts of nitrogen management and organic matter application on nitrous oxide emissions and soil organic carbon from spring maize fields in the North China Plain. Soil Tillage Res. 2020, 196, 104441. [Google Scholar]
- Guo, J.; Wang, Y.; Lai, J.; Pan, C.; Zhang, L. Spatiotemporal distribution of nitrogen biogeochemical processes in the coastal regions of northern Beibu Gulf, South China Sea. Chemosphere 2019, 239, 124803. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lee Burras, C.; Kravchenko, Y.S.; Duran, A.; Huffman, T.; Morras, H.; Studdert, G.; Zhang, X.; Cruse, R.M.; Yuan, X. Overview of Mollisols in the world: Distribution, land use and management. Can. J. Soil Sci. 2012, 92, 383–402. [Google Scholar] [CrossRef]
- Liu, B.; Xia, H.; Jiang, C.; Riaz, M.; Yang, L.; Chen, Y.; Fan, X.; Xia, X. 14 year applications of chemical fertilizers and crop straw effects on soil labile organic carbon fractions, enzyme activities and microbial community in rice-wheat rotation of middle China. Sci. Total Environ. 2022, 841, 156608. [Google Scholar] [CrossRef]
- Wang, R.; Wang, J.; Zhu, K.; Xiu, W.; Zhao, J.; Yang, D.; Li, G.; Liu, H. Abundance and diversity of diazotrophic nifH gene in the fluvo-aquic soil under different growth stages of maize. J. Tianjin Norm. Univ. (Nat. Sci. Ed.) 2018, 38, 35–41. [Google Scholar]
- Leptin, A.; Whitehead, D.; Anderson, C.R.; Cameron, K.C.; Lehto, N.J. Increased soil nitrogen supply enhances root-derived available soil carbon leading to reduced potential nitrification activity. Appl. Soil Ecol. 2021, 159, 103842. [Google Scholar] [CrossRef]
- Lyab, C.; Slabc, D.; Ygab, C.; Xia, X.; Jwab, C. Structure of rhizospheric microbial community and N cycling functional gene shifts with reduced N input in sugarcane-soybean intercropping in South China. Agric. Ecosyst. Environ. Pollut. 2021, 314, 107413. [Google Scholar]
- Yang, L.; Muhammad, I.; Chi, Y.X.; Liu, Y.X.; Wang, G.Y.; Wang, Y.; Zhou, X.B. Straw return and nitrogen fertilization regulate soil greenhouse gas emissions and global warming potential in dual maize cropping system. Sci. Total Environ. 2022, 853, 158370. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Gu, H.; Liu, J.; Wei, D.; Zhu, P.; Cui, X.; Zhou, B.; Chen, X.; Jin, J.; Liu, X.; et al. Metagenomics reveals divergent functional profiles of soil carbon and nitrogen cycling under long-term addition of chemical and organic fertilizers in the black soil region. Geoderma 2022, 418, 115846. [Google Scholar]
- Kelly, C.N.; Schwaner, G.W.; Cumming, J.R.; Driscoll, T.P. Metagenomic reconstruction of nitrogen and carbon cycling pathways in forest soil: Influence of different hardwood tree species. Soil Biol. Biochem. 2021, 156, 108226. [Google Scholar]
- Moormann, J.; Heinemann, B.; Hildebrandt, T.M. News about amino acid metabolism in plant–microbe interactions. Trends Biochem. Sci. 2022, 47, 839–850. [Google Scholar] [CrossRef]
- Xiao, W.; Zhang, Q.; Zhao, S.; Chen, D.; Gao, N.; Huang, M.; Ye, X. Citric acid secretion from rice roots contributes to reduction and immobilization of Cr(VI) by driving microbial sulfur and iron cycle in paddy soil. Sci. Total Environ. 2023, 854, 158832. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Cho, L.-H.; Tun, W.; Jeon, J.-S.; An, G. Sucrose signaling in higher plants. Plant Sci. 2021, 302, 110703. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lv, Y.; Lian, M.; Peng, F.; Xiao, Y. Effects of combined glycine and urea fertilizer application on the photosynthesis, sucrose metabolism, and fruit development of peach. Sci. Hortic. 2021, 289, 110504. [Google Scholar] [CrossRef]
- Tan, M.; Zong, R.; Lin, H.; Dhital, Y.P.; Ayantobo, O.O.; Chen, P.; Li, H.; Chen, R.; Wang, Z. Responses of soil nutrient and enzyme activities to long-term mulched drip irrigation (MDI) after the conversion of wasteland to cropland. Appl. Soil Ecol. 2023, 190, 104976. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, K.; Jiang, Q.; Liu, C.; Shan, J.; Teng, H. Mutual feeding mechanism of carbon, nitrogen and enzyme activity in Northeast China black soil under snow cover change. Appl. Soil Ecol. 2023, 190, 104991. [Google Scholar] [CrossRef]
- Sasse, J.; Martinoia, E.; Northen, T. Feed Your Friends: Do Plant Exudates Shape the Root Microbiome? Trends Plant Sci. 2018, 23, 25–41. [Google Scholar] [CrossRef] [Green Version]
- Wen, T.; Yu, G.-H.; Hong, W.-D.; Yuan, J.; Niu, G.-Q.; Xie, P.-H.; Sun, F.-S.; Guo, L.-D.; Kuzyakov, Y.; Shen, Q.-R. Root exudate chemistry affects soil carbon mobilization via microbial community reassembly. Fundam. Res. 2022, 2, 697–707. [Google Scholar] [CrossRef]
- Maurer, D.; Malique, F.; Alfarraj, S.; Albasher, G.; Horn, M.A.; Butterbach-Bahl, K.; Dannenmann, M.; Rennenberg, H. Interactive regulation of root exudation and rhizosphere denitrification by plant metabolite content and soil properties. Plant Soil 2021, 467, 107–127. [Google Scholar] [CrossRef]
- Wang, H.; Zheng, T.; Yang, X. Molecular ecology research progress for soil denitrification and research status for its infuencing factors. J. Agro-Environ. Sci. 2013, 32, 1915–1924. [Google Scholar]
- Liu, Y.; Chen, J.; Liu, Q.; Chen, L. Advances in studies of soil nitrification and denitrification and controlling factors. J. Sichuan For. Sci. Technol. 2006, 27, 36–41. [Google Scholar]
- Carrara, J.E.; Walter, C.A.; Hawkins, J.S.; Peterjohn, W.T.; Averill, C.; Brzostek, E.R. Interactions among plants, bacteria, and fungi reduce extracellular enzyme activities under long-term N fertilization. Glob. Chang. Biol. 2018, 24, 2721–2734. [Google Scholar] [CrossRef] [PubMed]
- Jiao, X.; Gao, C.; Lü, G.; Sui, Y. Effect of Long-Term Fertilization on Soil Enzyme Activities under Different Hydrothermal Conditions in Northeast China. Agric. Sci. China 2011, 10, 412–422. [Google Scholar] [CrossRef]

| Treatment | N Fertilizer kg/ha | P Fertilizer kg/ha | K Fertilizer kg/ha | Maize Straw kg/ha |
|---|---|---|---|---|
| CK | 0 | 0 | 0 | 0 |
| N | 150 | 0 | 0 | 0 |
| NPK | 150 | 75 | 75 | 0 |
| S | 0 | 0 | 0 | 5000 |
| PKS | 0 | 75 | 75 | 5000 |
| NPKS | 150 | 75 | 75 | 5000 |
| NITs U/g | NEA IU/mg | Nar IU/mg | Protease U/g | Urease U/g | |
|---|---|---|---|---|---|
| CK | 0.11 ± 0.00 b | 20.89 ± 1.95 e | 37.17 ± 3.37 a | 0.30 ± 0.01 d | 0.99 ± 0.01 c |
| N | 0.11 ± 0.01 b | 28.79 ± 0.75 c | 31.85 ± 0.88 b | 0.83 ± 0.17 c | 0.83 ± 0.02 d |
| NPK | 0.13 ± 0.00 a | 25.12 ± 0.62 d | 28.66 ± 0.27 c | 1.47 ± 0.19 b | 0.63 ± 0.00 e |
| S | 0.13 ± 0.00 a | 43.11 ± 4.59 a | 32.80 ± 1.36 b | 1.26 ± 0.02 b | 1.18 ± 0.09 a |
| PKS | 0.13 ± 0.00 a | 28.58 ± 3.31 b | 26.43 ± 0.37 c | 1.25 ± 0.09 b | 1.15 ± 0.01 a |
| NPKS | 0.13 ± 0.00 a | 35.94 ± 1.28 b | 27.52 ± 2.14 c | 1.78 ± 0.11 a | 1.06 ± 0.02 b |
| N Fixation | Denitrification | Nitrification | ANRA | DNRA | Anammox/ Hydroxylamine Oxidation | N Degradation | |
|---|---|---|---|---|---|---|---|
| CK | 25.99 ± 0.68 a | 17.79 ± 1.00 b | 1.26 ± 0.16 a | 16.98 ± 0.63 ab | 6.68 ± 0.51 b | 0.04 ± 0.00 b | 31.25 ± 0.80 a |
| N | 23.95 ± 2.20 a | 23.21 ± 3.52 a | 1.04 ± 0.32 ab | 15.63 ± 1.13 ab | 8.73 ± 1.28 a | 0.10 ± 0.03 a | 27.36 ± 0.73 b |
| NPK | 24.24 ± 1.20 a | 23.85 ± 1.68 a | 0.59 ± 0.21 c | 15.38 ± 0.83 b | 7.63 ± 0.66 ab | 0.08 ± 0.01 a | 28.24 ± 0.90 b |
| S | 24.53 ± 1.07 a | 17.71 ± 1.17 b | 0.93 ± 0.17 bc | 17.51 ± 0.99 a | 7.10 ± 0.72 ab | 0.02 ± 0.00 c | 32.20 ± 0.66 a |
| PKS | 24.47 ± 0.81 a | 18.34 ± 1.38 b | 1.09 ± 0.18 b | 17.16 ± 0.79 a | 6.99 ± 0.43 ab | 0.03 ± 0.01 bc | 31.93 ± 0.94 a |
| NPKS | 24.47 ± 1.69 a | 18.34 ± 2.32 b | 1.09 ± 0.14 b | 17.16 ± 0.05 ab | 6.99 ± 0.86 ab | 0.03 ± 0.01 bc | 31.93 ± 1.06 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Wang, C.; Wu, J.; Zhang, Y.; Li, Q.; Liu, S.; Gao, Y. The Effects of Localized Plant–Soil–Microbe Interactions on Soil Nitrogen Cycle in Maize Rhizosphere Soil under Long-Term Fertilizers. Agronomy 2023, 13, 2114. https://doi.org/10.3390/agronomy13082114
Li Y, Wang C, Wu J, Zhang Y, Li Q, Liu S, Gao Y. The Effects of Localized Plant–Soil–Microbe Interactions on Soil Nitrogen Cycle in Maize Rhizosphere Soil under Long-Term Fertilizers. Agronomy. 2023; 13(8):2114. https://doi.org/10.3390/agronomy13082114
Chicago/Turabian StyleLi, Yanan, Chengyu Wang, Junnan Wu, Yumang Zhang, Qi Li, Shuxia Liu, and Yunhang Gao. 2023. "The Effects of Localized Plant–Soil–Microbe Interactions on Soil Nitrogen Cycle in Maize Rhizosphere Soil under Long-Term Fertilizers" Agronomy 13, no. 8: 2114. https://doi.org/10.3390/agronomy13082114
APA StyleLi, Y., Wang, C., Wu, J., Zhang, Y., Li, Q., Liu, S., & Gao, Y. (2023). The Effects of Localized Plant–Soil–Microbe Interactions on Soil Nitrogen Cycle in Maize Rhizosphere Soil under Long-Term Fertilizers. Agronomy, 13(8), 2114. https://doi.org/10.3390/agronomy13082114






