Effect of Phosphorylation Sites Mutations on the Subcellular Localization and Activity of AGPase Bt2 Subunit: Implications for Improved Starch Biosynthesis in Maize
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Experiment Materials
2.2. iTRAQTM Labeling and Mass Spectrometry Analysis
2.3. Site-Directed Point Mutations
2.4. Subcellular Localization Analysis
2.5. Yeast Two-Hybrid Assay
2.6. Bimolecular Fluorescence Complementation
2.7. Protoplasm Preparation
2.8. Determination of AGPase Activity
3. Results
3.1. iTRAQTM Identification of Bt2 Phosphorylation Sites
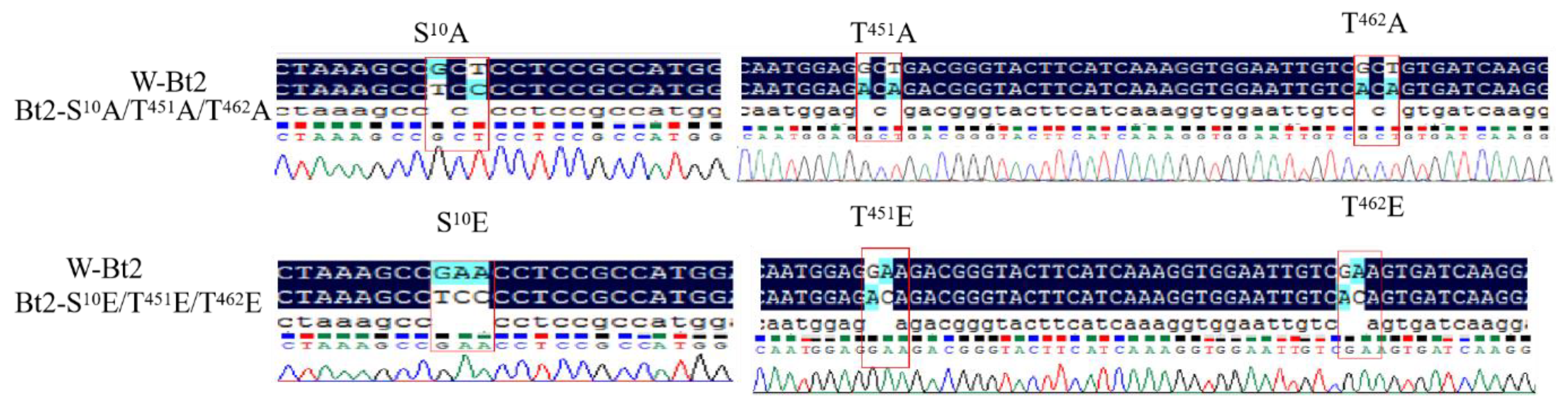
3.2. Site-Directed Point Mutations
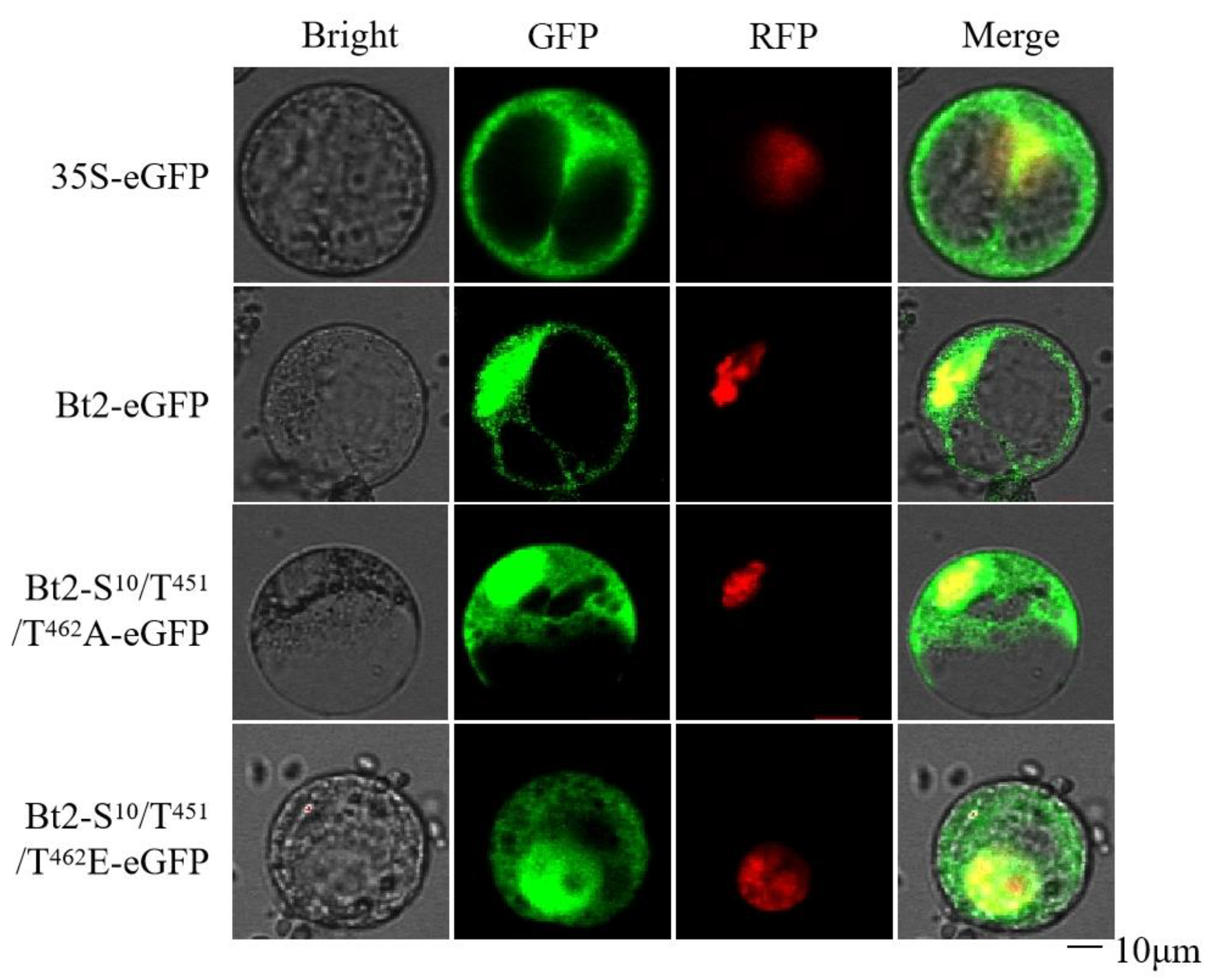
3.3. Mutations Effects on Subcellular Localization of AGPase Small Subunit Bt2
3.4. Interaction between Bt2 with Phosphorylation Sites Mutations and Sh2
3.5. Bimolecular Fluorescence Complementation Confirms Bt2 Interaction with Sh2
3.6. Effects of Site-Directed Point Mutations on AGPase Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Myburgh, J.A.; Finfer, S.; Bellomo, R.; Billot, L.; Cass, A.; Gattas, D.; Glass, P.; Lipman, J.; Liu, B.; McArthur, C. Hydroxyethyl starch or saline for fluid resuscitation in intensive care. N. Engl. J. Med. 2012, 367, 1901–1911. [Google Scholar] [CrossRef]
- Mojović, L.; Pejin, D.; Grujić, O.; Markov, S.; Pejin, J.; Rakin, M.; Vukašinović, M.; Nikolić, S.; Savić, D. Progress in the production of bioethanol on starch-based feedstocks. Chem. Ind. Chem. Eng. Q. 2009, 15, 211–226. [Google Scholar] [CrossRef]
- Asharuddin, S.M.; Othman, N.; Altowayti, W.A.H.; Bakar, N.A.; Hassan, A. Recent advancement in starch modification and its application as water treatment agent. Environ. Technol. Innov. 2021, 23, 101637. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, Y. Advantages Analysis of Corn Planting in China. J. Agric. Sci. Technol. China 2018, 20, 1–9. [Google Scholar]
- Willett, J. Starch in polymer compositions. In Starch; Elsevier: Amsterdam, The Netherlands, 2009; pp. 715–743. [Google Scholar]
- Shoaib, N.; Liu, L.; Ali, A.; Mughal, N.; Yu, G.; Huang, Y. Molecular Functions and Pathways of Plastidial Starch Phosphorylase (PHO1) in Starch Metabolism: Current and Future Perspectives. Int. J. Mol. Sci. 2021, 22, 10450. [Google Scholar] [CrossRef] [PubMed]
- Boehlein, S.K.; Shaw, J.R.; Stewart, J.D.; Hannah, L.C. Studies of the kinetic mechanism of maize endosperm ADP-glucose pyrophosphorylase uncovered complex regulatory properties. Plant Physiol. 2010, 152, 1056–1064. [Google Scholar] [CrossRef]
- Crofts, N.; Sugimoto, K.; Oitome, N.F.; Nakamura, Y.; Fujita, N. Differences in specificity and compensatory functions among three major starch synthases determine the structure of amylopectin in rice endosperm. Plant Mol. Biol. 2017, 94, 399–417. [Google Scholar] [CrossRef] [PubMed]
- Tickle, P.; Burrell, M.M.; Coates, S.A.; Emes, M.J.; Tetlow, I.J.; Bowsher, C.G. Characterization of plastidial starch phosphorylase in Triticum aestivum L. endosperm. J. Plant Physiol. 2009, 166, 1465–1478. [Google Scholar] [CrossRef] [PubMed]
- Satoh, H.; Shibahara, K.; Tokunaga, T.; Nishi, A.; Tasaki, M.; Hwang, S.K.; Okita, T.W.; Kaneko, N.; Fujita, N.; Yoshida, M.; et al. Mutation of the plastidial alpha-glucan phosphorylase gene in rice affects the synthesis and structure of starch in the endosperm. Plant Cell 2008, 20, 1833–1849. [Google Scholar] [CrossRef]
- Grimaud, F.; Rogniaux, H.; James, M.G.; Myers, A.M.; Planchot, V. Proteome and phosphoproteome analysis of starch granule-associated proteins from normal maize and mutants affected in starch biosynthesis. J. Exp. Bot. 2008, 59, 3395–3406. [Google Scholar] [CrossRef]
- Tetlow, I.J.; Beisel, K.G.; Cameron, S.; Makhmoudova, A.; Liu, F.; Bresolin, N.S.; Wait, R.; Morell, M.K.; Emes, M.J. Analysis of protein complexes in wheat amyloplasts reveals functional interactions among starch biosynthetic enzymes. Plant Physiol. 2008, 146, 1878–1891. [Google Scholar] [CrossRef]
- Hannah, L.C.; Shaw, J.R.; Giroux, M.J.; Reyss, A.; Prioul, J.-L.; Bae, J.-M.; Lee, J.-Y. Maize genes encoding the small subunit of ADP-glucose pyrophosphorylase. Plant Physiol. 2001, 127, 173–183. [Google Scholar] [CrossRef]
- Corbi, J.; Debieu, M.; Rousselet, A.; Montalent, P.; Le Guilloux, M.; Manicacci, D.; Tenaillon, M. Contrasted patterns of selection since maize domestication on duplicated genes encoding a starch pathway enzyme. Theor. Appl. Genet. 2011, 122, 705–722. [Google Scholar] [CrossRef]
- Cakir, B.; Tuncel, A.; Green, A.R.; Koper, K.; Hwang, S.-K.; Okita, T.W.; Kang, C. Substrate binding properties of potato tuber ADP-glucose pyrophosphorylase as determined by isothermal titration calorimetry. FEBS Lett. 2015, 589, 1444–1449. [Google Scholar] [CrossRef] [PubMed]
- Comparot-Moss, S.; Denyer, K. The evolution of the starch biosynthetic pathway in cereals and other grasses. J. Exp. Bot. 2009, 60, 2481–2492. [Google Scholar] [CrossRef]
- Bowsher, C.G.; Scrase-Field, E.F.; Esposito, S.; Emes, M.J.; Tetlow, I.J. Characterization of ADP-glucose transport across the cereal endosperm amyloplast envelope. J. Exp. Bot. 2007, 58, 1321–1332. [Google Scholar] [CrossRef]
- Kirchberger, S.; Leroch, M.; Huynen, M.A.; Wahl, M.; Neuhaus, H.E.; Tjaden, J. Molecular and biochemical analysis of the plastidic ADP-glucose transporter (ZmBT1) from Zea mays. J. Biol. Chem. 2007, 282, 22481–22491. [Google Scholar] [CrossRef] [PubMed]
- Tuncel, A. Allosteric Regulation of the Rice Endosperm ADP-Glucose Pyrophosphorylase; Washington State University: Washington, DC, USA, 2013. [Google Scholar]
- Geigenberger, P. Regulation of sucrose to starch conversion in growing potato tubers. J. Exp. Bot. 2003, 54, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Mangelsen, E.; Wanke, D.; Kilian, J.; Sundberg, E.; Harter, K.; Jansson, C. Significance of light, sugar, and amino acid supply for diurnal gene regulation in developing barley caryopses. Plant Physiol. 2010, 153, 14–33. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.M.; Fulton, D.C.; Chia, T.; Thorneycroft, D.; Chapple, A.; Dunstan, H.; Hylton, C.; Zeeman, S.C.; Smith, A.M. Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol. 2004, 136, 2687–2699. [Google Scholar] [CrossRef] [PubMed]
- Nakagami, H.; Sugiyama, N.; Mochida, K.; Daudi, A.; Yoshida, Y.; Toyoda, T.; Tomita, M.; Ishihama, Y.; Shirasu, K. Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 2010, 153, 1161–1174. [Google Scholar] [CrossRef]
- Rose, C.M.; Venkateshwaran, M.; Volkening, J.D.; Grimsrud, P.A.; Maeda, J.; Bailey, D.J.; Park, K.; Howes-Podoll, M.; den Os, D.; Yeun, L.H. Rapid phosphoproteomic and transcriptomic changes in the rhizobia-legume symbiosis. Mol. Cell. Proteom. 2012, 11, 724–744. [Google Scholar] [CrossRef] [PubMed]
- Dong, K.; Zhen, S.; Cheng, Z.; Cao, H.; Ge, P.; Yan, Y. Proteomic analysis reveals key proteins and phosphoproteins upon seed germination of wheat (Triticum aestivum L.). Front. Plant Sci. 2015, 6, 1017. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Lv, Y.; Shen, L.; Wang, Y.; Qing, Y.; Wu, N.; Li, Y.; Huang, H.; Zhang, N.; Liu, Y.; et al. The Proteomic Analysis of Maize Endosperm Protein Enriched by Phos-tag(tm) Reveals the Phosphorylation of Brittle-2 Subunit of ADP-Glc Pyrophosphorylase in Starch Biosynthesis Process. Int. J. Mol. Sci. 2019, 20, 986. [Google Scholar] [CrossRef]
- Ferrero, D.M.; Piattoni, C.V.; Asencion Diez, M.D.; Rojas, B.E.; Hartman, M.D.; Ballicora, M.A.; Iglesias, A.A. Phosphorylation of ADP-glucose pyrophosphorylase during wheat seeds development. Front. Plant Sci. 2020, 11, 1058. [Google Scholar] [CrossRef] [PubMed]
- Wiese, S.; Reidegeld, K.A.; Meyer, H.E.; Warscheid, B. Protein labeling by iTRAQ: A new tool for quantitative mass spectrometry in proteome research. Proteomics 2007, 7, 340–350. [Google Scholar] [CrossRef] [PubMed]
- Yokoe, H.; Meyer, T. Spatial dynamics of GFP-tagged proteins investigated by local fluorescence enhancement. Nat. Biotechnol. 1996, 14, 1252–1256. [Google Scholar] [CrossRef]
- Paiano, A.; Margiotta, A.; De Luca, M.; Bucci, C. Yeast two-hybrid assay to identify interacting proteins. Curr. Protoc. Protein Sci. 2019, 95, e70. [Google Scholar] [CrossRef] [PubMed]
- Bahaji, A.; Li, J.; Sánchez-López, Á.M.; Baroja-Fernández, E.; Muñoz, F.J.; Ovecka, M.; Almagro, G.; Montero, M.; Ezquer, I.; Etxeberria, E. Starch biosynthesis, its regulation and biotechnological approaches to improve crop yields. Biotechnol. Adv. 2014, 32, 87–106. [Google Scholar] [CrossRef]
- Cross, J.M.; Clancy, M.; Shaw, J.R.; Greene, T.W.; Schmidt, R.R.; Okita, T.W.; Hannah, L.C. Both subunits of ADP-glucose pyrophosphorylase are regulatory. Plant Physiol. 2004, 135, 137–144. [Google Scholar] [CrossRef]
- Greene, T.W.; Hannah, L.C. Enhanced stability of maize endosperm ADP-glucose pyrophosphorylase is gained through mutants that alter subunit interactions. Proc. Natl. Acad. Sci. USA 1998, 95, 13342–13347. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhang, S.; Zhao, Y.; Li, B.; Zhang, J. Over-expression of AGPase genes enhances seed weight and starch content in transgenic maize. Planta 2011, 233, 241–250. [Google Scholar] [CrossRef]
- Stark, D.M.; Timmerman, K.P.; Barry, G.F.; Preiss, J.; Kishore, G.M. Regulation of the amount of starch in plant tissues by ADP glucose pyrophosphorylase. Science 1992, 258, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-K.; Hwang, S.-K.; Han, M.; Eom, J.-S.; Kang, H.-G.; Han, Y.; Choi, S.-B.; Cho, M.-H.; Bhoo, S.H.; An, G. Identification of the ADP-glucose pyrophosphorylase isoforms essential for starch synthesis in the leaf and seed endosperm of rice (Oryza sativa L.). Plant Mol. Biol. 2007, 65, 531–546. [Google Scholar] [CrossRef]
- Rogowsky, P. Transcriptional and Metabolic Adjustments in AGPase Deficient bt2 Maize Kernels; American Society of Plant Biologists: Rockville, MD, USA, 2008. [Google Scholar]
- Koper, K. Glutamic Acid 358 Is Important for the Normal Allosteric Function and Heterotetramer Formation of Potato Adp-Glucose Pyrophosphorylase. Master’s Thesis, Fen Bilimleri Enstitüsü, Ankara, Turkey, 2013. [Google Scholar]
- Cohen, P. The regulation of protein function by multisite phosphorylation—A 25 year update. Trends Biochem. Sci. 2000, 25, 596–601. [Google Scholar] [CrossRef]
- Pysh, L.D.; Aukerman, M.J.; Schmidt, R.J. OHP1: A maize basic domain/leucine zipper protein that interacts with opaque2. Plant Cell 1993, 5, 227–236. [Google Scholar] [PubMed]
- Liu, X.; Wang, Z.; Wang, L.; Wu, R.; Phillips, J.; Deng, X. LEA 4 group genes from the resurrection plant Boea hygrometrica confer dehydration tolerance in transgenic tobacco. Plant Sci. 2009, 176, 90–98. [Google Scholar] [CrossRef]
- Zhang, X.; Dong, K.; Xu, K.; Zhang, K.; Jin, X.; Yang, M.; Zhang, Y.; Wang, X.; Han, C.; Yu, J. Barley stripe mosaic virus infection requires PKA-mediated phosphorylation of γb for suppression of both RNA silencing and the host cell death response. New Phytol. 2018, 218, 1570–1585. [Google Scholar] [CrossRef]
- Pooler, A.M.; Usardi, A.; Evans, C.J.; Philpott, K.L.; Noble, W.; Hanger, D.P. Dynamic association of tau with neuronal membranes is regulated by phosphorylation. Neurobiol. Aging 2012, 33, 431.e27–431.e38. [Google Scholar] [CrossRef]
- Tetlow, I.J.; Wait, R.; Lu, Z.; Akkasaeng, R.; Bowsher, C.G.; Esposito, S.; Kosar-Hashemi, B.; Morell, M.K.; Emes, M.J. Protein phosphorylation in amyloplasts regulates starch branching enzyme activity and protein-protein interactions. Plant Cell 2004, 16, 694–708. [Google Scholar] [CrossRef]
- Yu, G.; Shoaib, N.; Xie, Y.; Liu, L.; Mughal, N.; Li, Y.; Huang, H.; Zhang, N.; Zhang, J.; Liu, Y. Comparative Study of Starch Phosphorylase Genes and Encoded Proteins in Various Monocots and Dicots with Emphasis on Maize. Int. J. Mol. Sci. 2022, 23, 4518. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Gaoyang, Y.; Liu, L.; Shoaib, N.; Deng, Y.; Zhang, N.; Li, Y.; Huang, Y. The Structure, Function, and Regulation of Starch Synthesis Enzymes SSIII with Emphasis on Maize. Agronomy 2022, 12, 1359. [Google Scholar] [CrossRef]
- Figueroa, C.M.; Asencion Diez, M.D.; Ballicora, M.A.; Iglesias, A.A. Structure, function, and evolution of plant ADP-glucose pyrophosphorylase. Plant Mol. Biol. 2022, 108, 307–323. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, G.; Shoaib, N.; Yang, Y.; Liu, L.; Mughal, N.; Mou, Y.; Huang, Y. Effect of Phosphorylation Sites Mutations on the Subcellular Localization and Activity of AGPase Bt2 Subunit: Implications for Improved Starch Biosynthesis in Maize. Agronomy 2023, 13, 2119. https://doi.org/10.3390/agronomy13082119
Yu G, Shoaib N, Yang Y, Liu L, Mughal N, Mou Y, Huang Y. Effect of Phosphorylation Sites Mutations on the Subcellular Localization and Activity of AGPase Bt2 Subunit: Implications for Improved Starch Biosynthesis in Maize. Agronomy. 2023; 13(8):2119. https://doi.org/10.3390/agronomy13082119
Chicago/Turabian StyleYu, Guowu, Noman Shoaib, Yang Yang, Lun Liu, Nishbah Mughal, Yuewei Mou, and Yubi Huang. 2023. "Effect of Phosphorylation Sites Mutations on the Subcellular Localization and Activity of AGPase Bt2 Subunit: Implications for Improved Starch Biosynthesis in Maize" Agronomy 13, no. 8: 2119. https://doi.org/10.3390/agronomy13082119
APA StyleYu, G., Shoaib, N., Yang, Y., Liu, L., Mughal, N., Mou, Y., & Huang, Y. (2023). Effect of Phosphorylation Sites Mutations on the Subcellular Localization and Activity of AGPase Bt2 Subunit: Implications for Improved Starch Biosynthesis in Maize. Agronomy, 13(8), 2119. https://doi.org/10.3390/agronomy13082119






