Genetic Diversity of the Fall Armyworm Spodoptera frugiperda (J.E. Smith) in the Democratic Republic of the Congo
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of FAW Samples
2.2. DNA Extraction
2.3. PCR Amplification and Sequence Analysis
2.4. DNA Polymorphism Analysis
2.5. Haplotype Network Plot and Phylogenetic Analysis
2.6. Analysis of Molecular Variance (AMOVA)
3. Results
3.1. PCR Amplification and Sequence Analysis
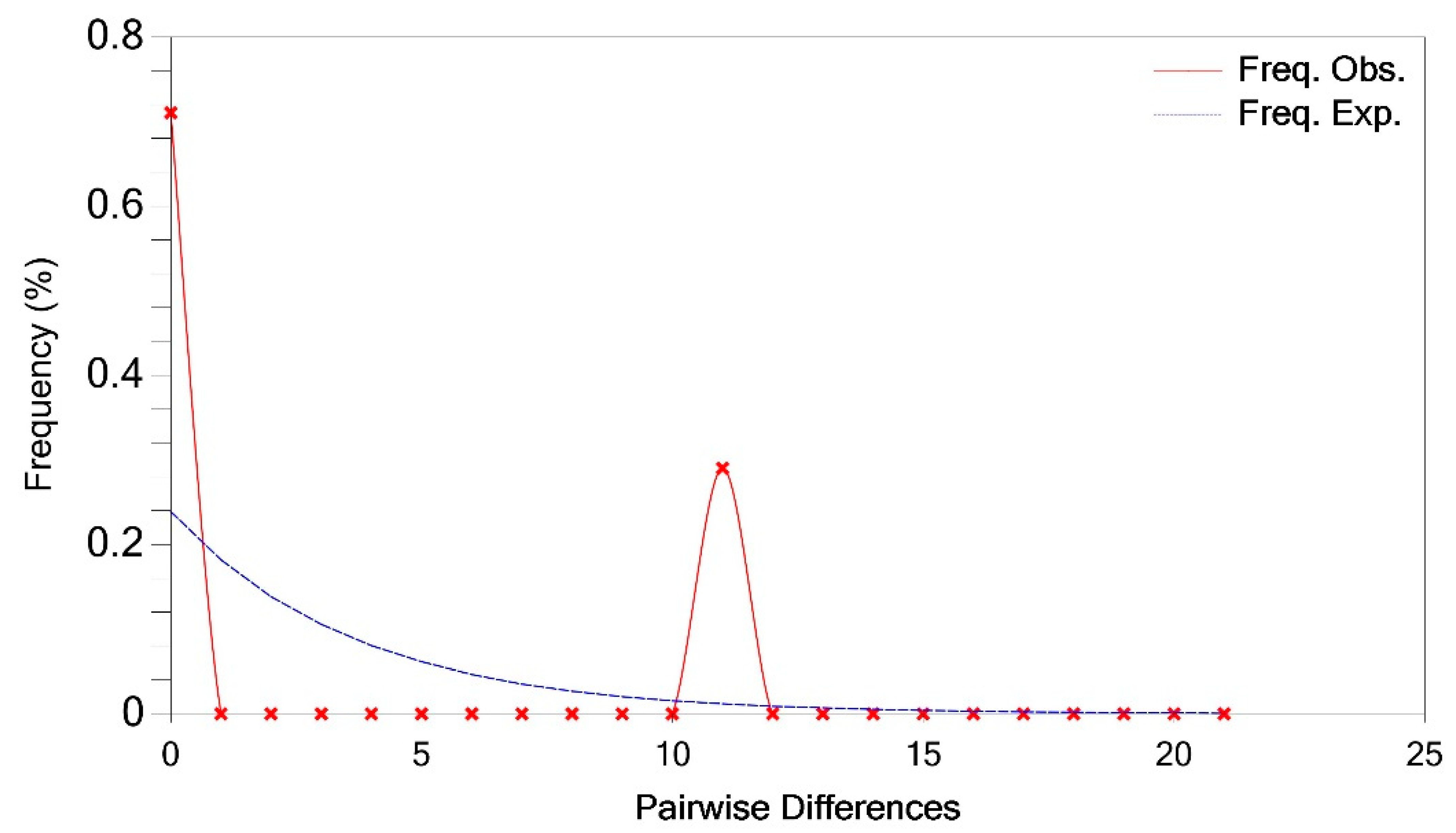
3.2. Polymorphism Analysis
3.3. Comparative Genetic Analyses of the FAW Population in the DRC and Three Geographic Regions
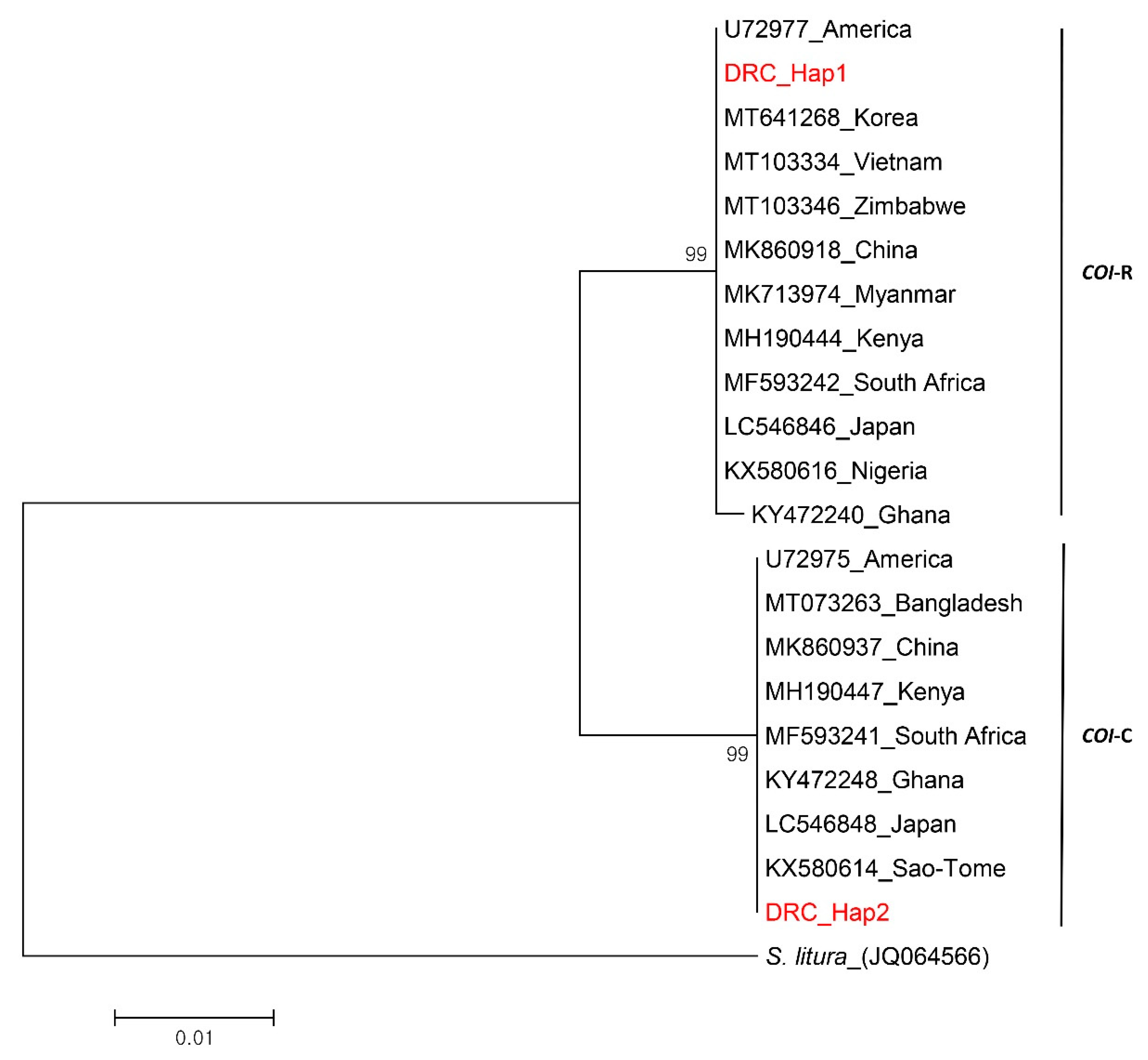
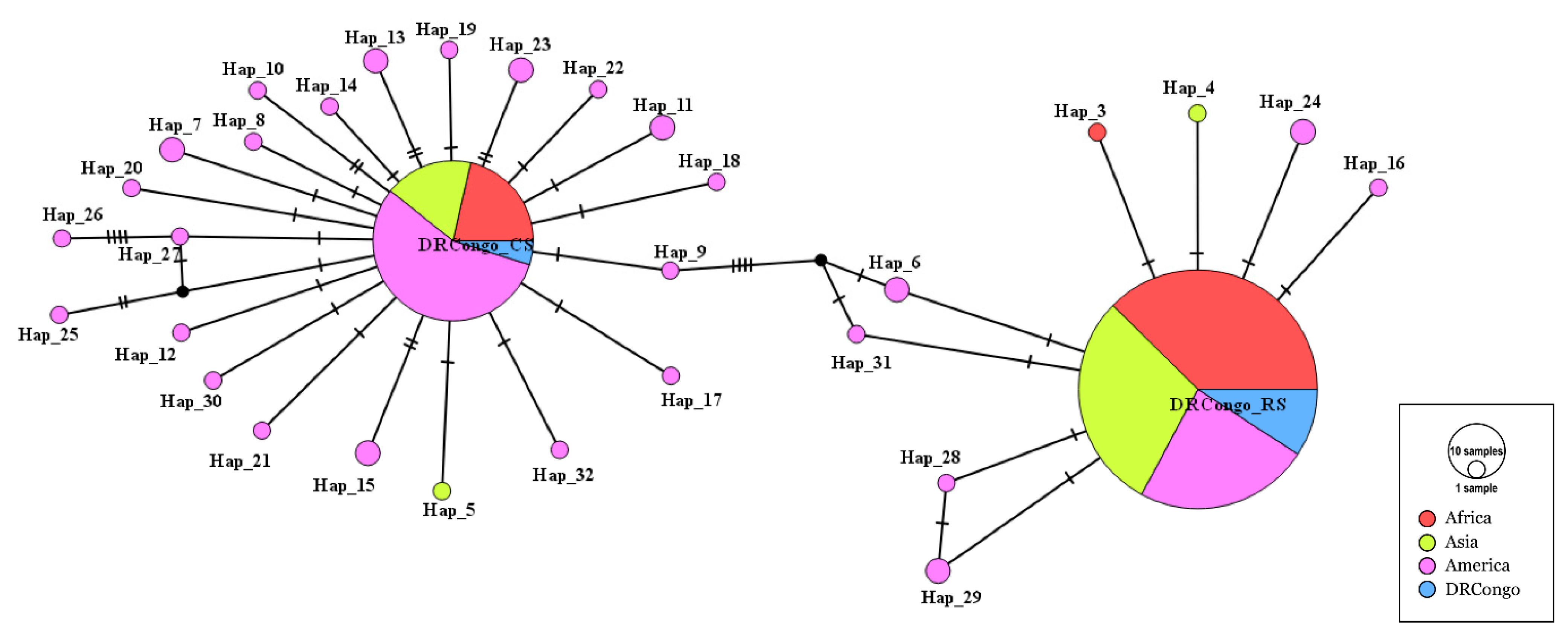
3.4. Comparative Phylogenetic and Haplotype Network Analysis
3.5. Population Structure of FAW
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| (A) COI gene sequences from America | |||
| No. | GenBank Accession | Location | Year Submitted |
| 1. | KX281221.1 | Canada | 2017 |
| 2. | U72978.1 | USA | 1996 |
| 3. | U72977.1 | USA | 1996 |
| 4. | U72976.1 | USA | 1996 |
| 5. | U72975.1 | USA | 1996 |
| 6. | U72974.1 | USA | 1996 |
| 7. | KT809294.1 | Brazil | 2018 |
| 8. | KT809293.1 | Brazil | 2018 |
| 9. | KT809292.1 | Brazil | 2018 |
| 10. | KT809291.1 | Brazil | 2018 |
| 11. | KT809290.1 | Brazil | 2018 |
| 12. | KT809289.1 | Brazil | 2018 |
| 13. | KT809288.1 | Brazil | 2018 |
| 14. | KT809287.1 | Brazil | 2018 |
| 15. | KT809286.1 | Brazil | 2018 |
| 16. | KT809285.1 | Brazil | 2018 |
| 17. | KT809284.1 | Brazil | 2018 |
| 18. | KT809283.1 | Brazil | 2018 |
| 19. | KT809282.1 | Brazil | 2018 |
| 20. | KT809281.1 | Brazil | 2018 |
| 21 | KT809280.1 | Brazil | 2018 |
| 22. | KT809279.1 | Brazil | 2018 |
| 23. | KT809278.1 | Brazil | 2018 |
| 24. | KT809277.1 | Brazil | 2018 |
| 25. | KT809276.1 | Brazil | 2018 |
| 26. | KT809275.1 | Brazil | 2018 |
| 27. | KT809274.1 | Brazil | 2018 |
| 28. | KT809273.1 | Brazil | 2018 |
| 29. | KT809272.1 | Brazil | 2018 |
| 30. | KT809271.1 | Brazil | 2018 |
| 31. | KT809270.1 | Brazil | 2018 |
| 32. | KT809269.1 | Brazil | 2018 |
| 33. | KT809268.1 | Brazil | 2018 |
| 34. | KT809267.1 | Brazil | 2018 |
| 35. | KT809266.1 | Brazil | 2018 |
| 36. | KT809265.1 | Brazil | 2018 |
| 37. | KT809264.1 | Brazil | 2018 |
| 38. | KT809263.1 | Brazil | 2018 |
| 39. | KT809262.1 | Brazil | 2018 |
| 40. | KT809261.1 | Brazil | 2018 |
| 41. | KT809260.1 | Brazil | 2018 |
| 42. | KT809259.1 | Brazil | 2018 |
| 43. | KT809258.1 | Brazil | 2018 |
| 44. | KT809257.1 | Brazil | 2018 |
| 45. | KT809256.1 | Brazil | 2018 |
| 46. | KT809255.1 | Brazil | 2018 |
| 47. | KT809254.1 | Brazil | 2018 |
| 48. | KT809253.1 | Brazil | 2018 |
| 49. | KT809252.1 | Brazil | 2018 |
| 50. | KT809251.1 | Brazil | 2018 |
| 51. | KT809250.1 | Brazil | 2018 |
| 52. | KT809249.1 | Brazil | 2018 |
| 53. | KT809248.1 | Brazil | 2018 |
| 54. | KT809247.1 | Brazil | 2018 |
| 55. | KT809246.1 | Brazil | 2018 |
| 56. | KT809245.1 | Brazil | 2018 |
| 57. | KT809244.1 | Brazil | 2018 |
| 58. | KT809243.1 | Brazil | 2018 |
| 59. | KT809242.1 | Brazil | 2018 |
| 60. | KT809241.1 | Brazil | 2018 |
| 61. | KT809240.1 | Brazil | 2018 |
| 62. | KT809239.1 | Brazil | 2018 |
| 63. | KT809238.1 | Brazil | 2018 |
| 64. | KT809237.1 | Brazil | 2018 |
| 65. | KT809236.1 | Brazil | 2018 |
| 66. | KT809235.1 | Brazil | 2018 |
| 67. | KJ634298.1 | Suriname | 2014 |
| 68. | KJ634297.1 | Honduras | 2014 |
| 69. | MK318422.1 | Mexico | 2019 |
| 70. | MK318420.1 | Mexico | 2019 |
| 71. | MK318377.1 | Puerto Rico | 2019 |
| 72. | MK318373.1 | Puerto Rico | 2019 |
| 73. | MK318372.1 | Mexico | 2019 |
| 74. | MK318311.1 | Mexico | 2019 |
| 75. | MK318297.1 | Dominican | 2019 |
| 76. | GU439151.1 | Ontario | 2018 |
| 77. | GU439150.1 | Puslinch | 2018 |
| 78. | GU439149.1 | Puslinch | 2018 |
| 79. | GU439148.1 | Puslinch | 2018 |
| 80. | GU439147.1 | Puslinch | 2018 |
| 81. | GU090724.1 | Puslinch | 2018 |
| 82. | GU090723.1 | Puslinch | 2018 |
| 83. | GU095403.1 | New Brunswick | 2018 |
| 84. | GU094756.1 | Puslinch | 2018 |
| 85. | GU094755.1 | Puslinch | 2018 |
| 86. | GU094754.1 | Puslinch | 2018 |
| 87. | KJ388147.1 | Quebec | 2018 |
| 88. | HM102314.1 | USA | 2016 |
| 89. | KJ641998.1 | Guano | 2015 |
| 90. | KJ641997.1 | Guano | 2015 |
| 91. | KF624877.1 | Roraima | 2014 |
| 92. | KF624876.1 | Roraima | 2014 |
| 93. | JQ559528.1 | Costa Rica | 2012 |
| 94. | JQ554012.1 | Costa Rica | 2012 |
| 95. | JQ572603.1 | Costa Rica | 2012 |
| 96. | JQ571459.1 | Costa Rica | 2012 |
| 97. | JQ547900.1 | Costa rica | 2012 |
| 98. | JQ577923.1 | Costa Rica | 2012 |
| 99. | JF854747.1 | Campina Grande | 2012 |
| 100. | JF854746.1 | Morretes | 2012 |
| 101. | JF854745.1 | Morretes | 2012 |
| 102. | JF854744.1 | Campina Grande | 2012 |
| 103. | JF854743.1 | Morretes | 2012 |
| 104. | JF854741.1 | Morretes | 2012 |
| 105. | JF854740.1 | Morretes | 2012 |
| 106. | HQ964527.1 | Massachusetts | 2012 |
| 107. | HQ964487.1 | Massachusetts | 2012 |
| 108. | HQ964486.1 | Massachusetts | 2012 |
| 109. | HQ964485.1 | Massachusetts | 2012 |
| 110. | HQ964443.1 | Massachusetts | 2012 |
| 111. | HQ964441.1 | Massachusetts | 2012 |
| 112. | HQ964442.1 | Massachusetts | 2012 |
| 113. | HQ964440.1 | Massachusetts | 2012 |
| 114. | HQ964439.1 | Massachusetts | 2012 |
| 115. | HQ964394.1 | Massachusetts | 2012 |
| 116. | HQ964393.1 | Massachusetts | 2012 |
| 117. | HQ964352.1 | Massachusetts | 2012 |
| 118. | HQ964351.1 | Massachusetts | 2012 |
| 119. | GU159435.1 | Costa Rica | 2012 |
| 120. | GU159434.1 | Costa Rica | 2012 |
| 121. | GU159433.1 | Costa Rica | 2012 |
| 122. | GU159432.1 | Costa Rica | 2012 |
| 123. | GU159431.1 | Costa Rica | 2012 |
| 124. | GU159430.1 | Costa Rica | 2012 |
| 125. | GU159429.1 | Costa Rica | 2012 |
| 126. | GU658451.1 | Alvaro Obregon | 2019 |
| (B) COI gene sequences from Africa | |||
| No. | GenBank Accession | Location | Year Submitted |
| 1. | MF593258.1 | South Africa | 2018 |
| 2. | MF593257.1 | South Africa | 2018 |
| 3. | MF593256.1 | South Africa | 2018 |
| 4. | MF593255.1 | South Africa | 2018 |
| 5. | MF593254.1 | South Africa | 2018 |
| 6. | MF593253.1 | South Africa | 2018 |
| 7. | MF593252.1 | South Africa | 2018 |
| 8. | MF593251.1 | South Africa | 2018 |
| 9. | MF593250.1 | South Africa | 2018 |
| 10. | MF593249.1 | South Africa | 2018 |
| 11. | MF593248.1 | South Africa | 2018 |
| 12. | MF593247.1 | South Africa | 2018 |
| 13. | MF593246.1 | South Africa | 2018 |
| 14. | MF593245.1 | South Africa | 2018 |
| 15. | MF593244.1 | South Africa | 2018 |
| 16. | MF593243.1 | South Africa | 2018 |
| 17. | MF593242.1 | South Africa | 2018 |
| 18. | MF593241.1 | South Africa | 2018 |
| 19 | MK493020.1 | South Africa | 2019 |
| 20. | MK493019.1 | South Africa | 2019 |
| 21. | MK493018.1 | South Africa | 2019 |
| 22. | MK493017.1 | South Africa | 2019 |
| 23. | MK493016.1 | South Africa | 2019 |
| 24. | MT933058 | Tanzania | 2020 |
| MT103348 | Tanzania | ||
| 25. | MT103346.1 | Zimbabwe | 2020 |
| MT103347 | Zimbabwe | ||
| 26. | KX580619.1 | Nigeria | 2016 |
| 27. | KX580618.1 | Nigeria | 2016 |
| 28. | KX580617.1 | Nigeria | 2016 |
| 29. | KX580616.1 | Nigeria | 2016 |
| 30. | KX580615.1 | Sao-Tome, | 2016 |
| 31. | KX580614.1 | Sao-Tome | 2016 |
| 32. | MT641267.1 | Uganda | 2020 |
| 33. | MF278659.1 | Tanzania | 2018 |
| 34. | MF278658.1 | Tanzania | 2018 |
| 35. | MF278657.1 | Tanzania | 2018 |
| 36. | MH190448.1 | Kenya | 2018 |
| 37. | MH190447.1 | Kenya | 2018 |
| 38. | MH190446.1 | Kenya | 2018 |
| 39. | MH190445.1 | Kenya | 2018 |
| 40. | MH190444.1 | Kenya | 2018 |
| 41. | KY472255.1 | Ghana | 2017 |
| 42. | KY472254.1 | Ghana | 2017 |
| 43. | KY472253.1 | Ghana | 2017 |
| 44. | KY472252.1 | Ghana | 2017 |
| 45. | KY472251.1 | Ghana | 2017 |
| 46. | KY472250.1 | Ghana | 2017 |
| 47. | KY472249.1 | Ghana | 2017 |
| 48. | KY472248.1 | Ghana | 2017 |
| 49. | KY472245.1 | Ghana | 2017 |
| 50. | KY472244.1 | Ghana | 2017 |
| 51. | KY472242.1 | Ghana | 2017 |
| 52. | KY472241.1 | Ghana | 2017 |
| 53. | KY472240.1 | Ghana | 2017 |
| 54. | MG993205.1 | Malawi: Sande | 2018 |
| 55. | MF197867.1 | Uganda | 2018 |
| 56. | MK493006.1 | Kenya | 2019 |
| 57. | MK493000.1 | Kenya | 2019 |
| 58. | MK492996.1 | Kenya | 2019 |
| 59. | MK493010.1 | Kenya | 2019 |
| 60. | MK493009.1 | Kenya | 2019 |
| 61. | MK493008.1 | Kenya | 2019 |
| 62. | MK493007.1 | Kenya | 2019 |
| 63. | MK493004.1 | Kenya | 2019 |
| 64. | MK493003.1 | Kenya | 2019 |
| 65. | MK493002.1 | Kenya | 2019 |
| 66. | MK493001.1 | Kenya | 2019 |
| 67. | MK492999.1 | Kenya | 2019 |
| 68. | MK492998.1 | Kenya | 2019 |
| 69. | MK492997.1 | Kenya | 2019 |
| 70. | MK492995.1 | Kenya | 2019 |
| 71. | MK492994.1 | Kenya | 2019 |
| 72. | MK492993.1 | Kenya | 2019 |
| 73. | MK492992.1 | Kenya | 2019 |
| 74. | MK492991.1 | Kenya | 2019 |
| 75. | MK492990.1 | Kenya | 2019 |
| 76. | MK492989.1 | Kenya | 2019 |
| 77. | MK492988.1 | Kenya | 2019 |
| 78. | MK492987.1 | Kenya | 2019 |
| 79. | MK492986.1 | Kenya | 2019 |
| 80. | MK492985.1 | Kenya | 2019 |
| 81. | MK492984.1 | Kenya | 2019 |
| 82. | MK492983.1 | Kenya | 2019 |
| 83. | MK492982.1 | Kenya | 2019 |
| 84. | MK492981.1 | Kenya | 2019 |
| 85 | MK492972.1 | Uganda | 2018 |
| 86 | MK492971.1 | Uganda | |
| 87 | MK492970.1 | Uganda | 2022 |
| 88 | MK492969.1 | Uganda | 2022 |
| 89 | MK492958.1 | Tanzania | 2020 |
| (C) COI gene sequences from Asia | |||
| No. | GenBank Accession | Location | Year Submitted |
| 1. | MT103344.1 | Bangladesh: Dhaka | 2020 |
| 2. | MT103343.1 | Bangladesh: Dhaka | 2020 |
| 3. | MT103342.1 | South Korea: Gyeongsan | 2020 |
| 4. | MT103341.1 | Viet Nam: Ninh binh | 2020 |
| 5. | MT103340.1 | Viet Nam: Ninh binh | 2020 |
| 6. | MT103339.1 | Viet Nam: Ha noi | 2020 |
| 7. | MT103338.1 | Viet Nam: Vinh phuc | 2020 |
| 8. | MT103336.1 | Viet Nam: Hanoi | 2020 |
| 9. | MT103335.1 | Viet Nam: Vinh Phuc | 2020 |
| 10. | MT103334.1 | Viet Nam: Ninh Binh | 2020 |
| 11. | MT641270.1 | South Korea: Gyeongsan | 2020 |
| 12. | MT641269.1 | South Korea: Jeju | 2020 |
| 13. | MT641268.1 | South Korea: Campus | 2020 |
| 14. | LC546868.1 | Japan: Aomori | 2020 |
| 15. | LC546867.1 | Japan: Aomori | 2020 |
| 16. | LC546866.1 | Japan: Iwate | 2020 |
| 17. | LC546865.1 | Japan: Kanagawa | 2020 |
| 18. | LC546864.1 | Japan: Chiba | 2020 |
| 19. | LC546863.1 | Japan: Fukushima | 2020 |
| 20. | LC546862.1 | Japan: Ibaraki | 2020 |
| 21 | LC546861.1 | Japan: Ibaraki | 2020 |
| 22. | LC546860.1 | Japan: Miyazaki | 2020 |
| 23. | LC546859.1 | Japan: Miyazaki | 2020 |
| 24. | LC546858.1 | Japan: Miyazaki | 2020 |
| 25. | LC546857.1 | Japan: Okinawa | 2020 |
| 26. | LC546856.1 | Japan: Okinawa | 2020 |
| 27. | LC546855.1 | Japan: Okinawa | 2020 |
| 28. | LC546854.1 | Japan: Kagoshima | 2020 |
| 29. | LC546853.1 | Japan: Kagoshima | 2020 |
| 30. | LC546852.1 | Japan: Kagoshima | 2020 |
| 31. | LC546851.1 | Japan: Kagoshima | 2020 |
| 32. | LC546850.1 | Japan: Kagoshima | 2020 |
| 33. | LC546849.1 | Japan: Kagoshima | 2020 |
| 34. | LC546848.1 | Japan: Kagoshima | 2020 |
| 35. | LC546847.1 | Japan: Kagoshima | 2020 |
| 36. | LC546846.1 | Japan: Kagoshima | 2020 |
| 37. | MK913648.1 | Viet Nam: Nghe An | 2019 |
| 38. | MK913647.1 | Viet Nam: Nghe An | 2019 |
| 39. | MK913646.1 | Viet Nam: Ha Noi | 2019 |
| 40. | MK860942.1 | China: Tengchong, Yunnan | 2019 |
| 41. | MK860941.1 | China: Tengchong, Yunnan | 2019 |
| 42. | MK860940.1 | China: Tengchong, Yunnan | 2019 |
| 43. | MK860939.1 | China: Tengchong, Yunnan | 2019 |
| 44. | MK860938.1 | China: Tengchong, Yunnan | 2019 |
| 45. | MK860937.1 | China: Tengchong, Yunnan | 2019 |
| 46. | MK860936.1 | China: Ruili, Yunnan | 2019 |
| 47. | MK860935.1 | China: Ruili, Yunnan | 2019 |
| 48. | MK860934.1 | China: Ruili, Yunnan | 2019 |
| 49. | MK860933.1 | China: Ruili, Yunnan | 2019 |
| 50. | MK860932.1 | China: Ruili, Yunnan | 2019 |
| 51. | MK860931.1 | China: Ruili, Yunnan | 2019 |
| 52. | MK860930.1 | China: Ruili, Yunnan | 2019 |
| 53. | MK860927.1 | China: Ruili, Yunnan | 2019 |
| 54. | MK860926.1 | China: Ruili, Yunnan | 2019 |
| 55. | MK860925.1 | China: Ruili, Yunnan | 2019 |
| 56. | MK860924.1 | China: Ruili, Yunnan | 2019 |
| 57. | MK860923.1 | China: Mangshi, Yunnan | 2019 |
| 58. | MK860922.1 | China: Mangshi, Yunnan | 2019 |
| 59. | MK860921.1 | China: Mangshi, Yunnan | 2019 |
| 60. | MK860920.1 | China: Mangshi, Yunnan | 2019 |
| 61. | MK860919.1 | China: Mangshi, Yunnan | 2019 |
| 62. | MK860918.1 | China: Mangshi, Yunnan | 2019 |
| 63. | MK713974.1 | Myanmar | 2019 |
| 64. | MN075831.1 | China | 2019 |
| 65. | MN075830.1 | China | 2019 |
| 66. | MK913645.1 | Viet Nam: Ninh Binh | 2019 |
| 67. | MT073263.1 | Bangladesh: Gazipur | 2020 |
| 68. | MT180097.1 | Pakistan | 2020 |
| 69. | OP132904.1 | South Korea | 2020 |
| 70. | MT073264.1 | Bangladesh: Bogura | 2020 |
| 71. | MT073266.1 | Bangladesh: Jamalpur | 2020 |
| 72. | MT073265.1 | Bangladesh: Rangpur | 2020 |
References
- Navik, O.; Shylesha, A.N.; Patil, J.; Venkatesan, T.; Lalitha, Y.; Ashika, T.R. Damage, distribution and natural enemies of invasive fall armyworm Spodoptera frugiperda (J.E. smith) under rainfed maize in Karnataka, India. Crop Prot. 2021, 143, 105536. [Google Scholar] [CrossRef]
- Day, R.; Abrahams, P.; Bateman, M.; Beale, T.; Clottey, V.; Cock, M.; Colmenarez, Y.; Corniani, N.; Early, R.; Godwin, J.; et al. Fall armyworm: Impacts and implications for Africa. Outlooks Pest Manag. 2017, 28, 196–201. [Google Scholar] [CrossRef]
- Goergen, G.; Kumar, P.L.; Sankung, S.B.; Togola, A.; Tamò, M. First report of outbreaks of the fall armyworm Spodoptera frugiperda (J.E. Smith) (Lepidoptera, Noctuidae), a new alien invasive pest in West and Central Africa. PLoS ONE 2016, 11, e0165632. [Google Scholar] [CrossRef] [PubMed]
- Qi, G.J.; Ma, J.; Wan, J.; Ren, Y.L.; McKirdy, S.; Hu, G.; Zhang, Z.F. Source regions of the first immigration of fall armyworm, Spodoptera frugiperda (Lepidoptera: Noctuidae) invading Australia. Insects 2021, 12, 1104. [Google Scholar] [CrossRef] [PubMed]
- Rane, R.; Walsh, T.K.; Lenancker, P.; Gock, A.; Dao, T.H.; Nguyen, V.L.; Khin, T.N.; Amalin, D.; Chittarath, K.; Faheem, M.; et al. Complex multiple introductions drive fall armyworm invasions into Asia and Australia. Sci. Rep. 2023, 13, 660. [Google Scholar] [CrossRef] [PubMed]
- Malekera, M.J.; Acharya, R.; Mostafiz, M.; Hwang, H.; Bhusal, N. Temperature-dependent development models describing the effects of temperature on the development of the fall armyworm Spodoptera frugiperda (J.E. Smith). Insects 2022, 13, 1084. [Google Scholar] [CrossRef] [PubMed]
- Du Plessis, H.; Schlemmer, M.L.; Van den Berg, J. The effect of temperature on the development of Spodoptera frugiperda (Lepidoptera: Noctuidae). Insects 2020, 11, 228. [Google Scholar] [CrossRef]
- Montezano, D.G.; Specht, A.; Sosa-Gómez, D.R.; Roque-Specht, V.F.; Sousa-Silva, J.C.; Paula-Moraes, S.V.; Peterson, J.A.; Hunt, T.E. Host Plants of Spodoptera frugiperda (Lepidoptera: Noctuidae) in the Americas. African Entomol. 2018, 26, 286–300. [Google Scholar] [CrossRef]
- Vickery, R.A. Studies on the Fall Army Worm in the Gulf Coast District of Texas; United States Department of Agriculture: Washington, DC, USA, 1929. [Google Scholar]
- Capinera, J.L. Fall Armyworm, Spodoptera frugiperda (J.E. Smith) (Insecta: Lepidoptera: Noctuidae); University of Florida IFAS Extension: Gainesville, FL, USA, 2002. [Google Scholar]
- Prowell, D.P.; McMichael, M.; Silvain, J.F. Multilocus genetic analysis of host use, introgression, and speciation in host strains of fall armyworm (Lepidoptera: Noctuidae). Ann. Entomol. Soc. Am. 2004, 97, 1034–1044. [Google Scholar] [CrossRef]
- Kergoat, G.J.; Prowell, D.P.; Le Ru, B.P.; Mitchell, A.; Dumas, P.; Clamens, A.L.; Condamine, F.L.; Silvain, J.F. Disentangling dispersal, vicariance and adaptive radiation patterns: A case study using armyworms in the pest genus Spodoptera (Lepidoptera: Noctuidae). Mol. Phylogenet. Evol. 2012, 65, 855–870. [Google Scholar] [CrossRef]
- Kenis, M.; Benelli, G.; Biondi, A.; Calatayud, P.-A.; Day, R.; Desneux, N.; Harrison, R.D.; Kriticos, D.; Rwomushana, I.; van den Berg, J.; et al. Invasiveness, biology, ecology, and management of the fall armyworm, Spodoptera frugiperda. Entomol. Gen. 2022, 43, 187–241. [Google Scholar] [CrossRef]
- Meagher, R.L.; Nagoshi, R.N.; Armstrong, J.S.; Niogret, J.; Epsky, N.D.; Flanders, K.L. Captures and host strains of fall armyworm (Lepidoptera: Noctuidae) males in traps baited with different commercial pheromone blends. Florida Entomol. 2013, 96, 729–740. [Google Scholar] [CrossRef]
- Nagoshi, R.N. The fall armyworm triose phosphate isomerase (Tpi) gene as a marker of strain identity and interstrain mating. Ann. Entomol. Soc. Am. 2010, 103, 283–292. [Google Scholar] [CrossRef]
- Levy, H.C.; Garcia-Maruniak, A.; Maruniak, J.E. Strain identification of Spodoptera frugiperda (Lepidoptera: Noctuidae) insects and cell line: PCR-RFLP of cytochrome oxidase C subunit I gene. Florida Entomol. 2003, 85, 186–190. [Google Scholar] [CrossRef]
- Nagoshi, R.N.; Goergen, G.; Tounou, K.A.; Agboka, K.; Koffi, D.; Meagher, R.L. Analysis of strain distribution, migratory potential, and invasion history of fall armyworm populations in northern sub-Saharan Africa. Sci. Rep. 2018, 8, 3710. [Google Scholar] [CrossRef] [PubMed]
- Nagoshi, R.N.; Htain, N.N.; Boughton, D.; Zhang, L.; Xiao, Y.; Nagoshi, B.Y.; Mota-Sanchez, D. Southeastern Asia fall armyworms are closely related to populations in Africa and India, consistent with common origin and recent migration. Sci. Rep. 2020, 10, 1421. [Google Scholar] [CrossRef]
- Nagoshi, R.N.; Goergen, G.; Plessis, H.D.; van den Berg, J.; Meagher, R. Genetic comparisons of fall armyworm populations from 11 countries spanning sub-Saharan Africa provide insights into strain composition and migratory behaviors. Sci. Rep. 2019, 9, 8311. [Google Scholar] [CrossRef]
- Tay, W.T.; Rane, R.; Padovan, A.; Walsh, T.; Elfekih, S.; Downes, S.; Nam, K.; d’Alençon, E.; Zhang, J.; Wu, Y.; et al. Whole genome sequencing of global Spodoptera frugiperda populations: Evidence for complex, multiple introductions across the Old World. bioRxiv 2020. bioRxiv:2020.06.12.147660. [Google Scholar]
- Yainna, S.; Tay, W.T.; Fiteni, E.; Legeai, F.; Clamens, A.L.; Gimenez, S.; Frayssinet, M.; Asokan, R.; Kalleshwaraswamy, C.M.; Deshmukh, S.; et al. Genomic balancing selection is key to the invasive success of the fall armyworm. bioRxiv 2020. [Google Scholar] [CrossRef]
- Acharya, R.; Akintola, A.A.; Malekera, M.J.; Kamulegeya, P.; Nyakunga, K.B.; Mutimbu, M.K.; Shrestha, Y.K.; Hemayet, J.S.M.; Hoat, T.X.; Dao, H.T.; et al. Genetic relationship of fall armyworm (Spodoptera frugiperda) populations that invaded Africa and Asia. Insects 2021, 12, 439. [Google Scholar] [CrossRef] [PubMed]
- Nagoshi, R.N.; Dhanani, I.; Asokan, R.; Mahadevaswamy, H.M.; Kalleshwaraswamy, C.M.; Sharanabasappa; Meagher, R.L. Genetic characterization of fall armyworm infesting South Africa and India indicate recent introduction from a common source population. PLoS ONE 2019, 14, e0217755. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Nayyar, N.; Gracy, R.G.; Ashika, T.R.; Mohan, G.; Swathi, R.S.; Mohan, M.; Chaudhary, M.; Bakthavatsalam, N.; Venkatesan, T. Population structure and genetic diversity of invasive fall armyworm after 2 years of introduction in India. Sci. Rep. 2021, 11, 7760. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Laura Juárez, M.; Gabriela Murúa, M.; Gabriela García, M.; Ontivero, M.; Teresa Vera, M.; Vilardi, J.C.; Groot, A.T.; Castagnaro, A.P.; Gastaminza, G.; Willink, E. Host association of Spodoptera frugiperda (Lepidoptera: Noctuidae) corn and rice strains in Argentina, Brazil, and Paraguay. J. Econ. Entomol. 2012, 105, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Nagoshi, R.N.; Fleischer, S.; Meagher, R.L.; Hay-Roe, M.; Khan, A.; Murúa, M.G.; Silvie, P.; Vergara, C.; Westbrook, J. Fall armyworm migration across the lesser antilles and the potential for genetic exchanges between north and south American populations. PLoS ONE 2017, 12, e0171743. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef]
- Omuut, G.; Mollel, H.G.; Kanyesigye, D.; Akohoue, F.; Adumo Aropet, S.; Wagaba, H.; Otim, M.H. Genetic analyses and detection of point mutations in the acetylcholinesterase-1 gene associated with organophosphate insecticide resistance in fall armyworm (Spodoptera frugiperda) populations from Uganda. BMC Genom. 2023, 24, 22. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783. [Google Scholar] [CrossRef]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinforma. 2005, 1, 117693430500100. [Google Scholar] [CrossRef]
- Nagoshi, R.N.; Meagher, R.L.; Flanders, K.; Gore, J.; Jackson, R.; Lopez, J.; Armstrong, J.S.; Buntin, G.D.; Sansone, C.; Leonard, B.R. Using haplotypes to monitor the migration of fall armyworm (Lepidoptera: Noctuidae) corn-strain populations from Texas and Florida. J. Econ. Entomol. 2008, 101, 742–749. [Google Scholar] [CrossRef] [PubMed]
- Murúa, M.G.; Nagoshi, R.N.; Santos, D.A.D.; Hay-Roe, M.M.; Meagher, R.L.; Vilardi, J.C. Demonstration using field collections that Argentina fall armyworm populations exhibit strain-specific host plant preferences. J. Econ. Entomol. 2015, 108, 2305–2315. [Google Scholar] [CrossRef] [PubMed]
- Nagoshi, R.N. Evidence that a major subpopulation of fall armyworm found in the Western Hemisphere is rare or absent in Africa, which may limit the range of crops at risk of infestation. PLoS ONE 2019, 14, e0208966. [Google Scholar] [CrossRef]
- Kolbe, J.J.; Glor, R.E.; Schettino, L.R.; Lara, A.C.; Larson, A.; Losos, J.B. Genetic variation increases during biological invasion by a Cuban lizard. Nature 2004, 431, 177–181. [Google Scholar] [CrossRef] [PubMed]
- The World Bank Group. Climate Risk Profile: Congo, Democratic Republic; The World Bank Group: Washington, DC, USA, 2021. [Google Scholar]

| No. | Sample ID | Province/Territory/Village | Location | Collection Date (Day/Month/Year) | Accession Number | Genetic Group | ||
|---|---|---|---|---|---|---|---|---|
| COI | Tpi | COI | Tpi | |||||
| 1 | Congo11 | Sud-Kivu/Kabare/ Katana | 2°22′51″ N 28°82′35″ E | 29 November 2018 | MT103350 | MT894220 | COI-RS | Tpi-Ca1a |
| 2 | Congo42 | Sud-Kivu/Walungu/Nduba | 2°63′73″ N 28°69′63″ E | 15 December 2018 | MT103349 | MT894225 | COI-RS | Tpi-Ca1a |
| 3 | Congo3 | Sud-Kivu/Kalehe/Bunyakiri | 1°99′49″ N 28°54′62″ E | 29 November 2018 | OQ612484 | OQ632453 | COI-RS | Tpi-Ca1a |
| 4 | Congo41 | Sud-Kivu/Uvira/Sange | 3°06′10″ N 29°08′55″ E | 15 December 2018 | MT933055 | MT894224 | COI-RS | Tpi-Ca2b |
| 5 | Congo31 | Sud-Kivu/Uvira/Luvungi | 2°89′15″ N 28°97′12″ E | 15 December 2018 | MT933054 | MT894223 | COI-RS | Tpi-Ca2a |
| 6 | Congo21 | Sud-Kivu/Kalehe/Minova | 1°74′73″ N 28°98′78″ E | 29 November 2018 | MT933053 | MT894222 | COI-RS | Tpi-Ca2a |
| 7 | Congo12 | Sud-Kivu/ Kabare/Miti | 2°33′06″ N 28°76′69″ E | 29 November 2018 | MT933052 | MT894221 | COI-RS | Tpi-Ca2b |
| 8 | K1 | Lomami/Kabinda/Kabinda | 6°07′48″ S 24°28′48″ E | 18 July 2020 | OP132901 | OQ468459 | COI-RS | Tpi-Ca1a |
| 9 | Gem1 | Sud-ubangi/Gemena/Gemena1 | 3°14′56″ N 19°46′36″ E | 15 July 2020 | OP132892 | OQ468451 | COI-RS | Tpi-Ca1a |
| 10 | Bkd | Sud-ubangi/Gemena/Bokunda | 3°12′39″N 19°46′29″ E | 15 July 2020 | OP132899 | OQ468460 | COI-RS | Tpi-Ca1a |
| 11 | Bsg1 | Sud-ubangi/Gemena/Bosengwen | 3°13′50″N 19°42′57″ E | 18 July 2020 | OP132898 | OQ468458 | COI-CS | Tpi-Ca1a |
| 12 | Bbw1 | Sud-ubangi/Gemena/Bombawuli | 3°13′48″ N 19°53′51″ E | 18 July 2020 | OP132896 | OQ468455 | COI-RS | Tpi-Ca1a |
| 13 | Mtf1 | Tanganyika/Kalemie/Kalemie | 5°52′08″ S 29°10′14″ E | 21 July 2020 | OP132894 | OQ468453 | COI-RS | Tpi-Ca1a |
| 14 | Tshb1 | Tshuapa/Boende/Boende1 | 0°17′13″ S 20°52′24″ E | 18 July 2020 | OP132895 | OQ468454 | COI-RS | Tpi-Ca1a |
| 15 | Blk1 | Tshuapa/Boende/Baliko | 0°18′05″ S 20°52′30″ E | 18 July 2020 | OP132897 | OQ468456 | COI-CS | Tpi-Ca1a |
| 16 | Bde1 | Tshuapa/Boende/Boende3 | 0°16′39″ S 20°53′05″ E | 15 July 2020 | OP132898 | OQ468457 | COI-RS | Tpi-Ca1a |
| 17 | Isi1 | Haut-Uélé/Isiro/Isiro | 2°45′57″ N 27°36′32″ E | 8 August 2020 | OP132893 | OQ468452 | COI-RS | Tpi-Ca1a |
| 18 | M1 | Kongo central/Matadi/Matadi | 5°47′58″ S 13°26′26″ E | 18 July 2020 | OP132900 | OQ632454 | COI-RS | Tpi-Ca1a |
| 19 | Kst1 | Kongo central/Kisantu/Kisantu1 | 5°13′82″ S, 15°09′08″ E | 15 December 2022 | OQ427278 | OQ468462 | COI-RS | Tpi-Ca2a |
| 20 | Kst2 | Kongo central/Kisantu/Kisantu2 | 5°13′82″ S, 15°09′08″ E | 15 December 2022 | OQ427279 | OQ468466 | COI-CS | Tpi-Ca1a |
| 21 | Kst3 | Kongo central/Kisantu/Kisantu3 | 5°13′82″ S, 15°09′08″ E | 15 December 2022 | OQ427280 | OQ857569 | COI-RS | Tpi-Ca1a |
| 22 | Plaba1 | Kinshasa/Plateau de Bateke1 | 4°20′72″ S, 15°84′48″ E | 20 December 2022 | OQ427282 | OQ468463 | COI-RS | Tpi-Ca1a |
| 23 | Plaba2 | Kinshasa/Plateau de Bateke2 | 4°20′72″ S, 15°84′48″ E | 20 December 2022 | OQ427284 | OQ468464 | COI-CS | Tpi-Ca1a |
| 24 | Kimw1 | Kinshasa/Kimwenza1 | 4°47′11″ S, 15°30′14″ E | 20 December 2022 | OQ427281 | OQ468461 | COI-RS | Tpi-Ca1a |
| 25 | Kimw2 | Kinshasa/Kimwenza2 | 4°47′11″ S, 15°30′14″ E | 20 December 2022 | OQ427283 | OQ468465 | COI-RS | Tpi-Ca1a |
| DRC | Africa | America | Asia | Total | |
|---|---|---|---|---|---|
| No. of sequences | 25 | 89 | 126 | 72 | 308 |
| No. of sites | 483 | 483 | 482 | 483 | 482 |
| No. of polymorphic sites | 7 | 8 | 34 | 9 | 37 |
| No. of mutations | 7 | 8 | 38 | 9 | 41 |
| No. of haplotypes | 2 | 3 | 29 | 4 | 32 |
| Haplotype diversity | 0.324 | 0.344 | 0.742 | 0.378 | 0.562 |
| Nucleotides diversity | 0.00469 | 0.00478 | 0.00855 | 0.00520 | 0.00735 |
| Fu’s Fs statistic | 6.012 | 6.837 | −9.966 | 5.134 | −9.841 |
| Fu and Li’s D × test statistic | 1.29627 | 0.47452 | −3.82406 ** | −0.08303 | −5.46527 ** |
| Fu and Li’s F × test statistic | 1.14734 | 0.79287 | −3.33095 ** | 0.30287 | −4.28883 ** |
| Tajima’s D | 0.53489 | 1.13421 | −1.25518 | 0.92310 | −1.28326 |
| Group | Source | df | SS | Variance Component | Total Variance | p-Value |
|---|---|---|---|---|---|---|
| All | Among groups | 3 | 71.411 | 0.2369 | 12.70 | 0.0001 |
| Among populations within groups | 22 | 94.082 | 0.2900 | 15.54 | ||
| Within populations | 283 | 379.008 | 1.3392 | 71.76 | ||
| Total | 308 | 544.502 | 1.86629 | |||
| DRC and Africa | Among groups | 1 | 0.019 | −0.04839 | −4.33 | 0.17595 |
| Among populations within groups | 12 | 19.429 | 0.07543 | 6.75 | ||
| Within populations | 96 | 104.744 | 1.09108 | 97.58 | ||
| Total | 109 | 124.191 | 1.11811 | |||
| America and DRC | Among groups | 1 | 18.325 | 0.25957 | 10.94 | 0.0001 |
| Among populations within groups | 11 | 86.942 | 0.63554 | 26.79 | ||
| Within populations | 142 | 209.759 | 1.47718 | 62.27 | ||
| Total | 154 | 315.026 | 2.37228 | |||
| Asia and DRC | Among groups | 1 | 0.154 | −0.06807 | −4.44 | 0.1700 |
| Among populations within groups | 10 | 22.870 | 0.11554 | 7.54 | ||
| Within populations | 81 | 120.245 | 1.48451 | 96.90 | ||
| Total | 92 | 143.269 | 1.53197 | |||
| Africa and America | Among groups | 1 | 5.473 | 0.03769 | 11.17 | 0.0001 |
| Among populations within groups | 12 | 10.107 | 0.04366 | 12.94 | ||
| Within populations | 206 | 52.757 | 0.25610 | 75.89 | ||
| Total | 219 | 68.336 | 0.33745 | |||
| America and Asia | Among groups | 1 | 38.821 | 0.25114 | 11.51 | 0.0001 |
| Among populations within groups | 11 | 94.217 | 0.54236 | 24.85 | ||
| Within populations | 190 | 263.859 | 1.38873 | 63.64 | ||
| Total | 202 | 396.897 | 2.18223 | |||
| Africa and Asia | Among groups | 1 | 0.132 | −0.04126 | −3.48 | 0.0400 |
| Among populations within groups | 12 | 26.895 | 0.11284 | 9.52 | ||
| Within populations | 147 | 163.694 | 1.11356 | 93.96 | ||
| Total | 160 | 190.720 | 1.18514 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malekera, M.J.; Mamba, D.M.; Bushabu, G.B.; Murhula, J.C.; Hwang, H.-S.; Lee, K.-Y. Genetic Diversity of the Fall Armyworm Spodoptera frugiperda (J.E. Smith) in the Democratic Republic of the Congo. Agronomy 2023, 13, 2175. https://doi.org/10.3390/agronomy13082175
Malekera MJ, Mamba DM, Bushabu GB, Murhula JC, Hwang H-S, Lee K-Y. Genetic Diversity of the Fall Armyworm Spodoptera frugiperda (J.E. Smith) in the Democratic Republic of the Congo. Agronomy. 2023; 13(8):2175. https://doi.org/10.3390/agronomy13082175
Chicago/Turabian StyleMalekera, Matabaro Joseph, Damas Mamba Mamba, Gauthier Bope Bushabu, Justin Cishugi Murhula, Hwal-Su Hwang, and Kyeong-Yeoll Lee. 2023. "Genetic Diversity of the Fall Armyworm Spodoptera frugiperda (J.E. Smith) in the Democratic Republic of the Congo" Agronomy 13, no. 8: 2175. https://doi.org/10.3390/agronomy13082175
APA StyleMalekera, M. J., Mamba, D. M., Bushabu, G. B., Murhula, J. C., Hwang, H. -S., & Lee, K. -Y. (2023). Genetic Diversity of the Fall Armyworm Spodoptera frugiperda (J.E. Smith) in the Democratic Republic of the Congo. Agronomy, 13(8), 2175. https://doi.org/10.3390/agronomy13082175









