Abstract
Drought stress presents a significant threat to the growth and development of maize. It is important to study the genes and mechanisms that contribute to drought tolerance. In this study, we identified ZmHDT103 (that encodes a histone deacetylase) by conducting a homologous sequence comparison and found that the expression of ZmHDT103 in maize seedlings is responsive to treatment with polyethylene glycol (PEG). We utilized CRISPR/Cas9 gene editing technology to generate three distinct knockout lines and obtained the ChinaMU mutant of the ZmHDT103 gene. Under drought conditions, the seedlings of ZmHDT103 mutants exhibited significantly lower water loss rate (WLR), relative electrolytic leakage (REL), hydrogen peroxide (H2O2) level, and malonaldehyde (MDA) level than those of their wild-type (WT) counterparts. Additionally, the seedlings of ZmHDT103 mutants exhibited significantly higher levels of abscisic acid (ABA), relative water content (RWC), peroxidase (POD), and proline (Pro) than those of the WT control. These findings indicate that ZmHDT103 acts as a negative regulator of drought tolerance in maize.
1. Introduction
Maize (Zea mays L.) is one of the most important crops in the world [1]. It is estimated that maize production needs to increase by 67% by 2050 to meet the growing population and food demand [2]. However, global warming exacerbates drought stress for crops. Drought stress is a major abiotic stress factor that profoundly affects the growth and development of maize, ultimately affecting crop yield in two primary ways. Firstly, the inadequate soil water content leads to a decrease in crop yield or even its complete loss [3]. Secondly, crops can regulate the stomatal openings in their leaves, which reduces water transpiration and CO2 uptake. This regulation ultimately affects the crop yield by reducing the rate of photosynthesis and the synthesis of organic matter during drought conditions [4].
The structure of chromatin can be influenced by various factors, including histone modifications, DNA methylation, and chromatin remodeling [5]. In particular, histone acetylation is often associated with an open chromatin structure, enabling the binding of transcription factors and other regulators to initiate gene transcription. The level of histone acetylation is controlled by histone acetyltransferases (HATs) and histone deacetylases (HDACs) [6]. The HDT gene family, belonging to the subfamily of HDACs, is a plant-specific family that has been implicated in various aspects of plant biology, including development, hormone signaling, and response to abiotic stress. Previous studies have suggested the involvement of HDT genes in drought stress in other plant species [7]. For example, mutant lines hd2c-1 and hd2c-3 of Arabidopsis showed increased sensitivity to ABA and NaCl, as well as reduced tolerance to salt stress, compared to wild-type plants during germination [8]. The mutation of HDA19 in Arabidopsis enhances the plant’s capacity to tolerate a range of abiotic stresses, such as salt, drought, and high temperature [9]. The AtHDT4 negatively regulates drought tolerance in Arabidopsis [10]. In maize, ZmHDA108 regulates maize nutrition and reproductive development, whereas ZmHDA101 influences maize seed development by modulating histone acetylation levels [11,12,13,14]. The expression levels of the ZmHDAC genes family in maize were determined using the stress-protective phytohormone salicylic acid (SA), and most of the ZmHDAC genes were downregulated under SA treatment. There are also studies indicating that under low-temperature stress, the expression of ZmHDACs and the levels of histones H4ac, H3K9ac, and H4K5ac are reduced [15]. These findings suggest the potential involvement of ZmHDAC genes in maize in response to abiotic stresses.
The CRISPR/Cas9 gene editing technology has revolutionized plant molecular breeding by offering simplicity, cost-effectiveness, and high efficiency in editing genes. The CRISPR/Cas9 technology was used to knockout specific genes, such as the AtPDS3 and NbPDS genes in Arabidopsis [16]. In rice, the OsPDS-SP1 and OsBADH2 genes were successfully knocked out, resulting in genetically modified rice with whitening and dwarfing phenotypes [17]. Additionally, the knockout of the ZmHKT1 gene in maize resulted in increased sensitivity to salt stress, while the overexpression of this gene improved salt tolerance, highlighting the positive regulation of ZmHKT1 in enhancing maize tolerance to salt stress [18]. Overall, the simplicity, cost-effectiveness, and high efficiency of the CRISPR/Cas9 gene editing technology have paved the way for significant advancements in plant molecular breeding. This technology has the potential to improve crop traits and enhance agricultural productivity.
In response to abiotic stress, plants employ active growth suppression as an adaptive strategy to enhance their chances of survival [19]. Stress triggers an elevation in reactive oxygen species (ROS) levels in crops, which disrupt normal plant metabolism and lead to lipid peroxidation, protein denaturation, and DNA damage [20]. The antioxidant system, comprising low-molecular-weight antioxidants and antioxidant enzymes, plays a vital role in controlling ROS levels to alleviate their adverse effects. The plant’s antioxidant enzyme system can efficiently eliminate ROS generated by mild drought stress [21,22]. For example, hydrogen peroxide (H2O2) can induce cellular oxidative damage [23], but peroxidases (POD), as antioxidant enzymes, catalyze the reduction of H2O2 to H2O, thereby mitigating the oxidative state of plants [24]. Additionally, changes in physiological indicators such as relative water content (RWC), malondialdehyde (MDA), and proline (Pro) are important parameters for evaluating drought tolerance in plants. Plants with stronger drought resistance often maintain higher RWC levels [25]. Under drought stress, lipid peroxidation occurs in plant membranes, resulting in the production of malondialdehyde (MDA). Excessive MDA content damages plant cell membranes, leading to decreased drought tolerance [26]. Proline (Pro), as a plant osmotic regulator, actively accumulates and maintains cell osmotic pressure. Higher Pro levels under drought stress enable cells to maintain normal water uptake, thereby enhancing plant drought resistance [27]. Abscisic acid (ABA) is commonly regarded as a stress hormone in plants, and it controls stomatal closure and transpiration rate. Research has indicated that a slight increase in ABA levels in plants under insufficient soil moisture can promote root growth, resulting in an increase in the root-to-shoot ratio [28]. Conversely, under severe soil moisture deficiency, ABA inhibits root and shoot growth but promotes lateral root growth, known as drought-induced root growth [29].
The objective of this study was to investigate the role of the ZmHDT103 gene in drought tolerance. We obtained CRISPR/Cas9 mutants of ZmHDT103 using CRISPR/Cas9 gene editing technology and ChinaMU mutant. Subsequently, we assessed the drought tolerance of ZmHDT103 mutants in an artificial climate chamber. We then investigate the physiological consequences of these ZmHDT103 knocking-out lines. Our findings demonstrated the crucial role of ZmHDT103 in regulating drought tolerance in maize.
2. Materials and Methods
2.1. Phylogenetic Analysis
The amino acid sequence of ZmHDT103 was downloaded from the Phytozome database (https://phytozome-next.jgi.doe.gov/, accessed on 1 February 2021) and used as a query to compare with homologous genes in other plants that have been previously studied. Multiple sequence alignment was performed using Clustal W, and a phylogenetic tree was constructed using the neighbor-joining method in MEGA-X software 10.0.5.
2.2. Generation Mutants of ZmHDT103
To design specific target sequences for CRISPR/Cas9 targeting ZmHDT103, we used the guide design resources available on https://zlab.bio/guide-design-resources, accessed on 2 May 2021. Two guide RNAs (gRNAs) were designed within the coding sequence of ZmHDT103. The transgenic operation was performed in our laboratory using the maize inbred line Zong31 (Z31) as the recipient and the vector PBUE411 [30] for CRISPR/Cas9 mutant generation. To identify CRISPR/Cas9 homozygous mutant plants, we extracted DNA from each transformed plant and confirmed the editing site through Sanger sequencing. Homozygous progenies were generated through self-crossing. Additionally, we obtained a ChinaMU mutant of ZmHDT103 from the maize mutant database [31]. We designed primers using the website, https://bioinfo.ut.ee/primer3/, accessed on 23 January 2022, to target regions 200–300 bp upstream and downstream of the mutation site. The primers used for CRISPR/Cas9 mutant identification are as follows: Crispr-hdt103-F: 5′CATGCATGAACTTGTTGGTCTT3′ (Tm = 60 °C) and Crispr-hdt103-R: 5′TCCTCGTCTTCATCTTCAGAATC3′ (Tm = 59.8 °C). The primers used for ChinaMU mutant identification are as follows: hdt103-F: 5′TTGGTACAAGCCCAATGATG3′ (Tm = 59.4 °C); hdt103-R: 5′TTCGCATGGACCTAAAAACA3′ (Tm = 59.2); and MU: 5′GAAGCCAACGCCAWCGCCTCYATTTCGTCGAAT3′ (Tm = 70 °C).
2.3. Mutant Plant Growth and Phenotyping
A greenhouse with a 16 h light/8 h dark photoperiod at 28 °C was used for planting all materials. In the drought stress treatment, ZmHDT103 transgenic plant lines and wild-type plants were planted in large pots (length × width × height = 60 cm × 25 cm × 15 cm) with the wild type serving as a control. Small pots (length × width × height = 30 cm × 25 cm × 13 cm) were used to plant ChinaMU mutant lines, with the wild type serving as a control. Each pot was filled with equal volumes of well-mixed soil, and 32 seeds were planted per line. A total of 2 L of water was evenly sprinkled, and after 20 days of water control, noticeable phenotypic differences were observed between the Z31 inbred line and the transgenic plants. An additional 2 L of water was then evenly distributed. In the well-watered (WW) treatment, after planting the seeds for a week, 2 L of water was evenly sprinkled every 3–4 days. For the watered, drought, and rewatered phenotypes, photographs were taken of the plants after 13, 20–22, and 3–5 days of rehydration, respectively.
For the PEG (polyethylene glycol) stress experiment, the germinated seeds of the B73 inbred line were transferred to the Hoagland nutrient solution in a greenhouse with a 16 h light/8 h dark photoperiod at 28 °C. The plants were grown until they reached the three-leaf stage. Subsequently, the seedlings were subjected to PEG6000 treatment (20% w/v). The leaf tissues of the maize seedlings were collected at 0 h, 1 h, 3 h, 6 h, 12 h, and 24 h after the treatment. The collected samples were immediately frozen in liquid nitrogen and used for RNA isolation.
2.4. Quantitative Real-Time PCR Analysis
Plant tissue RNA was isolated using the RNA Easy Fast Plant Tissue kit (TIANGEN, Beijing, China), and the samples were treated with RNase-free DNase I (Lany) to remove genomic DNA, following the manufacturer’s instructions. The concentration and purity of the RNA were determined using a spectrophotometer (Nanodrop 2000 C, Gene Company Limited, Hong Kong/Beijing, China) and Agarose Gel Electrophoresis (BEIJING JUNYI-DONGFANG ELECTROPHORESIS, Beijing, China). First-strand cDNA was synthesized from 1 μg of DNase I-treated RNA using the HiScript III 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China). RT-qPCR was performed using the SYBR Green Fast qPCR Mix (Vazyme, Nanjing, China) and a Bio-Rad CFX96 machine (Analytikjena, Beijing, China). The expression levels were calculated using the 2−∆∆CT method, with GAPDH serving as the reference gene for normalization. Three biological replicates and three technical replicates were used in the RT-qPCR experiment, and Microsoft Excel software 2021 was used for data analysis. For the qRT-PCR primer design, we selected coding sequences that spanned introns, and the amplicons ranged from 150 to 200 bp in size. The primers used for qRT-PCR are as follows: qRT-hdt103-F: 5′CTGAAGAAGGCGATGATGATTC3′ (Tm = 61.1 °C); qRT-hdt103-R: 5′AGATAGAGGAGTTTTCAGCACG3′ (Tm = 57.8 °C); GAPDH-F: 5′AGGATATCAAGAAAGCTATTAAGGC3′ (Tm = 57.8 °C); and GAPDH-R: 5′GTAGCCCCACTCGTTGTCG3′ (Tm = 61.7 °C).
2.5. Measurement of Water Loss Rate
At the three-leaf stage, whole second leaves were carefully removed from ZmHDT103 knockout plants, ChinaMU mutant, and their respective wild types. These detached leaves were then gently placed on flat filter paper and subjected to controlled conditions. The weight of the leaves was meticulously measured every hour, ensuring that each sample was 6 times. The water loss rate (WLR) was calculated using the following equation: WLR (%) = (fresh weight − desiccated weight)/fresh weight × 100. Six biological replicates were measured for each material. We used Microsoft Excel software for data analysis, including calculating mean and standard deviation, and performing the significance test.
2.6. Measurement Contents of Endogenous ABA
The endogenous abscisic acid (ABA) contents were measured in the ZmHDT103 knockout mutants, ChinaMU mutant, and their respective wild types. Approximately 1 g of aerial tissue was harvested from 12-day-old seedlings that were cultivated under normal conditions, including a 16 h light and 18 h dark photoperiod at 28 °C. The measurement of ABA contents was carried out using the Plant abscisic acid ELISA kit (Product code: MM-1185O1-96T). Three biological replicates were measured for each material, with each containing three technical replicates. Microsoft Excel software was used for data analysis.
2.7. Measurement of Survival Rate, Relative Electrolytic Leakage, and Water Content
For the survival rate, we counted the number of seedlings emerging during normal watering and the number of surviving plants after rehydration. For the relative electrolytic leakage (REL) and the relative water content (RWC), the seedlings showed obvious phenotypes under drought stress. Approximately 0.1 g of veined leaves were placed in a centrifuge tube filled with 15 mL of deionized water at room temperature for 12 h. The conductivity R1 of the extract was measured using a conductivity meter. The leaves were then soaked in boiling water for 30 min, and the conductivity R2 of the extract was measured when it was cooled to room temperature. The REL was calculated using the following equation: REL (%) = R1/R2 × 100. The leaves were rinsed with deionized water, any surface moisture was wiped off, and the fresh weight (W1) was measured. Then, the leaves were immersed in distilled water for 24 h, any remaining surface moisture was removed using filter paper, and the saturation weight (W2) was measured. Next, the leaves were transferred to a 105 °C oven for 30 min, and then, the temperature was adjusted to 80 °C, and drying was continued until no further change in weight was observed. Finally, the dry weight (W3) was measured. The RWC was calculated using the following equation: RWC (%) = (W1 − W3)/(W2 − W3) × 100. Three biological replicates were measured for each material, with each containing three technical replicates. Microsoft Excel software was used for data analysis.
2.8. Estimation of Contents of H2O2, MDA, POD, and Pro
The contents of hydrogen peroxide (H2O2), malonaldehyde (MDA), peroxides (POD), and proline (Pro) were measured in knockout and ChinaMU mutants. Approximately 0.1 g of fresh tissue per sample was harvested from the transgenic lines and the wild type when it showed obvious phenotypes under drought stress. The tissue was added into a 2.0 mL centrifuge tube containing steel balls, immediately frozen in liquid nitrogen, and placed at −80 °C for the measurement of enzyme contents. The contents of H2O2, MDA, POD, and Pro were determined using the Hydrogen Peroxide Assay Kit, the Malondialdehyde Assay Kit, the Peroxidase Assay Kit, and the Proline Assay Kit, respectively (Comingbio, Suzhou, China). Three biological replicates were measured for each material, with each containing three technical replicates. Microsoft Excel software was used for data analysis.
3. Results
3.1. Phylogenetic Analysis and Drought Stress Responsiveness of ZmHDT103
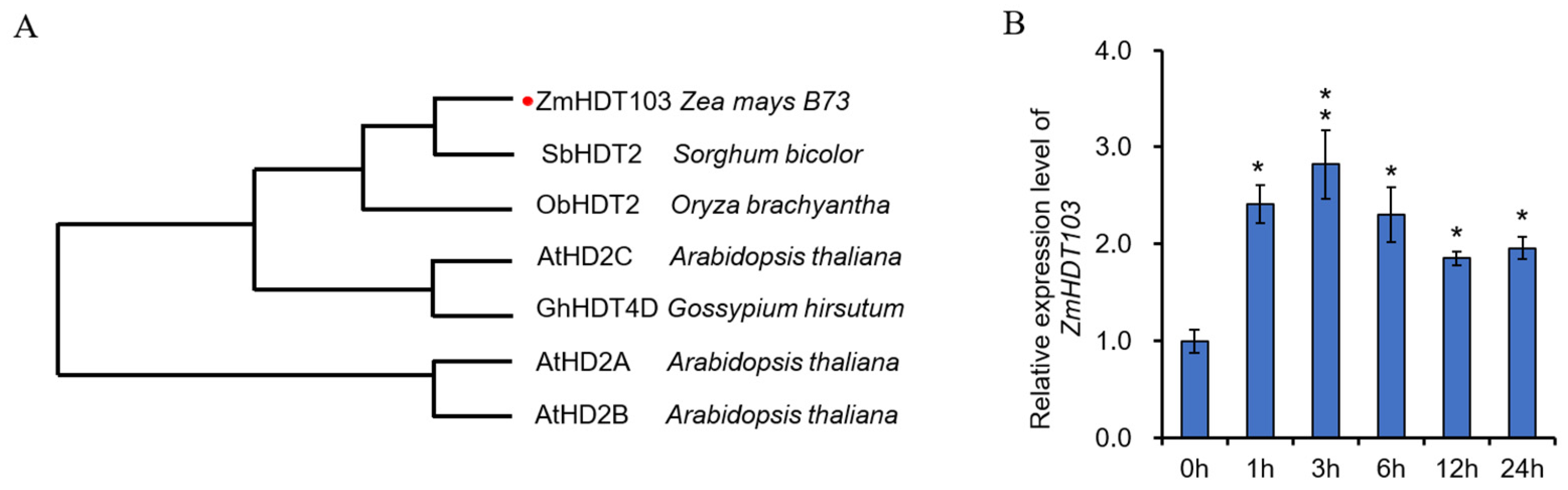
In order to identify the potential drought-responsive HDT genes in maize, we conducted a homologous sequence comparison with other crops. Through this analysis, we identified the ZmHDT103 gene as a candidate involved in drought stress in maize. The ZmHDT103 gene (GenBank Accession number: AQK87994) encodes a full-length protein consisting of 259 amino acids, with a molecular mass of 28.09 kDa and a deduced isoelectric point of 4.63. To investigate the evolutionary relationships between ZmHDT103 and other HDT proteins in different crops, a phylogenetic analysis was performed. The analysis revealed that ZmHDT103 showed the highest similarity to SbHDT2 and formed a different branch from ObHDT2 in Oryza brachyantha (Figure 1A). Sequence alignments indicated that ZmHDT103 exhibited high identities with the homologous sequences of SbHDT2, ObHDT2, AtHD2C, GhHDT4D, AtHD2A, and AtHD2B (Figure S1). Moreover, SbHDT2 controlled cell growth under abiotic stress and showed increased acetylation levels when exposed to abiotic stresses [32]. These results indicated that ZmHDT103 may be involved in the response of maize to abiotic stresses.
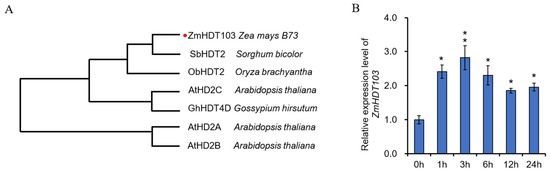
Figure 1.
Sequence analysis of ZmHDT103 protein and drought stress responsiveness of ZmHDT103. (A) Phylogenetic analysis of ZmHDT103 and its closely related HDT proteins from other species was constructed with MEGA-X using a bootstrap value of 1000. The accession numbers of selected HDTs are as follows: from Sorghum bicolor, SbHDT2 (XP_002441441.1); from Oryza brachyantha, ObHDT2 (XP_006654887.1); from Gossypium hirsutum, GhHDT4D (Ghir_D11G035640); and from Arabidopsis thaliana, AtHD2A (ARB50223.1), AtHD2B (T52287), and AtHD2C (AAF70197). The red dots represent the location of the ZmHDT103 gene. (B) Expression patterns of ZmHDT103 under PEG treatment. ZmHDT103 expression in response to 20% (w/v) PEG treatment over 24 h. The maize GAPDH gene was used as the internal reference gene for normalization. Data are represented as means ± SDs of three replicates. * and ** indicate significance at p < 0.05 (t-test) and 0.01, respectively.
We conducted an analysis on the drought response of ZmHDT103 using polyethylene glycol (PEG) treatment. The response of ZmHDT103 to PEG was examined in maize seedlings. The results from qRT-PCR showed that the expression of ZmHDT103 was upregulated in response to drought stress for 1–3 h. However, the expression of ZmHDT103 gradually decreased between 6–24 h. Interestingly, after PEG treatment, the expression of ZmHDT103 increased compared to the control at 0 h (Figure 1B). Therefore, ZmHDT103 may serve as an important indicator of the drought stress response in maize.
3.2. Knockout of ZmHDT103 Enhances Drought Tolerance
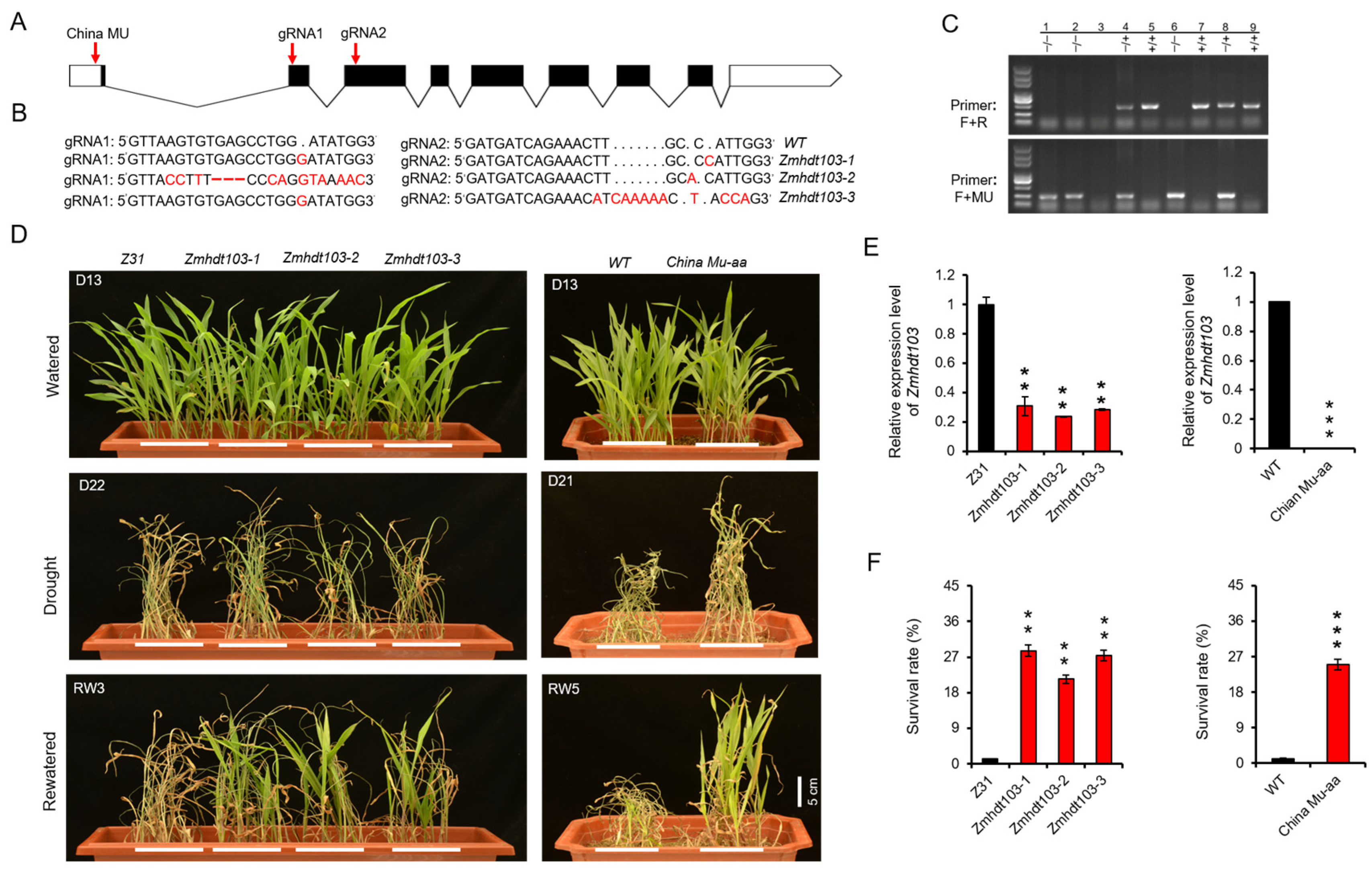
To elucidate the function of ZmHDT103, we employed CRISPR/Cas9 gene editing technology to generate three independent knockout lines (Zmhdt103-1, Zmhdt103-2, and Zmhdt103-3). For the construction of the CRISPR/Cas9 vector, we designed two target sites in the second and third exons of ZmHDT103 (Figure 2A). Subsequently, the vector was transformed into the inbred line Z31 to generate the mutant lines. From seven transgenic lines of ZmHDT103, we obtained three homozygous individual transformants by sequencing. These mutants are referred to as Zmhdt103-1, Zmhdt103-2, and Zmhdt103-3. Zmhdt103-1 has a 1 bp insertion in both the first gRNA1 and the second gRNA2. Zmhdt103-2 has an 11 bp substitution and a 3 bp deletion in the first gRNA1, as well as a 1 bp insertion in the second gRNA2. Zmhdt103-3 has a 1 bp insertion in the first gRNA1, as well as a 6 bp substitution and a 5 bp insertion in the second gRNA2 (Figure 2B). Additionally, we obtained the ChinaMU (http://chinamu.jaas.ac.cn/Default.aspx, accessed on 15 April 2021) mutant of the ZmHDT103 gene through self-cross and PCR identification (Figure 2C).
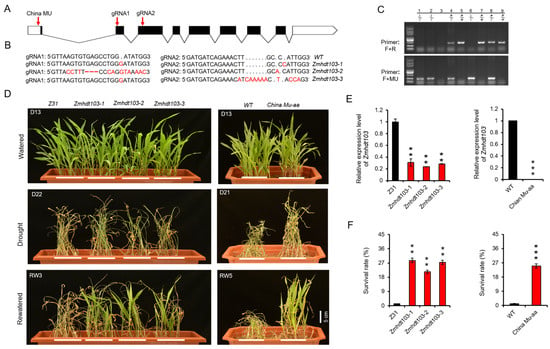
Figure 2.
ZmHDT103 expression and drought tolerance analyses. (A) ZmHDT103 structure with the CRISPR/Cas9 target sites and ChinaMU mutant site is shown, the white box indicates 5′ and 3′UTR sequence, the black box indicates the exon sequence, and the broken line indicates the intron sequence. (B) Insertion, substitution, and deletion sites of three mutations are shown in comparison with the wild type, the red letters indicates base insertion or substitution, and the red short line indicates deletion. (C) The ChinaMU mutant identification, ‘hdt103-F’ and ‘hdt103-R’ represent primers on both sides of the insertion site, and ‘MU’ represents specific primers inserted by MU. 1–9 represents the identified individual plant number, −/− indicates homozygous insertion, +/− indicates heterozygous insertion, and +/+ indicates no insertion. (D) Phenotypes of ZmHDT103 mutants in response to drought treatments. (E) Expression patterns of ZmHDT103 mutants. (F) Survival rates of ZmHDT103 mutants after being rewatered for 3–5 days. Data are represented as means ± SDs of three replicates. ** <0.01 and *** <0.001 by t-test, respectively.
We conducted a study in the greenhouse to investigate the susceptibility of transgenic seedlings to drought stress. Through the qRT-PCR analysis, we observed a downregulation of ZmHDT103 expression in ZmHDT103 knockout plants and ChinaMU mutants (Figure 2E). No significant phenotypic changes were observed in ZmHDT103 knockout plants and ChinaMU mutants compared to their respective WT plants after 13 days of watering (Figure 2D). However, after 21–22 days, the WT plants died, while most of the knockout plants and ChinaMU mutants became dehydrated and appeared dead-like (Figure 2D). After 3–5 days of rewatering, the ZmHDT103 knockout plants and ChinaMU mutants quickly recovered and most of them survived. In contrast, the WT plants were nearly dead and did not recover despite watering (Figure 2D). The survival rates in ZmHDT103 knockout plants and ChinaMU mutants were significantly higher than in the control plants (Figure 2F). The above results demonstrate that ZmHDT103 gene negatively regulates drought resistance of maize seedlings.
3.3. Measurement of WLR and ABA in ZmHDT103 Mutants
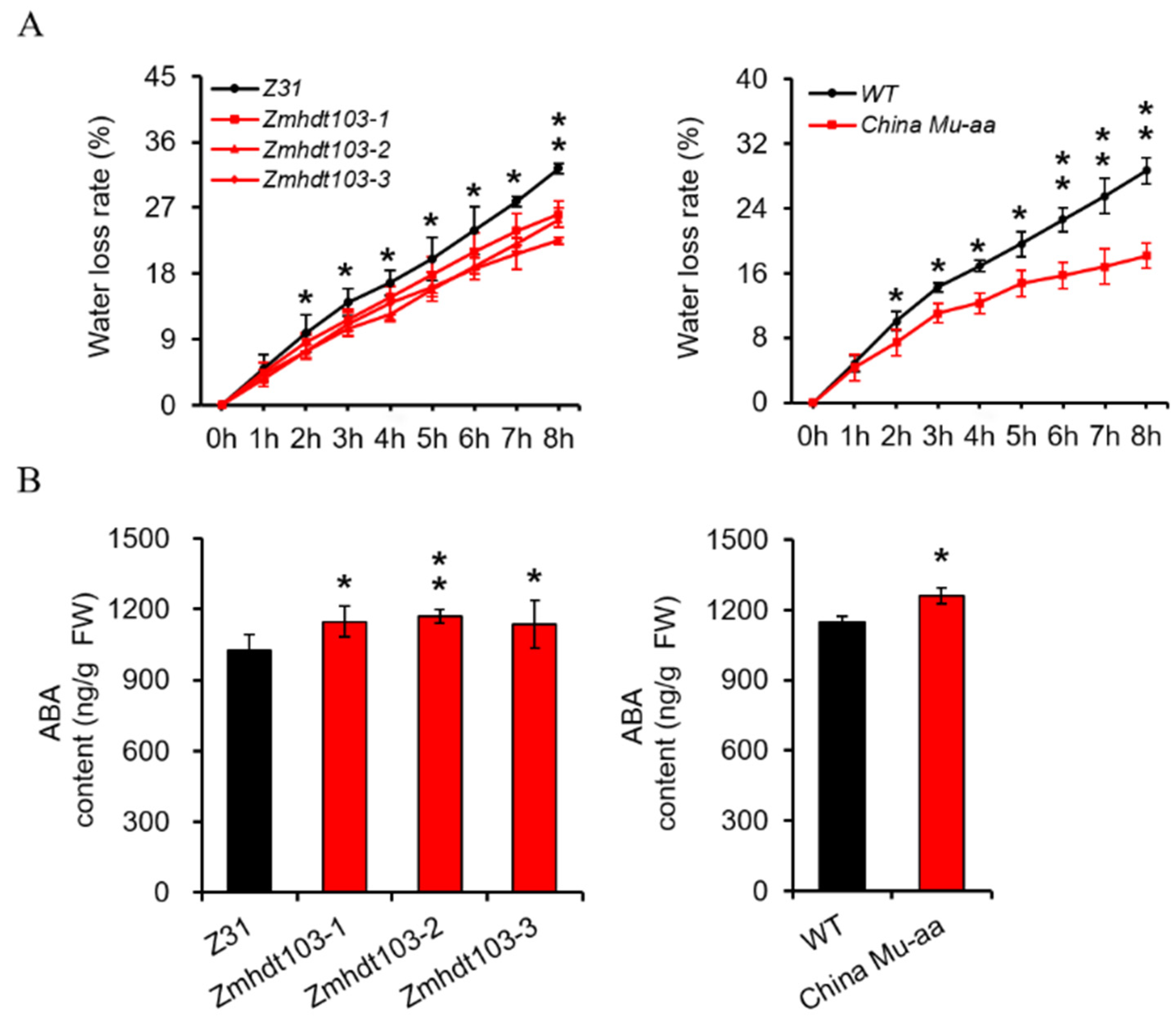
It is widely recognized that plants with a higher capacity to retain water are more likely to survive during periods of water scarcity. Hence, we conducted an investigation to determine whether mutations in the ZmHDT103 gene have any effect on the rate of water loss (WLR) in mutant plants. The WLR of detached leaves was measured. Consistent with their phenotype under drought stress conditions, both ZmHDT103 knockout plants and ChinaMU mutants exhibited a significantly lower WLR compared to their respective WT plants (Figure 3A). The accumulation of ABA is critical for the drought response as it positively contributes to drought tolerance by regulating various processes. In our study, we assessed the levels of endogenous ABA in ZmHDT103 knockout mutants and ChinaMU mutant seedlings. Interestingly, we observed an increase in ABA levels in the aboveground portion of 12-day-old seedlings compared to the corresponding wild-type plants (Figure 3B). This explains how ZmHDT103 knockout plants and ChinaMU mutants displayed greater insensitivity to drought stress compared to the wild type.
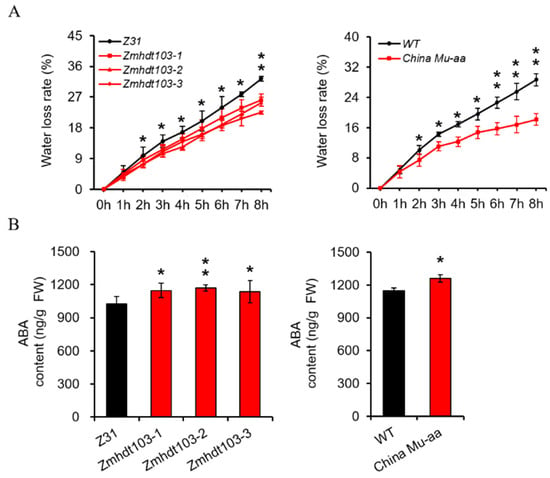
Figure 3.
ZmHDT103 affects the water loss rate and the ABA content of maize seedlings. (A) Water loss rate of detached leaves of ZmHDT103 knockout mutants and ChinaMU mutant seedlings at different time points. (B) Endogenous ABA content of ZmHDT103 knockout mutants and ChinaMU mutant in the aerial part of 12-day-old seedlings. Data are represented as means ± SDs of three replicates. * <0.05 and ** <0.01 by t-test, respectively.
3.4. Physiological Active Substances of Measurement in ZmHDT103 Mutants
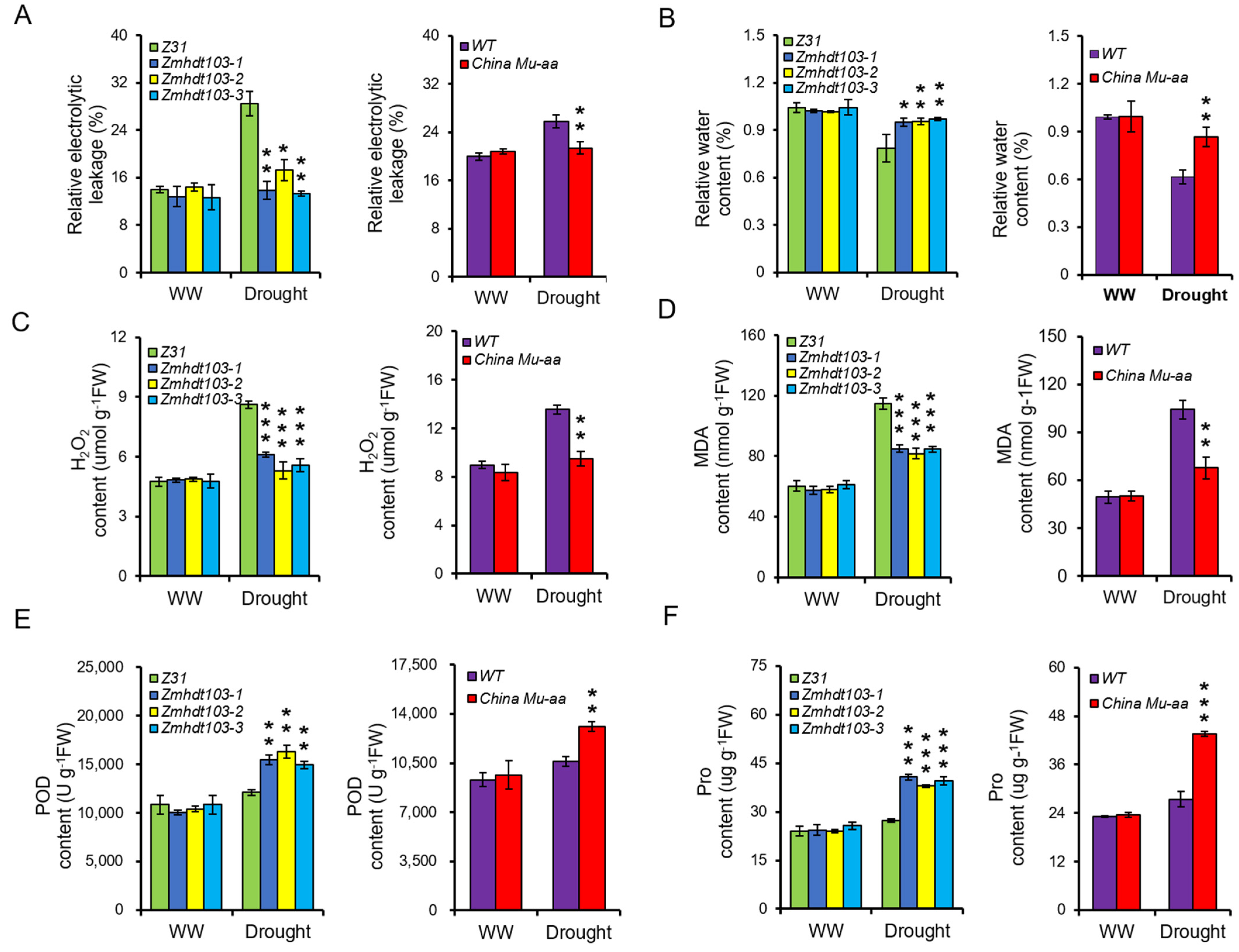
The physiological changes measured in this study included relative electrolyte leakage (REL), relative water content (RWC), and the activity of antioxidant enzymes in leaves. Under WW conditions, no obvious differences were observed between the WT plants and ZmHDT103 knockout mutants and ChinaMU mutant plants. However, significant differences were observed under drought conditions, and the ZmHDT103 knockout mutants and ChinaMU mutant plants exhibited lower REL and higher RWC than the WT plants (Figure 4A,B). Additionally, the activities of antioxidant enzyme systems, including hydrogen peroxide (H2O2), malondialdehyde (MDA), peroxidase (POD), and proline (Pro), were measured. Under WW conditions, no obvious changes in H2O2 and MDA contents were observed in ZmHDT103 knockout mutants, ChinaMU mutant plants, and WT plants. However, under drought conditions, ZmHDT103 knockout mutants and ChinaMU mutant plants had lower H2O2 and MDA contents than control plants (Figure 4C,D). Furthermore, POD and Pro contents were measured. No significant differences in POD and Pro contents were observed between ZmHDT103 knockout mutants, ChinaMU mutant plants, and control plants under WW conditions. However, under drought conditions, ZmHDT103 knockout mutants and ChinaMU mutant plants exhibited higher levels of POD and Pro contents than control plants (Figure 4E,F). Based on these results, the consistently observed physiological phenotypes in the ZmHDT103 knockout plants and ChinaMU plants suggested that ZmHDT103 is a negative regulator of drought tolerance.
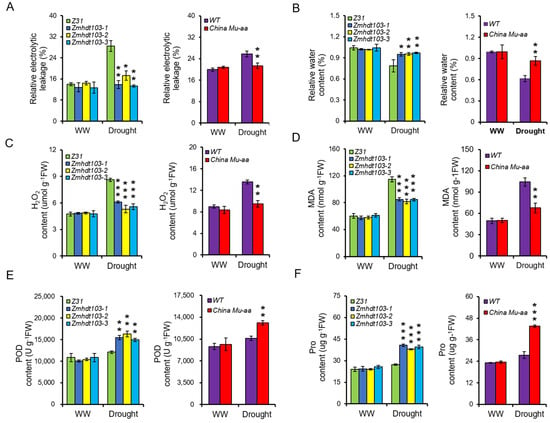
Figure 4.
Physiological changes in ZmHDT103 mutant lines. (A) Relative electrolytic leakage in ZmHDT103 transgenic plants and ChinaMU mutant under WW (well-watered) and drought conditions. (B) Relative water content in ZmHDT103 transgenic plants and ChinaMU mutant under WW and drought conditions. (C–F) Changes in H2O2 (hydrogen peroxide), MDA (malonaldehyde), POD (peroxidase), and Pro contents in ZmHDT103 transgenic plants and ChinaMU mutant under WW and drought conditions. Data are represented as means ± SDs of three replicates. * <0.05, ** <0.01, and *** <0.001 by t-test, respectively.
4. Discussion
The application of key genes in crop breeding is the main strategy to enhance drought resistance in crops [33]. Several studies have demonstrated that HDACs are involved in plant resistance to abiotic stress [34]. By combining GWAS with transcriptome analysis, it was established that HD2B was a genetic factor that affects seed dormancy in Arabidopsis thaliana [35]. The increased expression of PvHDA6 during cold treatment in common beans indicated that PvHDA6 was a cold stress-responsive gene involved in plant abiotic stress [36]. PtHDT902 gene regulates salt stress tolerance in Poplar and Arabidopsis [37]. However, few HDAC genes have been functionally investigated in maize. In this study, maize inbred seedlings were treated with PEG to determine whether the ZmHDT103 gene is involved in drought stress. We found that the expression level of the ZmHDT103 gene significantly increased, with the highest expression level observed after 3 h of treatment, thus proving the involvement of the ZmHDT103 gene in drought stress (Figure 1B). We used CRISPR/Cas9 technology to verify the involvement of ZmHDT103 gene in drought stress at the maize seedling stage, and we also obtained the ChinaMU mutant of ZmHDT103. A drought rehydration experiment was conducted in an artificial climate chamber to verify the effect of ZmHDT103 in drought resistance. The growth of the CRISPR/Cas9 knockout mutants of ZmHDT103 and ChinaMU did not show significant difference from their wild type counterparts under 13 days of drought stress. The wild type plants died after 22 days of drought stress, and the mutants of ZmHDT103 exhibited a wilting phenotype. These results indicate that the ZmHDT103 acts as negative regulator in maize drought tolerance.
Previous studies have reported that the overexpression of HDACs can alter plant drought resistance [38,39,40,41]. For instance, the overexpression of HDA705 reduced the level of ABA and salt stress resistance during seed germination in rice [38]. Similarly, the overexpression of the HDT701 gene in rice reduced tolerance to ABA, salt stress, and osmotic stress during seed germination [39]. The knockout mutants of OsHDA710 displayed high survival rates under salt stress at the seedling stage [40]. In apple, the MdHDA6 gene negatively regulated drought tolerance by catalyzing deacetylation on histones associated with drought-responsive genes. Transgenic apple plants overexpressing MdHDA6 showed reduced drought tolerance, while those with downregulated MdHDA6 expression exhibited increased drought resistance compared to non-transgenic apple plants [41]. Therefore, the knockout or overexpression of HDAC genes supported that the close homologs of ZmHDT103 negatively regulated plant drought tolerance and reflected the phenotypic consequences and physiological changes caused by ZmHDT103 mutation in this study.
WLR and ABA contents are important evaluation indexes for drought stress. ABA accumulates rapidly upon drought stress, causing stomatal closure to limit transpiration water loss [42]. Additionally, ABA stimulates the expression of a number of stress-responsive genes [43]. In this study, ABA content was higher in ZmHDT103 mutant seedlings compared to that of the respective wild type under drought stress (Figure 3B), and the leaf WLR of maize seedlings in ZmHDT103 mutants was significantly lower than that of WT (Figure 3A), indicating that HDT103 acts upstream of genes controlling ABA content and ABA-mediated stomatal movement. It is generally believed that drought stress reduces RWC content in drought-tolerant plants, but some studies have also found that the RWC of plant leaves does not decrease under drought conditions [44]. For example, by evaluating changes in RWC during drought stress in five snap bean cultivars, it was observed that the cultivars exhibiting higher RWC levels demonstrated greater drought tolerance than those with lower RWC levels [45]. In this study, under drought conditions, the mutant exhibited significantly higher RWC compared to the control materials, while there was no significant difference under WW conditions (Figure 4B), further supporting that ZmHDT103 is an upstream regulator of stomatal movement. The REL reflects changes in plant membrane permeability, and the contents of MDA can reflect the level of membrane lipid peroxidation and the degree of injury [46]. In this study, REL and MDA were significantly lower in the ZmHDT103 mutants compared to those in the respective wild type under drought conditions, and there was no significant difference observed under WW conditions (Figure 4A,D). The osmotic regulation mechanism is an important physiological mechanism for crop resistance to drought stress [47]. Proline is a plant osmoregulation substance that actively accumulates and maintains cell osmotic pressure during drought stress [48]. In this study, the content of Pro was significantly higher in the ZmHDT103 mutant compared to the respective wild type under drought conditions, and there was no significant difference under WW conditions (Figure 4F). Overall, the results suggest that ZmHDT103 plays a role in regulating stomatal movement and ABA content under drought stress.
Reactive oxygen species (ROS) play dual roles in plant growth and development, acting both as signaling molecules and as harmful substances that cause oxidative damage to plant tissues [49]. ROS are produced when plants experience drought stress, causing a disturbance in the dynamic balance of ROS redox homeostasis and resulting in the accumulation of excessive ROS. The excessive ROS can have detrimental effects on plant genetic materials, leading to mutations, chromosomal aberrations, and even cell death [50]. These ROS can be eliminated by the antioxidant enzyme systems in plants [51]. The antioxidant enzymes in plants include superoxide dismutase, peroxidase, ascorbate peroxidase, catalase, glutathione peroxidase, glutathione reductase, and glutathione S-transferase [52]. For example, H2O2 can cause cell destruction through oxidation, and POD catalyzes the oxidation reaction of various reducing agents, including H2O2, converting it to H2O and reducing the oxidation state in plants [53]. Khan et al. analyzed the response of two maize varieties to drought stress using the antioxidant system. Under drought stress, the activity of antioxidant enzymes such as POD, SOD, and CAT showed a significant increase with the elevation of H2O2 levels. Based on these results, the drought-tolerant varieties can be selected due to their well-developed antioxidant system [54]. In addition, previous studies have indicated that epigenetic modifications, including DNA methylation and histone deacetylation, regulated the expression of specific stress-responsive genes and enhanced the antioxidant capacity of plants [55]. For example, Arabidopsis hd2ahd2b mutant plants exhibited enhanced drought resistance compared to the respective wild type. When subjected to drought stress, the hd2ahd2b mutant plants demonstrated reduced levels of reactive oxygen species (ROS), a decrease in stomatal aperture size, and an upregulation of genes associated with drought resistance [56]. In this study, the H2O2 content was significantly lower and the POD content was higher in the ZmHDT103 mutant compared to those in the respective wild type under drought conditions. However, there was no significant difference under WW conditions (Figure 4C,E). Therefore, the mutation of ZmHDT103 leads to reduced H2O2 accumulation in maize seedlings under drought conditions, resulting in less cell damage and improved drought tolerance. In conclusion, ZmHDT103 acts as a negative regulator of maize drought tolerance and represents a potential target for breeding drought-resistant maize varieties.
However, further experiments are needed to uncover how ZmHDT103 regulated the contents of H2O2, MDA, ABA, POD, and Pro. Notably, histone deacetylase can remove the acetyl groups in lysine residues of core histone and change the chromatin structure [57]. This biological process may influence a great number of genes that are involved in different biological and physiological processes. The mutant lines obtained in this study are invaluable materials for unraveling the downstream genes and can be used as the starting materials to investigate how ZmHDT103 regulate the content of antioxidants and other molecules, and how these molecules are related to maize’s drought tolerance. Moreover, the favorable alleles of ZmHDT103 and the downstream genes related to drought tolerance can be screened from diversity panels containing natural materials and introduced into hybrid varieties to enhance their stress tolerance.
5. Conclusions
Our findings suggest that ZmHDT103 negatively regulates drought tolerance in maize seedlings. The ZmHDT103 mutants exhibited improved drought tolerance, as evidenced by reductions in WLR, REL, H2O2, and MDA. Additionally, the mutants showed increased levels of ABA, RWC, POD, and Pro. These results contribute to our understanding of the genetic mechanisms underlying drought tolerance in maize and highlight the potential of ZmHDT103 as a target for enhancing drought resistance in maize breeding.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy14010134/s1, Figure S1: Multiple amino acid sequence alignment of ZmHDT103 with its homologous amino acid sequences.
Author Contributions
Conceptualization, J.Z. and H.Z.; methodology, X.W.; software, X.W.; validation, Y.W. and Y.G.; formal analysis, X.W.; investigation, Y.W.; resources, J.Z.; data curation, X.W. and Y.W.; writing—original draft preparation, X.W., H.Z., and J.Z.; writing—review and editing, Y.P. and J.Z.; visualization, Y.P.; supervision, Y.P.; project administration, J.Z.; funding acquisition, J.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Natural Science Foundation of Xinjiang Uygur Autonomous Region (grant no. 2022D01D87), the National Key Research and Development Program of China (grant no. 2021YFD1200705), and the Hainan Yazhou Bay Seed Laboratory (B23CJ0208).
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhang, S.; Bai, J.; Zhang, G.; Xia, Z.; Wu, M.; Lu, H. Negative effects of soil warming, and adaptive cultivation strategies of maize: A review. Sci. Total Environ. 2023, 862, 160738. [Google Scholar] [CrossRef] [PubMed]
- Ray, D.K.; Mueller, N.D.; West, P.C.; Foley, J.A. Yield Trends Are Insufficient to Double Global Crop Production by 2050. PLoS ONE 2013, 8, e66428. [Google Scholar] [CrossRef] [PubMed]
- Leng, G.; Hall, J. Crop yield sensitivity of global major agricultural countries to droughts and the projected changes in the future. Sci. Total Environ. 2019, 654, 811–821. [Google Scholar] [CrossRef] [PubMed]
- Prado, K.; Maurel, C. Regulation of leaf hydraulics: From molecular to whole plant levels. Front. Plant Sci. 2013, 4, 255. [Google Scholar] [CrossRef] [PubMed]
- Allis, C.D.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Peserico, A.; Simone, C. Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J. Biomed. Biotechnol. 2011, 2011, 371832. [Google Scholar] [CrossRef] [PubMed]
- Hollender, C.; Liu, Z. Histone deacetylase genes in Arabidopsis development. J. Integr. Plant Biol. 2008, 50, 875–885. [Google Scholar] [CrossRef]
- Luo, M.; Wang, Y.Y.; Liu, X.; Yang, S.; Lu, Q.; Cui, Y.; Wu, K. HD2C interacts with HAD6 and is involved in ABA and salt stress response in Arabidopsis. J. Exp. Bot. 2012, 63, 3297–3306. [Google Scholar] [CrossRef]
- Ueda, M.; Matsui, A.; Nakamura, T.; Abe, T.; Sunaoshi, Y.; Shimada, H.; Seki, M. Versatility of HDA19-deficiency in increasing the tolerance of Arabidopsis to different environmental stresses. Plant Signal Behav. 2018, 13, e1475808. [Google Scholar] [CrossRef]
- Zhao, B.; Shao, Z.; Wang, L.; Zhang, F.; Chakravarty, D.; Zong, W.; Dong, J.; Song, L.; Qiao, H. MYB44-ENAP1/2 restricts HDT4 to regulate drought tolerance in Arabidopsis. PLoS Genet. 2022, 18, e1010473. [Google Scholar] [CrossRef]
- Varotto, S.; Locatelli, S.; Canova, S.; Pipal, A.; Motto, M.; Rossi, V. Expression profile and cellular localization of maize Rpd3-type histone deacetylases during plant development. Plant Physiol. 2003, 133, 606–617. [Google Scholar] [CrossRef] [PubMed]
- Rossi, V.; Locatelli, S.; Varotto, S.; Donn, G.; Pirona, R.; Henderson, D.A.; Hartings, H.; Motto, M. Maize histone deacetylase hda101 is involved in plant development, gene transcription, and sequence-specific modulation of histone modification of genes and repeats. Plant Cell 2007, 19, 1145–1162. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Liu, X.; Xin, M.; Du, J.; Hu, Z.; Peng, H.; Rossi, V.; Sun, Q.; Ni, Z.; Yao, Y. Genome-Wide Mapping of Targets of Maize Histone Deacetylase HDA101 Reveals Its Function and Regulatory Mechanism during Seed Development. Plant Cell. 2016, 28, 629–645. [Google Scholar] [CrossRef] [PubMed]
- Forestan, C.; Farinati, S.; Rouster, J.; Lassagne, H.; Lauria, M.; Dal Ferro, N.; Varotto, S. Control of Maize Vegetative and Reproductive Development, Fertility, and rRNAs Silencing by HISTONE DEACETYLASE 108. Genetics 2018, 208, 1443–1466. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, L.; Zhao, L.; Li, J.; He, S.; Zhou, K.; Yang, F.; Huang, M.; Jiang, L.; Li, L. Trichostatin A selectively suppresses the cold-induced transcription of the ZmDREB1 gene in maize. PLoS ONE 2011, 6, e22132. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Li, J.F.; Norville, J.E.; Aach, J.; McCormack, M.; Zhang, D.; Bush, J.; Church, G.M.; Sheen, J. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 2013, 31, 688–691. [Google Scholar] [CrossRef]
- Jiang, Z.; Song, G.; Shan, X.; Wei, Z.; Liu, Y.; Jiang, C.; Jiang, Y.; Jin, F.; Li, Y. Association Analysis and Identification of ZmHKT1;5 Variation With Salt-Stress Tolerance. Front. Plant Sci. 2018, 9, 1485. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Zhang, S.; He, Y.; Sen, B.; Wang, G. Reactive oxygen species and their applications toward enhanced lipid accumulation in oleaginous microorganisms. Bioresour. Technol. 2020, 307, 123234. [Google Scholar] [CrossRef]
- Li, J.; Liu, J.; Zhu, T.; Zhao, C.; Li, L.; Chen, M. The Role of Melatonin in Salt Stress Responses. Int. J. Mol. Sci. 2019, 20, 1735. [Google Scholar] [CrossRef] [PubMed]
- Mishra, N.; Jiang, C.; Chen, L.; Paul, A.; Chatterjee, A.; Shen, G. Achieving abiotic stress tolerance in plants through antioxidative defense mechanisms. Front. Plant Sci. 2023, 14, 1110622. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Li, S. Novel insight into functions of ascorbate peroxidase in higher plants: More than a simple antioxidant enzyme. Redox Biol. 2023, 64, 102789. [Google Scholar] [CrossRef] [PubMed]
- Moloi, S.J.; Ngara, R. The roles of plant proteases and protease inhibitors in drought response: A review. Front. Plant Sci. 2023, 14, 1165845. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, D.; Qian, J.; Di, B.; Zhang, G.; Ren, Z. Electrical impedance spectroscopy (EIS) in plant roots research: A review. Plant Methods 2021, 17, 118. [Google Scholar] [CrossRef] [PubMed]
- Spormann, S.; Nadais, P.; Sousa, F.; Pinto, M.; Martins, M.; Sousa, B.; Fidalgo, F.; Soares, C. Accumulation of Proline in Plants under Contaminated Soils-Are We on the Same Page? Antioxidants 2023, 12, 668. [Google Scholar] [CrossRef]
- Moriwaki, T.; Miyazawa, Y.; Kobayashi, A.; Takahashi, H. Molecular mechanisms of hydrotropism in seedling roots of Arabidopsis thaliana (Brassicaceae). Am. J. Bot. 2013, 100, 25–34. [Google Scholar] [CrossRef]
- Vartanian, N.; Marcotte, L.; Giraudat, J. Drought Rhizogenesis in Arabidopsis thaliana (Differential Responses of Hormonal Mutants). Plant Physiol. 1994, 104, 761–767. [Google Scholar] [CrossRef]
- Xing, H.L.; Dong, L.; Wang, Z.P.; Zhang, H.Y.; Han, C.Y.; Liu, B.; Wang, X.C.; Chen, Q.J. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014, 14, 327. [Google Scholar] [CrossRef]
- Liang, L.; Zhou, L.; Tang, Y.; Li, N.; Song, T.; Shao, W.; Zhang, Z.; Cai, P.; Feng, F.; Ma, Y.; et al. A Sequence-Indexed Mutator Insertional Library for Maize Functional Genomics Study. Plant Physiol. 2019, 181, 1404–1414. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Fang, Y.; Jiang, J.; Chen, M.; Fu, X.; Yang, Z.; Luo, L.; Wu, Q.; Yang, Q.; Wang, L.; et al. Characterization of histone deacetylases and their roles in response to abiotic and PAMPs stresses in Sorghum bicolor. BMC Genomics 2022, 23, 28. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, C.D.; Debray, K.; Herwegh, D.; Develtere, W.; Impens, L.; Schaumont, D.; Vandeputte, W.; Aesaert, S.; Coussens, G.; De Boe, Y.; et al. BREEDIT: A multiplex genome editing strategy to improve complex quantitative traits in maize. Plant Cell 2023, 35, 218–238. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Liu, X.; Luo, M.; Yang, S.; Wu, K. Involvement of histone modifications in plant abiotic stress responses. J. Integr. Plant Biol. 2013, 55, 892–901. [Google Scholar] [CrossRef]
- Yano, R.; Takebayashi, Y.; Nambara, E.; Kamiya, Y.; Seo, M. Combining association mapping and transcriptomics identify HD2B histone deacetylase as a genetic factor associated with seed dormancy in Arabidopsis thaliana. Plant J. 2013, 74, 815–828. [Google Scholar] [CrossRef] [PubMed]
- Hayford, R.K.; Ligaba-Osena, A.; Subramani, M.; Brown, A.; Melmaiee, K.; Hossain, K.; Kalavacharla, V.K. Characterization and Expression Analysis of Common Bean Histone Deacetylase 6 during Development and Cold Stress Response. Int. J. Genomics 2017, 2017, 2502691. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Liang, X.; Lv, S.; Guan, T.; Jiang, T.; Cheng, Y. Histone deacetylase gene PtHDT902 modifies adventitious root formation and negatively regulates salt stress tolerance in poplar. Plant Sci. 2020, 290, 110301. [Google Scholar] [CrossRef]
- Zhao, J.; Li, M.; Gu, D.; Liu, X.; Zhang, J.; Wu, K.; Zhang, X.; Teixeira da Silva, J.A.; Duan, J. Involvement of rice histone deacetylase HDA705 in seed germination and in response to ABA and abiotic stresses. Biochem. Biophys. Res. Commun. 2016, 470, 439–444. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, J.; Zhang, W.; Wu, K.; Zheng, F.; Tian, L.; Liu, X.; Duan, J. Expression and functional analysis of the plant-specific histone deacetylase HDT701 in rice. Front. Plant Sci. 2014, 5, 764. [Google Scholar] [CrossRef]
- Ullah, F.; Xu, Q.; Zhao, Y.; Zhou, D.X. Histone deacetylase HDA710 controls salt tolerance by regulating ABA signaling in rice. J. Integr. Plant Biol. 2020. online ahead of print. [Google Scholar] [CrossRef]
- Li, W.; Deng, M.; Wang, S.; Wang, C.; Guo, M.; Song, Y.; Guo, J.; Yan, J.; Ma, F.; Guan, Q.; et al. HISTONE DEACETYLASE 6 interaction with ABSCISIC ACID-INSENSITIVE 5 decreases apple drought tolerance. Plant Physiol. 2023, 193, 2711–2733. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.; Inoue, T.; Hiraide, M.; Khatun, N.; Jahan, A.; Kuwata, K.; Katagiri, S.; Umezawa, T.; Yotsui, I.; Sakata, Y.; et al. Activation of SnRK2 by Raf-like kinase ARK represents a primary mechanism of ABA and abiotic stress responses. Plant Physiol. 2021, 185, 533–546. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, S.; Devi, P.; HanumanthaRao, B.; Jha, U.C.; Sharma, K.D.; Prasad, P.V.V.; Kumar, S.; Siddique, K.H.M.; Nayyar, H. Physiological and Molecular Approaches for Developing Thermotolerance in Vegetable Crops: A Growth, Yield and Sustenance Perspective. Front. Plant Sci. 2022, 13, 878498. [Google Scholar] [CrossRef] [PubMed]
- Omae, H.; Kumar, A.; Kashiwaba, K.; Shono, M. Assessing drought tolerance of snap bean (Phaseolus vulgaris) from genotypic differences in leaf water relations, shoot growth and photosynthetic parameters. Plant Prod. Sci. 2007, 10, 28–35. [Google Scholar] [CrossRef]
- Kim, Y.H.; Khan, A.L.; Waqas, M.; Lee, I.J. Silicon Regulates Antioxidant Activities of Crop Plants under Abiotic-Induced Oxidative Stress: A Review. Front. Plant Sci. 2017, 8, 510. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Yang, Y. How Plants Tolerate Salt Stress. Curr. Issues Mol. Biol. 2023, 45, 5914–5934. [Google Scholar] [CrossRef]
- Jazy, H.D. Investigation on some biochemical components accumulation in three bread wheat cultivars (Triticum aestivum L.) under drought stress. Tech. J. Eng. Appl. Sci. 2013, 3, 2752–2754. [Google Scholar]
- Mittler, R.; Vanderauwera, S.; Suzuki, N.; Miller, G.; Tognetti, V.B.; Vandepoele, K.; Gollery, M.; Shulaev, V.; Van Breusegem, F. ROS signaling: The new wave? Trends Plant Sci. 2011, 16, 300–309. [Google Scholar] [CrossRef]
- Nadarajah, K.K. ROS Homeostasis in Abiotic Stress Tolerance in Plants. Int. J. Mol. Sci. 2020, 21, 5208. [Google Scholar] [CrossRef]
- Lv, X.; Li, Y.; Chen, R.; Rui, M.; Wang, Y. Stomatal Responses of Two Drought-Tolerant Barley Varieties with Different ROS Regulation Strategies under Drought Conditions. Antioxidants 2023, 12, 790. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, A.; Halder, K.; Abdin, M.Z.; Majee, M.; Datta, A. Abiotic Stress Tolerance in Plants: Brassinosteroids Navigate Competently. Int. J. Mol. Sci. 2022, 23, 14577. [Google Scholar] [CrossRef] [PubMed]
- Moloi, M.J.; van der Merwe, R. Drought Tolerance Responses in Vegetable-Type Soybean Involve a Network of Biochemical Mechanisms at Flowering and Pod-Filling Stages. Plants 2021, 10, 1502. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.N.; Samran, D.; Khan, M.D.; Abdullah, J.; Nisar, A. Evaluation of maize varieties based on antioxidant system in response to drought stress. Int. J. Biol. Biotechnol. 2016, 13, 561–570. [Google Scholar]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Han, Y.; Haouel, A.; Georgii, E.; Priego-Cubero, S.; Wurm, C.J.; Hemmler, D.; Schmitt-Kopplin, P.; Becker, C.; Durner, J.; Lindermayr, C. Histone Deacetylases HD2A and HD2B Undergo Feedback Regulation by ABA and Modulate Drought Tolerance via Mediating ABA-Induced Transcriptional Repression. Genes 2023, 14, 1199. [Google Scholar] [CrossRef]
- Mehnert, J.M.; Kelly, W.K. Histone deacetylase inhibitors: Biology and mechanism of action. Cancer J. 2007, 13, 23–29. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).