Detection and Analysis of Chili Pepper Root Rot by Hyperspectral Imaging Technology
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Cultivation and Pathogen Inoculation
2.2. Hyperspectral Imaging System and Image Acquisition
2.2.1. Hyperspectral Imaging System
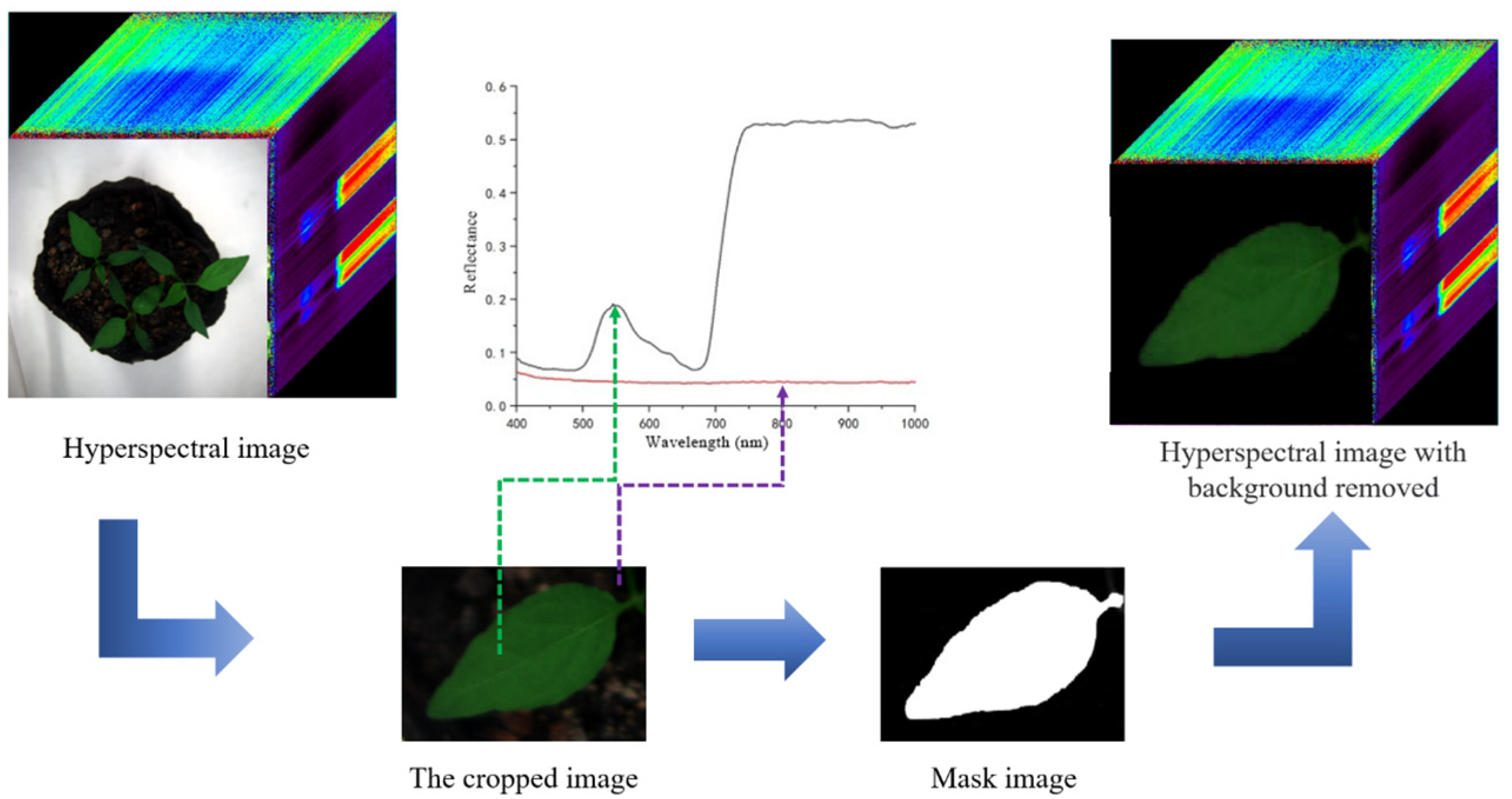
2.2.2. Hyperspectral Image Acquisition and Processing
2.3. Dataset Partitioning
2.4. Data Dimension Reduction Methods
2.4.1. Use SPA to Select Effective Wavelengths
2.4.2. Calculation of Spectral Index
2.5. Classification Model
2.5.1. PLS-DA Model
2.5.2. LSSVM Model
2.5.3. BP Neural Network
2.6. Model Accuracy Evaluation Method
3. Results
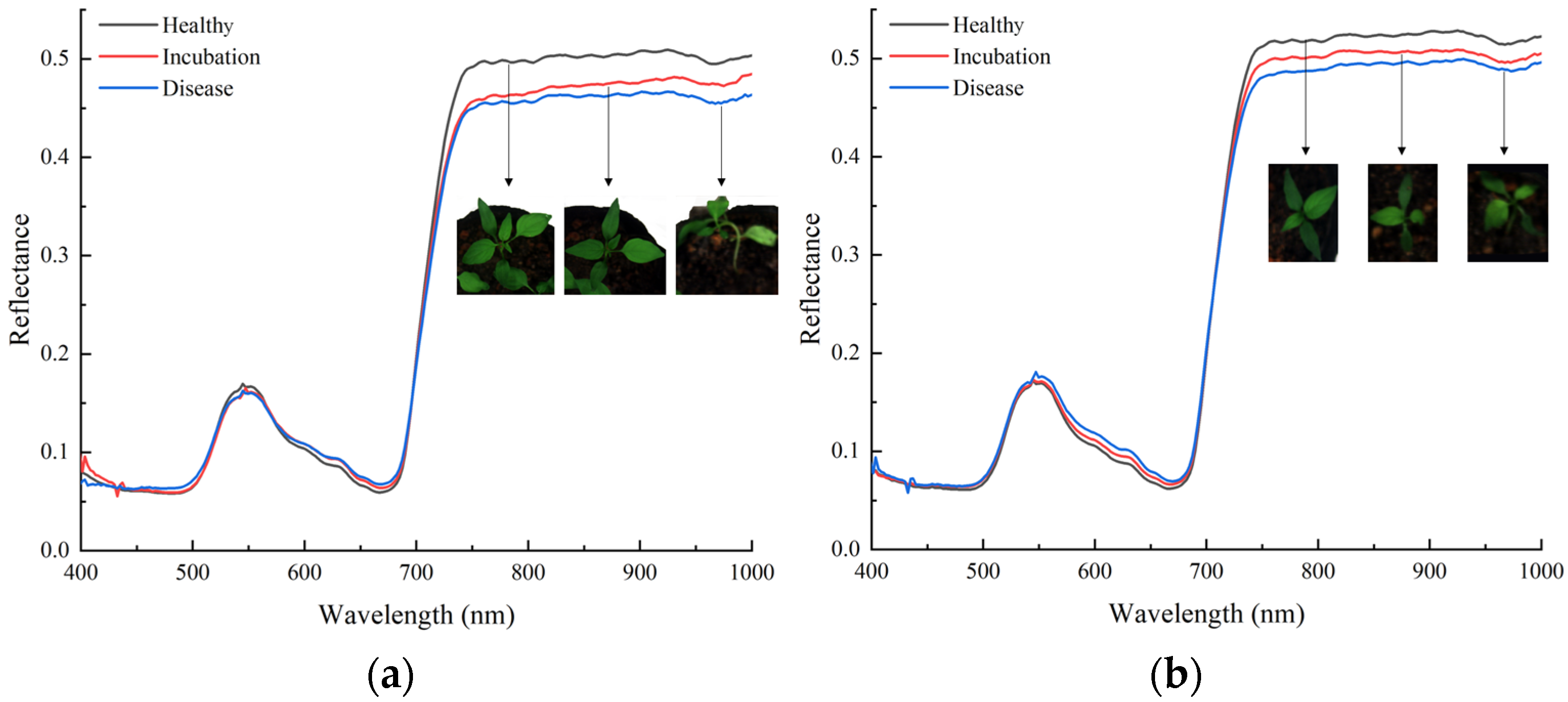
3.1. Analysis of Spectral Properties
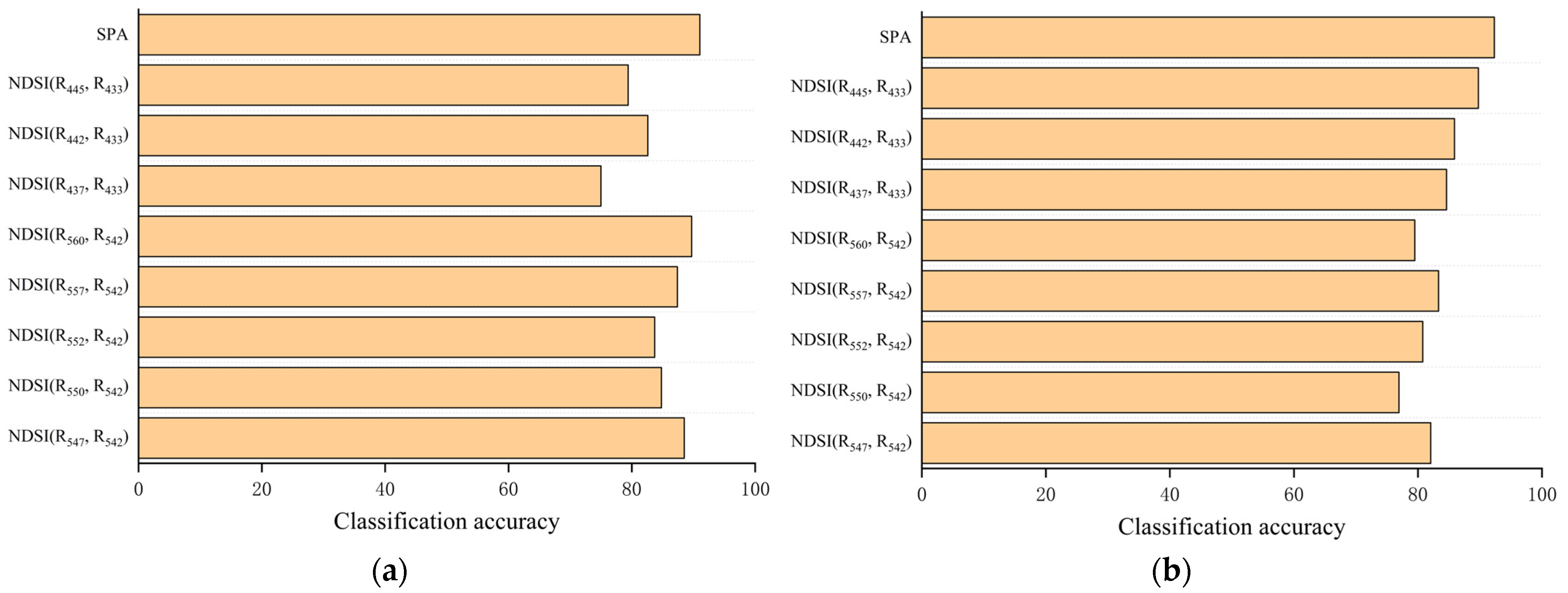
3.2. Effective Wavelength Selection
3.3. Correlation Analysis of Spectral Index
3.4. Early Disease Detection Model
3.4.1. PLS-DA Model Based on Effective Wavelength
3.4.2. LSSVM Model
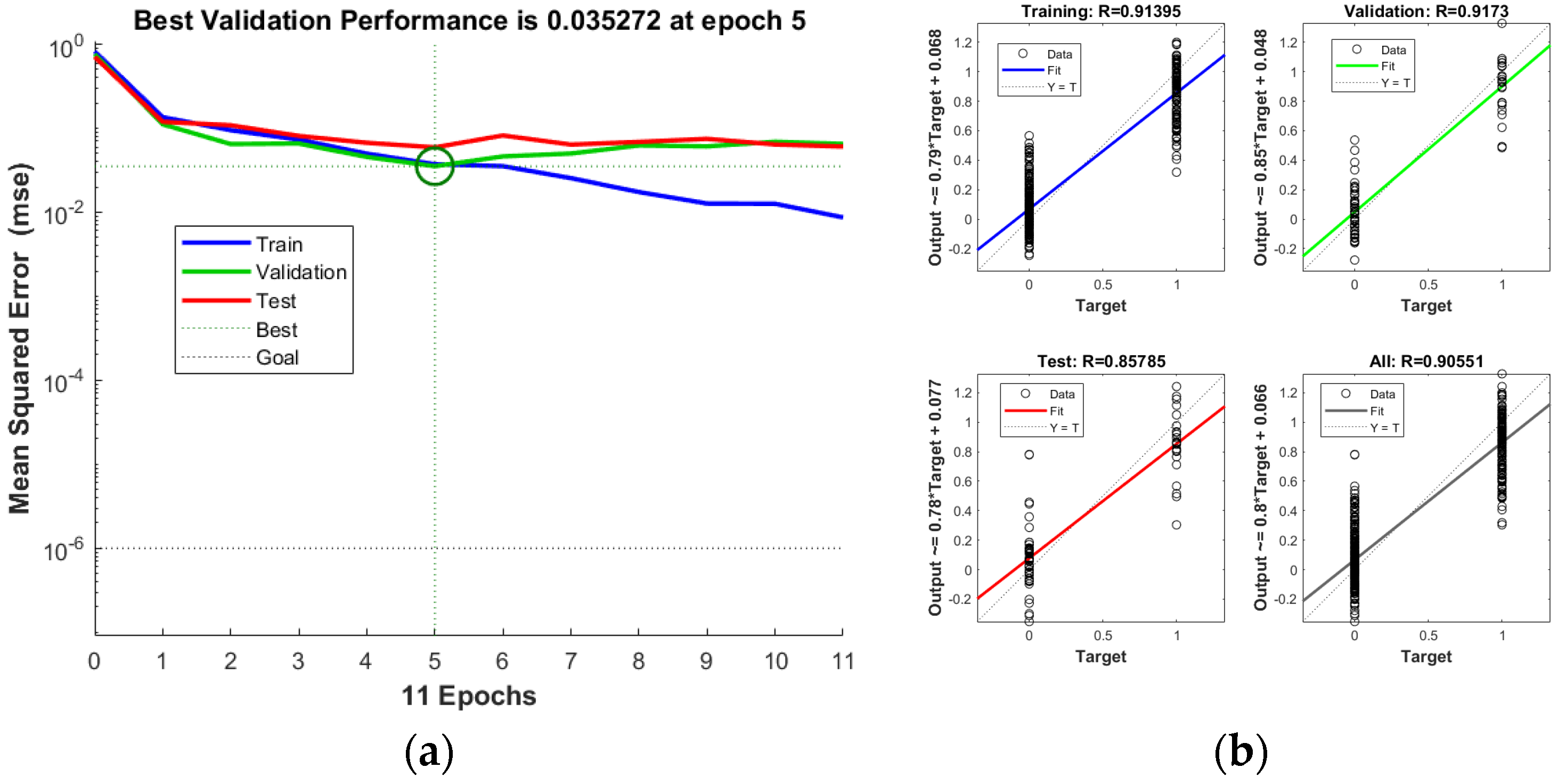
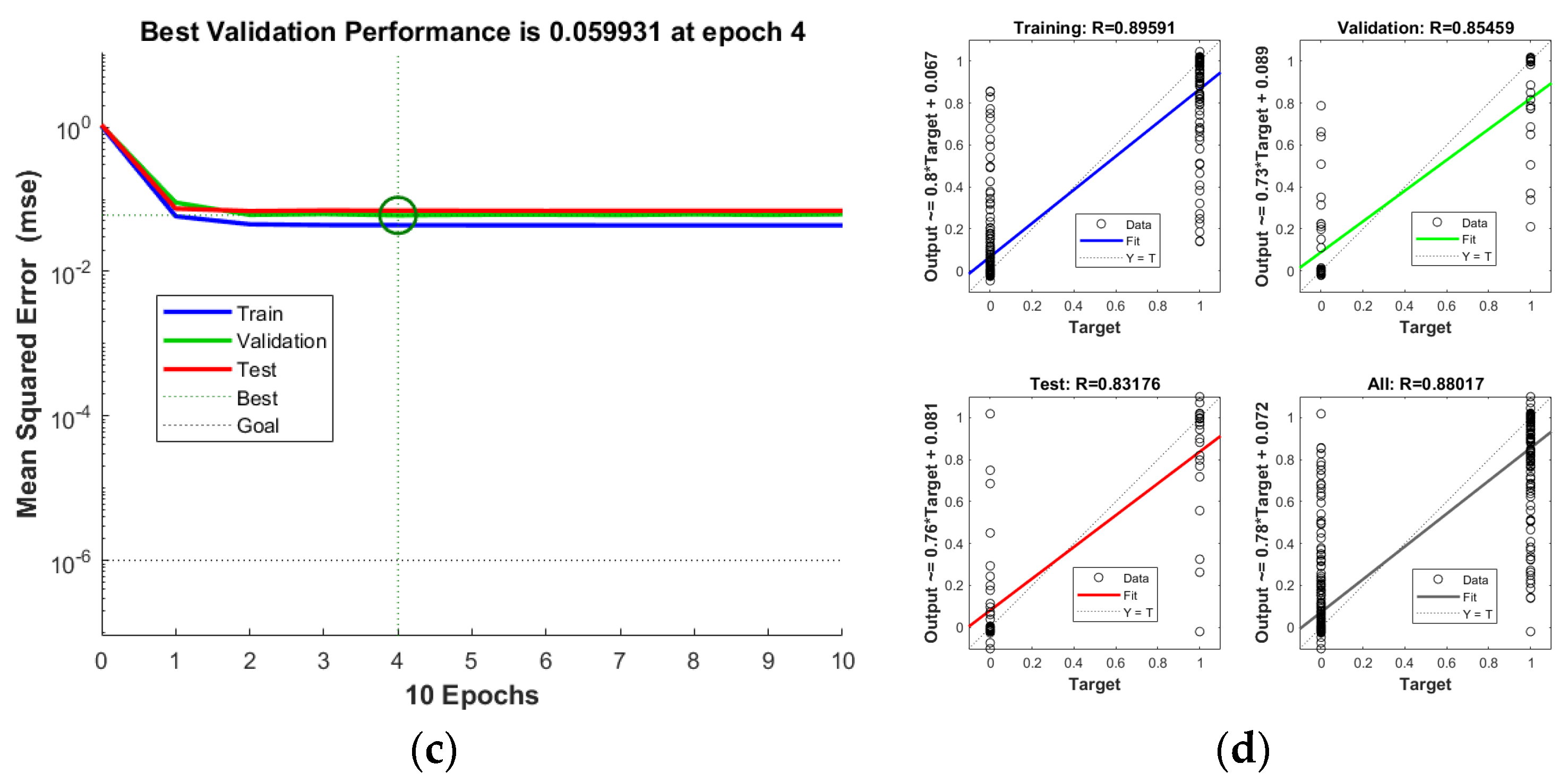
3.4.3. BP Neural Network
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rong, H.; Guo, D.; Yang, W.; Dai, W.; Yang, J.; Liou, X. Isolation and identification of the pathogen of capsicum root rot in Tibet. Acta Agric. Boreali Occi Dent. Sin. 2015, 24, 139–143. [Google Scholar]
- Gustafson, L.L.; Arzul, I.; Burge, C.A.; Carnegie, R.B.; Caceres-Martinez, J.; Creekmore, L.; Dewey, W.; Elston, R.; Friedman, C.S.; Hick, P.; et al. Optimizing surveillance for early disease detection: Expert guidance for Ostreid herpesvirus surveillance design and system sensitivity calculation. Prev. Vet. Med. 2021, 194, 105419. [Google Scholar] [CrossRef]
- Debnath, O.; Saha, H.N. An IoT-based intelligent farming using CNN for early disease detection in rice paddy. Microprocess. Microsyst. 2022, 94, 104631. [Google Scholar] [CrossRef]
- Gao, Z.; Khot, L.R.; Naidu, R.A.; Zhang, Q. Early detection of grapevine leafroll disease in a red-berried wine grape cultivar using hyperspectral imaging. Comput. Electron. Agric. 2020, 179, 105807. [Google Scholar] [CrossRef]
- Shao, Y.; Liu, Y.; Xuan, G.; Shi, Y.; Li, Q.; Hu, Z. Detection and analysis of sweet potato defects based on hyperspectral imaging technology. Infrared Phys. Technol. 2022, 127, 104403. [Google Scholar] [CrossRef]
- Sharma, M.; Jindal, V. Approximation techniques for apple disease detection and prediction using computer enabled technologies: A review. Remote Sens. Appl. Soc. Environ. 2023, 32, 101038. [Google Scholar] [CrossRef]
- Daniya, T.; Vigneshwari, S. Rider Water Wave-enabled deep learning for disease detection in rice plant. Adv. Eng. Softw. 2023, 182, 103472. [Google Scholar] [CrossRef]
- Khan, A.I.; Quadri, S.M.K.; Banday, S.; Latief Shah, J. Deep diagnosis: A real-time apple leaf disease detection system based on deep learning. Comput. Electron. Agric. 2022, 198, 107093. [Google Scholar] [CrossRef]
- Allen, B.; Dalponte, M.; Ørka, H.O.; Næsset, E.; Puliti, S.; Astrup, R.; Gobakken, T. UAV-Based Hyperspectral Imagery for Detection of Root, Butt, and Stem Rot in Norway Spruce. Remote Sens. 2022, 14, 3830. [Google Scholar] [CrossRef]
- Adam, E.; Deng, H.; Odindi, J.; Abdel-Rahman, E.M.; Mutanga, O. Detecting the Early Stage of Phaeosphaeria Leaf Spot Infestations in Maize Crop Using In Situ Hyperspectral Data and Guided Regularized Random Forest Algorithm. J. Spectrosc. 2017, 2017, 6961387. [Google Scholar] [CrossRef]
- Ba, W.; Jin, X.; Lu, J.; Rao, Y.; Zhang, T.; Zhang, X.; Zhou, J.; Li, S. Research on predicting early Fusarium head blight with asymptomatic wheat grains by micro-near infrared spectrometer. Spectrochim. Acta Part A: Mol. Biomol. Spectrosc. 2023, 287, 122047. [Google Scholar] [CrossRef]
- Chen, B.; Li, S.-K.; Wang, K.-R.; Wang, J.; Wang, F.-Y.; Xiao, C.-H.; Lai, J.-C.; Wang, N. Spectrum Characteristics of Cotton Canopy Infected with Verticillium Wilt and Applications. Agric. Sci. China 2008, 7, 561–569. [Google Scholar] [CrossRef]
- Abdulridha, J.; Ampatzidis, Y.; Qureshi, J.; Roberts, P. Laboratory and UAV-Based Identification and Classification of Tomato Yellow Leaf Curl, Bacterial Spot, and Target Spot Diseases in Tomato Utilizing Hyperspectral Imaging and Machine Learning. Remote Sens. 2020, 12, 2732. [Google Scholar] [CrossRef]
- Calamita, F.; Imran, H.A.; Vescovo, L.; Mekhalfi, M.L.; La Porta, N. Early Identification of Root Rot Disease by Using Hyperspectral Reflectance: The Case of Pathosystem Grapevine/Armillaria. Remote Sens. 2021, 13, 2537. [Google Scholar] [CrossRef]
- Wei, X.; He, J.; Zheng, S.; Ye, D. Modeling for SSC and firmness detection of persimmon based on NIR hyperspectral imaging by sample partitioning and variables selection. Infrared Phys. Technol. 2020, 105, 103099. [Google Scholar] [CrossRef]
- Galvão, R.K.H.; Araujo, M.C.U.; José, G.E.; Pontes, M.J.C.; Silva, E.C.; Saldanha, T.C.B. A method for calibration and validation subset partitioning. Talanta 2005, 67, 736–740. [Google Scholar] [CrossRef]
- Li, Y.; Fang, T.; Zhu, S.; Huang, F.; Chen, Z.; Wang, Y. Detection of olive oil adulteration with waste cooking oil via Raman spectroscopy combined with iPLS and SiPLS. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 189, 37–43. [Google Scholar] [CrossRef]
- Tian, H.; Zhang, L.; Li, M.; Wang, Y.; Sheng, D.; Liu, J.; Wang, C. Weighted SPXY method for calibration set selection for composition analysis based on near-infrared spectroscopy. Infrared Phys. Technol. 2018, 95, 88–92. [Google Scholar] [CrossRef]
- Sun, J.; Zhou, X.; Hu, Y.; Wu, X.; Zhang, X.; Wang, P. Visualizing distribution of moisture content in tea leaves using optimization algorithms and NIR hyperspectral imaging. Comput. Electron. Agric. 2019, 160, 153–159. [Google Scholar] [CrossRef]
- Song, Y.; Cao, S.; Chu, X.; Zhou, Y.; Xu, Y.; Sun, T.; Zhou, G.; Liu, X. Non-destructive detection of moisture and fatty acid content in rice using hyperspectral imaging and chemometrics. J. Food Compos. Anal. 2023, 121, 105397. [Google Scholar] [CrossRef]
- Datt, B. A New Reflectance Index for Remote Sensing of Chlorophyll Content in Higher Plants: Tests using Eucalyptus Leaves. J. Plant Physiol. 1999, 154, 30–36. [Google Scholar] [CrossRef]
- Daughtry, C.S.T.; Hunt, E.R.; McMurtrey, J.E. Assessing crop residue cover using shortwave infrared reflectance. Remote Sens. Environ. 2004, 90, 126–134. [Google Scholar] [CrossRef]
- Chen, T.; Zhang, J.; Chen, Y.; Wan, S.; Zhang, L. Detection of peanut leaf spots disease using canopy hyperspectral reflectance. Comput. Electron. Agric. 2019, 156, 677–683. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, J.; Wang, A. Comparison of various approaches for estimating leaf water content and stomatal conductance in different plant species using hyperspectral data. Ecol. Indic. 2022, 142, 109278. [Google Scholar] [CrossRef]
- Yao, X.; Zhu, Y.; Tian, Y.; Feng, W.; Cao, W. Exploring hyperspectral bands and estimation indices for leaf nitrogen accumulation in wheat. Int. J. Appl. Earth Obs. Geoinf. 2010, 12, 89–100. [Google Scholar] [CrossRef]
- Zhao, M.; Cang, H.; Chen, H.; Zhang, C.; Yan, T.; Zhang, Y.; Gao, P.; Xu, W. Determination of quality and maturity of processing tomatoes using near-infrared hyperspectral imaging with interpretable machine learning methods. LWT 2023, 183, 114861. [Google Scholar] [CrossRef]
- Peerbhay, K.Y.; Mutanga, O.; Ismail, R. Commercial tree species discrimination using airborne AISA Eagle hyperspectral imagery and partial least squares discriminant analysis (PLS-DA) in KwaZulu–Natal, South Africa. ISPRS J. Photogramm. Remote Sens. 2013, 79, 19–28. [Google Scholar] [CrossRef]
- Cucuzza, P.; Serranti, S.; Capobianco, G.; Bonifazi, G. Multi-level color classification of post-consumer plastic packaging flakes by hyperspectral imaging for optimizing the recycling process. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 302, 123157. [Google Scholar] [CrossRef]
- Cruz-Tirado, J.P.; Lima Brasil, Y.; Freitas Lima, A.; Alva Pretel, H.; Teixeira Godoy, H.; Barbin, D.; Siche, R. Rapid and non-destructive cinnamon authentication by NIR-hyperspectral imaging and classification chemometrics tools. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 289, 122226. [Google Scholar] [CrossRef]
- Zhang, W.; Pan, L.; Lu, L. Prediction of TVB-N content in beef with packaging films using visible-near infrared hyperspectral imaging. Food Control 2023, 147, 109562. [Google Scholar] [CrossRef]
- Xu, M.; Sun, J.; Yao, K.; Cai, Q.; Shen, J.; Tian, Y.; Zhou, X. Developing deep learning based regression approaches for prediction of firmness and pH in Kyoho grape using Vis/NIR hyperspectral imaging. Infrared Phys. Technol. 2022, 120, 104003. [Google Scholar] [CrossRef]
- Cheng, L.; Liu, G.; He, J.; Wan, G.; Ma, C.; Ban, J.; Ma, L. Non-destructive assessment of the myoglobin content of Tan sheep using hyperspectral imaging. Meat Sci. 2020, 167, 107988. [Google Scholar] [CrossRef]
- Wang, Y.; Song, S. Variety identification of sweet maize seeds based on hyperspectral imaging combined with deep learning. Infrared Phys. Technol. 2023, 130, 104611. [Google Scholar] [CrossRef]
- Wang, T.; Gao, M.; Cao, C.; You, J.; Zhang, X.; Shen, L. Winter wheat chlorophyll content retrieval based on machine learning using in situ hyperspectral data. Comput. Electron. Agric. 2022, 193, 106728. [Google Scholar] [CrossRef]
- Ma, J.; Cheng, J.; Wang, J.; Pan, R.; He, F.; Yan, L.; Xiao, J. Rapid detection of total nitrogen content in soil based on hyperspectral technology. Inf. Process. Agric. 2022, 9, 566–574. [Google Scholar] [CrossRef]
- Zhao, J.; Yan, H.; Huang, L. A joint method of spatial–spectral features and BP neural network for hyperspectral image classification. Egypt. J. Remote Sens. Space Sci. 2023, 26, 107–115. [Google Scholar] [CrossRef]
- Li, Z.; Ni, C.; Wu, R.; Zhu, T.; Cheng, L.; Yuan, Y.; Zhou, C. Online small-object anti-fringe sorting of tobacco stem impurities based on hyperspectral superpixels. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 302, 123084. [Google Scholar] [CrossRef]
- An, J.; Zhang, C.; Zhou, L.; Jin, S.; Zhang, Z.; Zhao, W.; Pan, X.; Zhang, W. Tensor based low rank representation of hyperspectral images for wheat seeds varieties identification. Comput. Electr. Eng. 2023, 110, 108890. [Google Scholar] [CrossRef]
- Tada, T.; Tanaka, C.; Katsube-Tanaka, T.; Shiraiwa, T. Effects of wounding and relative humidity on the incidence of Phytophthora root and stem rot in soybean seedlings. Physiol. Mol. Plant Pathol. 2021, 116, 101737. [Google Scholar] [CrossRef]
- Zhang, J.; Tian, Y.; Yan, L.; Wang, B.; Wang, L.; Xu, J.; Wu, K. Diagnosing the symptoms of sheath blight disease on rice stalk with an in-situ hyperspectral imaging technique. Biosyst. Eng. 2021, 209, 94–105. [Google Scholar] [CrossRef]


| Wavelength Combination | Spectral Index | Correlation Coefficient |
|---|---|---|
| R547, R542 | NDSI (R547, R542) | 0.911 |
| R550, R542 | NDSI (R550, R542) | 0.887 |
| R552, R542 | NDSI (R552, R542) | 0.894 |
| R557, R542 | NDSI (R557, R542) | 0.898 |
| R560, R542 | NDSI (R560, R542) | 0.909 |
| R437, R433 | NDSI (R437, R433) | 0.896 |
| R442, R433 | NDSI (R442, R433) | 0.883 |
| R445, R433 | NDSI (R445, R433) | 0.887 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shao, Y.; Ji, S.; Xuan, G.; Ren, Y.; Feng, W.; Jia, H.; Wang, Q.; He, S. Detection and Analysis of Chili Pepper Root Rot by Hyperspectral Imaging Technology. Agronomy 2024, 14, 226. https://doi.org/10.3390/agronomy14010226
Shao Y, Ji S, Xuan G, Ren Y, Feng W, Jia H, Wang Q, He S. Detection and Analysis of Chili Pepper Root Rot by Hyperspectral Imaging Technology. Agronomy. 2024; 14(1):226. https://doi.org/10.3390/agronomy14010226
Chicago/Turabian StyleShao, Yuanyuan, Shengheng Ji, Guantao Xuan, Yanyun Ren, Wenjie Feng, Huijie Jia, Qiuyun Wang, and Shuguo He. 2024. "Detection and Analysis of Chili Pepper Root Rot by Hyperspectral Imaging Technology" Agronomy 14, no. 1: 226. https://doi.org/10.3390/agronomy14010226






