Enhancing Resistance to Salinity in Wheat by Using Streptomyces sp. HU2014
Abstract
:1. Introduction
2. Materials and Methods
2.1. Salt Stress and HU2014 Inoculation on Wheat
2.2. Determination of Wheat Physiological Indicators
2.3. Soil Physiochemical Investigation
2.4. DNA Extraction and Sequencing, and Bioinformatics Analysis
2.5. Statistical Analysis
3. Results
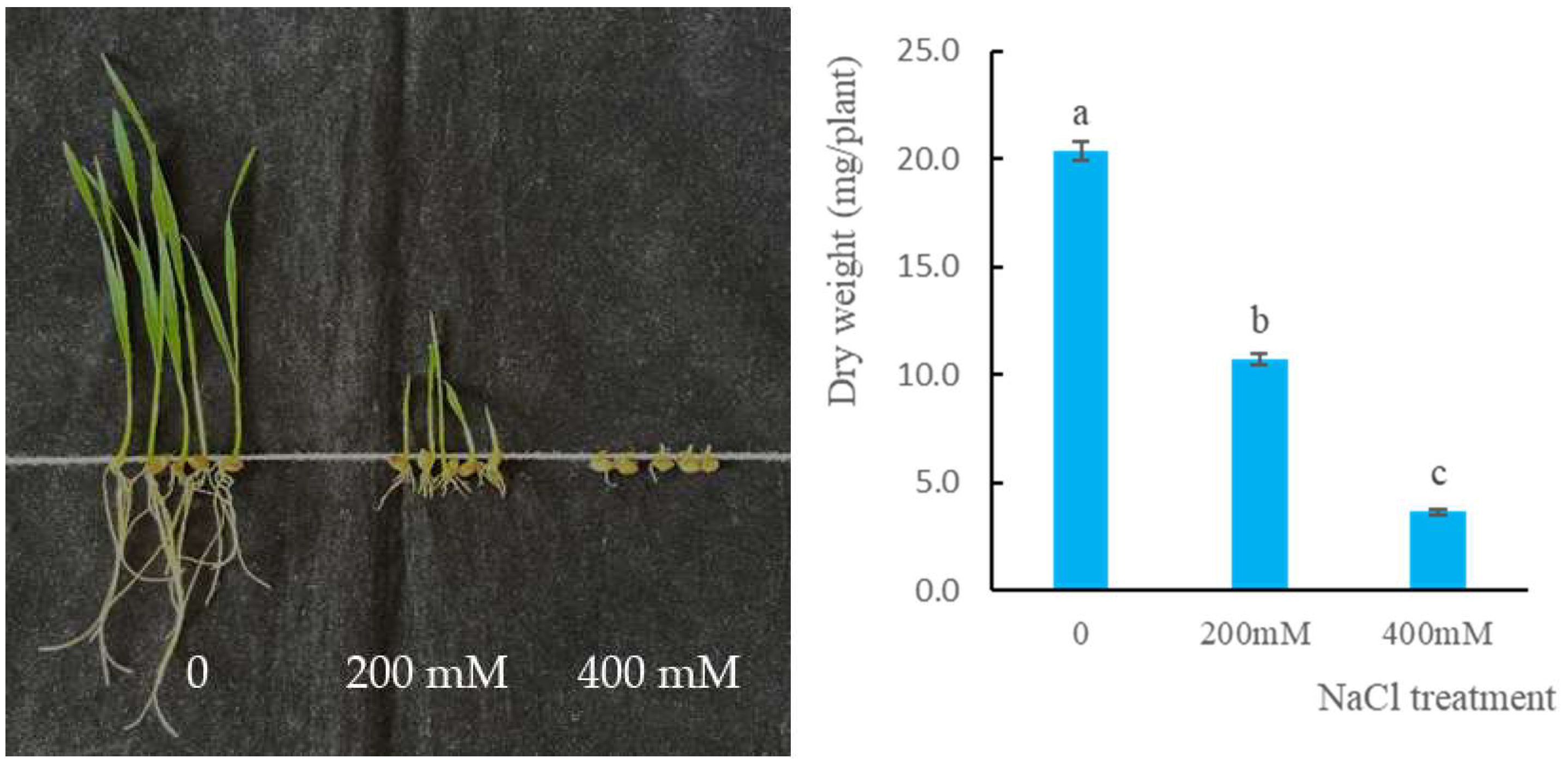
3.1. NaCl Resistance of ZM22
3.2. Soil Physicochemical Properties in Rhizosphere
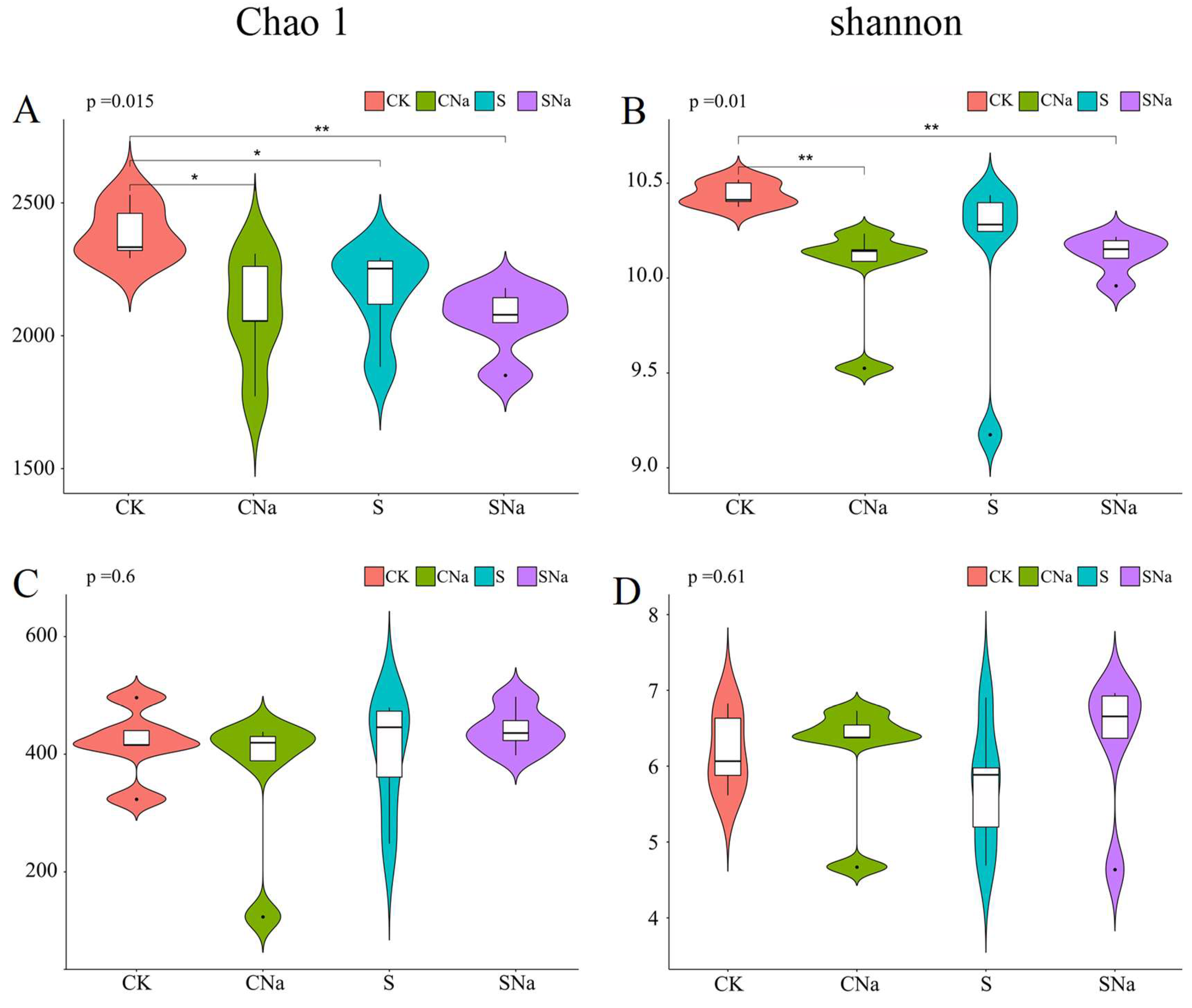
3.3. Alpha Diversity of Bacteria and Fungi Communities
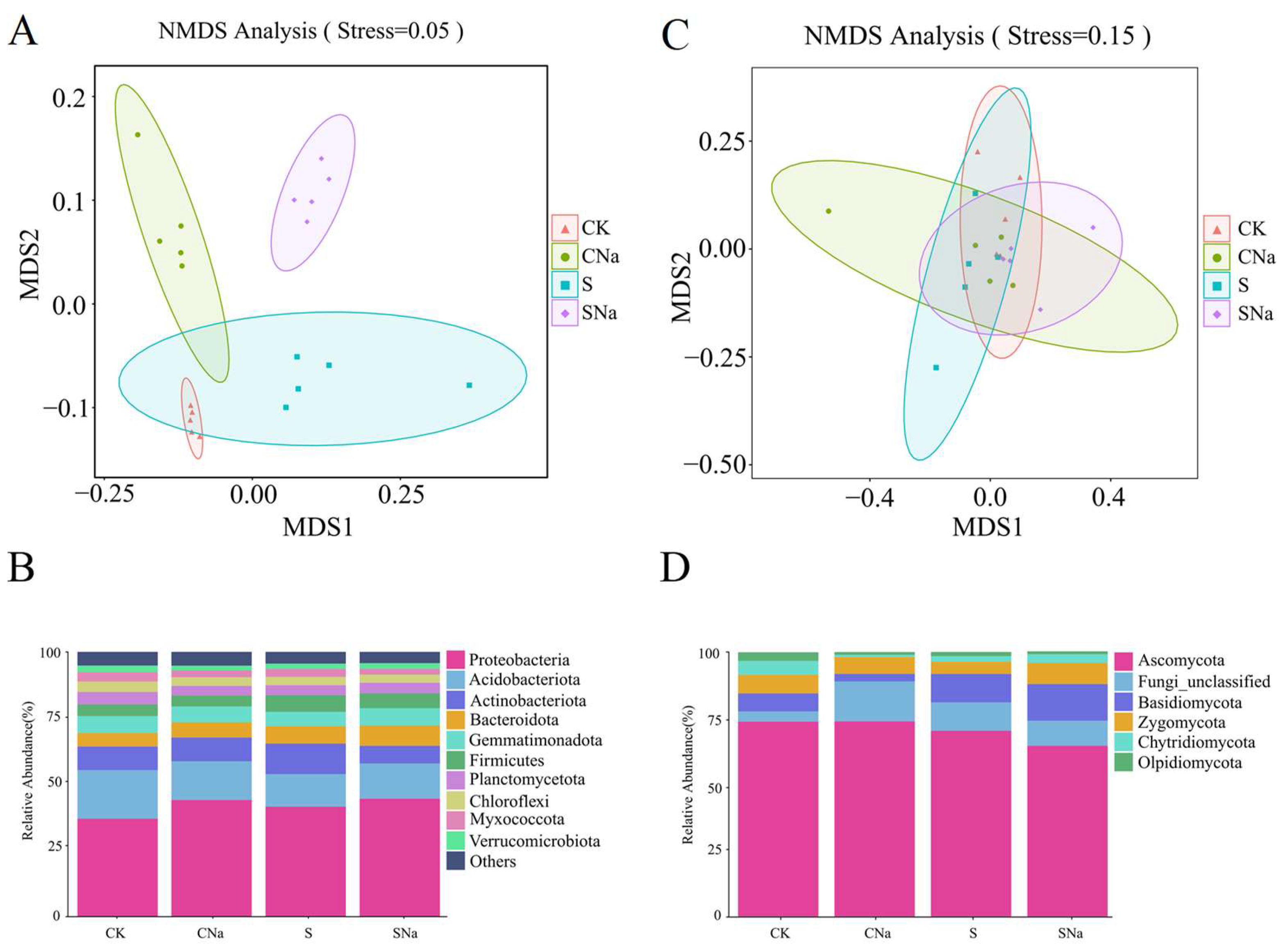
3.4. Distribution of Bacteria and Fungi Community Structures
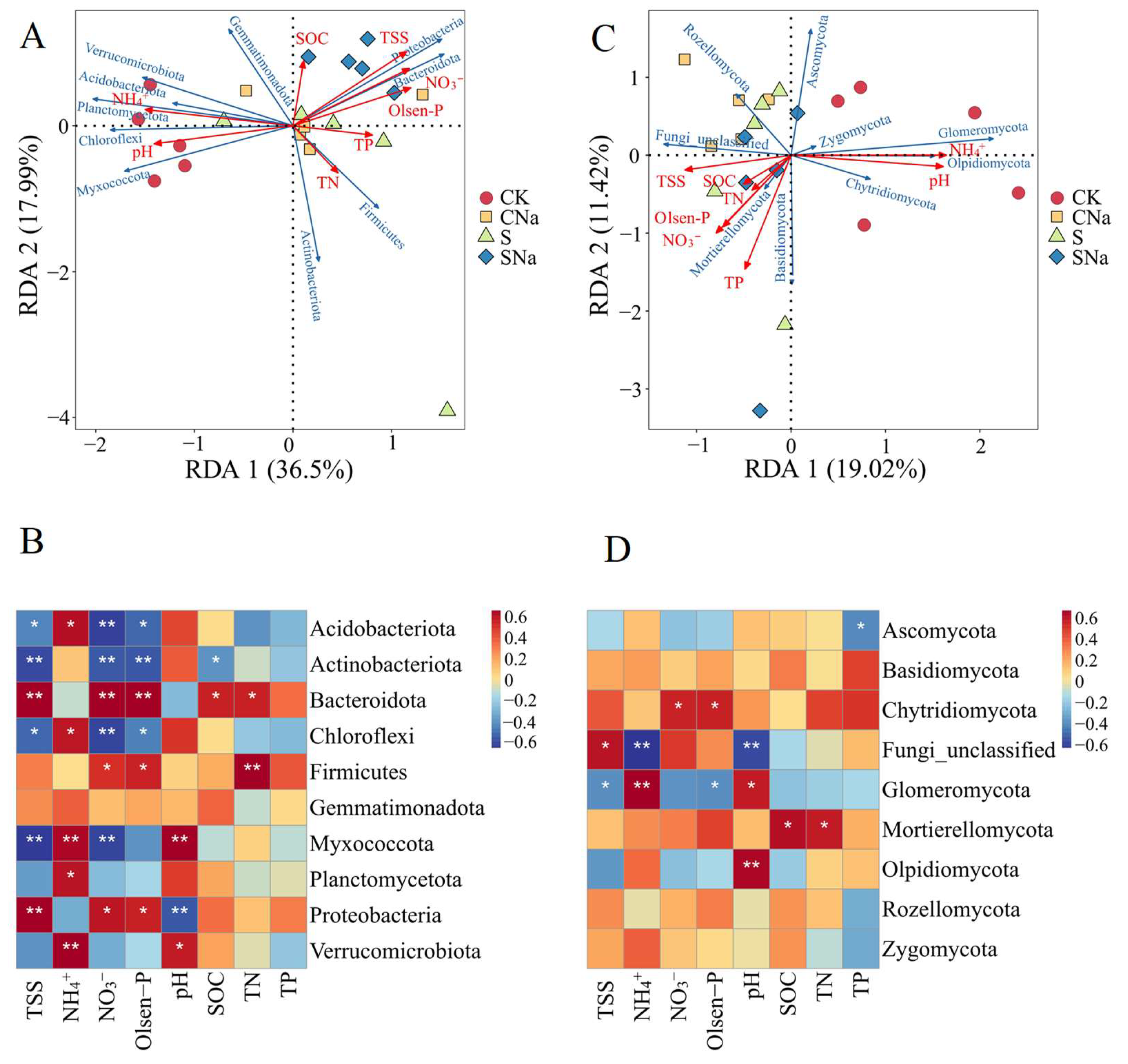
3.5. Relationship between Soil Properties and Microbial Communities
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wu, H.F.; Liu, X.L.; You, L.P.; Zhang, L.B.; Zhou, D.; Feng, J.H.; Zhao, J.M.; Yu, J.B. Effects of salinity on metabolic profiles, gene expressions, and antioxidant enzymes in halophyte. J. Plant Growth Regul. 2012, 31, 332–341. [Google Scholar] [CrossRef]
- Shabala, S. Learning from halophytes: Physiological basis and strategies to improve abiotic stress tolerance in crops. Ann. Bot. 2013, 112, 1209–1221. [Google Scholar] [CrossRef] [PubMed]
- Li, J.G.; Pu, L.J.; Han, M.F.; Zhu, M.; Zhang, R.S.; Xiang, Y.Z. Soil salinization research in China: Advances and prospects. J. Geogr. Sci. 2014, 24, 943–960. [Google Scholar] [CrossRef]
- Sairam, R.K.; Rao, K.V.; Srivastava, G.C. Differential response of wheat genotypes to long term salinity stress in relation to oxidative stress, antioxidant activity and osmolyte concentration. Plant Sci. 2002, 163, 1037–1046. [Google Scholar] [CrossRef]
- Hou, Y.L.; Zeng, W.Z.; Hou, M.L.; Wang, Z.; Luo, Y.; Lei, G.Q.; Zhou, B.; Huang, J.S. Responses of the soil microbial community to salinity stress in maize fields. Biology 2021, 10, 1114. [Google Scholar] [CrossRef]
- Wang, L.; Seki, K.; Miyazaki, T.; Ishihama, Y. The causes of soil alkalinization in the Songnen Plain of Northeast China. Paddy Water Environ. 2009, 7, 259–270. [Google Scholar] [CrossRef]
- Hanin, M.; Ebel, C.; Ngom, M.; Laplaze, L.; Masmoudi, K. New insights on plant salt tolerance mechanisms and their potential use for breeding. Front. Plant Sci. 2016, 7, 1787. [Google Scholar] [CrossRef]
- Song, Y.S.; Yang, W.J.; Fan, H.; Zhang, X.S.; Sui, N. TaMYB86B encodes a R2R3-type MYB transcription factor and enhances salt tolerance in wheat. Plant Sci. 2020, 300, 110624. [Google Scholar] [CrossRef]
- Anwar, A.; Zhang, S.; He, L.L.; Gao, J.W. Understanding the physiological and molecular mechanism of salinity stress tolerance in plants. Not. Bot. Horti. Agrobo. 2022, 50, 12595. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.X.; Yan, X.R.; Guo, J.P. Physiological and transcriptomic analyses of yellow horn (Xanthoceras sorbifolia) provide important insights into salt and saline-alkali stress tolerance. PLoS ONE 2020, 15, e0244365. [Google Scholar] [CrossRef]
- Guo, M.; Wang, X.S.; Guo, H.D.; Bai, S.Y.; Khan, A.; Wang, X.M.; Gao, Y.M.; Li, J.S. Tomato salt tolerance mechanisms and their potential applications for fighting salinity: A review. Front. Plant Sci. 2022, 13, 949541. [Google Scholar] [CrossRef] [PubMed]
- Tavakkoli, E.; Fatehi, F.; Coventry, S.; Rengasamy, P.; McDonald, G.K. Additive effects of Na+ and Cl− ions on barley growth under salinity stress. J. Exp. Bot. 2011, 62, 2189–2203. [Google Scholar] [CrossRef] [PubMed]
- Reginato, M.A.; Castagna, A.; Furlán, A.; Castro, S.; Ranieri, A.; Luna, V. Physiological responses of a halophytic shrub to salt stress by Na2SO4 and NaCl: Oxidative damage and the role of polyphenols in antioxidant protection. AoB Plants 2014, 6, plu042. [Google Scholar] [CrossRef] [PubMed]
- Ferreira-Silva, S.L.; Voigt, E.L.; Silva, E.N.; Maia, J.M.; Aragao, T.C.R.; Silveira, J.A.G. Partial oxidative protection by enzymatic and non-enzymatic components in cashew leaves under high salinity. Biol. Plant. 2012, 56, 172–176. [Google Scholar] [CrossRef]
- Sorkheh, K.; Shiran, B.; Rouhi, V.; Khodambashi, M.; Sofo, A. Salt stress induction of some key antioxidant enzymes and metabolites in eight Iranian wild almond species. Acta Physiol. Plant. 2012, 34, 203–213. [Google Scholar] [CrossRef]
- Dien, D.C.; Mochizuki, T.; Yamakawa, T. Effect of various drought stresses and subsequent recovery on proline, total soluble sugar and starch metabolisms in rice (Oryza sativa L.) varieties. Plant Prod. Sci. 2019, 22, 530–545. [Google Scholar] [CrossRef]
- Moghaddam, M.; Farhadi, N.; Panjtandoust, M.; Ghanati, F. Seed germination, antioxidant enzymes activity and proline content in medicinal plant Tagetes minuta under salinity stress. Plant Biosyst. 2020, 154, 835–842. [Google Scholar] [CrossRef]
- Fang, S.M.; Hou, X.; Liang, X.L. Response mechanisms of plants under saline-alkali stress. Front. Plant Sci. 2021, 12, 667458. [Google Scholar] [CrossRef]
- Omae, N.; Tsuda, K. Plant-microbiota interactions in abiotic stress environments. Mol. Plant Microbe Interact. 2022, 35, 511–526. [Google Scholar] [CrossRef]
- Meena, K.K.; Sorty, A.M.; Bitla, U.M.; Choudhary, K.; Gupta, P.; Pareek, A.; Singh, D.P.; Prabha, R.; Sahu, P.K.; Gupta, V.K.; et al. Abiotic stress responses and microbe-mediated mitigation in plants: The omics strategies. Front. Plant Sci. 2017, 8, 172. [Google Scholar] [CrossRef]
- Zhang, Y.Q.; Yang, F.; Ren, H.J.; Liu, J.L.; Mu, W.Y.; Wang, Y. Pseudomonas monteilii PN1: A great potential P-nitrophenol degrader with plant growth promoting traits under drought and saline-alkali stresses. Biotechnol. Lett. 2019, 41, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.J.; Liu, L.; Singh, R.P.; Meng, C.; Ma, S.Q.; Jing, C.L.; Li, Y.Q.; Zhang, C.S. Nodule and root zone microbiota of salt-tolerant wild soybean in coastal sand and saline-alkali soil. Front. Microbiol. 2020, 11, 523142. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.Y.; Hao, L.P.; Ji, C.; Zhou, Q.S.; Song, X.; Liu, Y.; Li, H.Y.; Li, C.H.; Gao, Q.X.; Li, J.T.; et al. The effect of salt-tolerant antagonistic bacteria CZ-6 on the rhizosphere microbial community of winter jujube (Ziziphus jujuba Mill. “Dongzao”) in saline-alkali land. Biomed Res. Int. 2021, 2021, 5171086. [Google Scholar]
- Pang, F.; Solanki, M.K.; Wang, Z. Streptomyces can be an excellent plant growth manager. World J. Microbiol. Biotechnol. 2022, 38, 193. [Google Scholar] [CrossRef] [PubMed]
- Azeem, M.A.; Shah, F.H.; Ullah, A.; Ali, K.; Jones, D.A.; Khan, M.E.H.; Ashraf, A. Biochemical Characterization of Halotolerant Bacillus safensis PM22 and Its Potential to Enhance Growth of Maize under Salinity Stress. Plants 2022, 11, 1721. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, A.; Karimi, E.; Dahaji, P.A.; Javid, M.G.; Dalvand, Y.; Askari, H. Plant growth promoting activity of an auxin and siderophore producing isolate of Streptomyces under saline soil conditions. World J. Microbiol. Biotechnol. 2012, 28, 1503–1509. [Google Scholar] [CrossRef] [PubMed]
- Afridi, M.S.; Mahmoode, T.; Salam, A.; Mukhtar, T.; Mehmood, S.; Ali, J.; Khatoon, Z.; Bibi, M.; Javed, T.; Sultan, T.; et al. Induction of tolerance to salinity in wheat genotypes by plant growth promoting endophytes: Involvement of ACC deaminase and antioxidant enzymes. Plant Physiol. Biochem. 2019, 139, 569–577. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Reich, P.B.; Trivedi, C.; Eldridge, D.J.; Abades, S.; Alfaro, F.D.; Bastida, F.; Berhe, A.A.; Cutler, N.A.; Gallardo, A.; et al. Multiple elements of soil biodiversity drive ecosystem functions across biomes. Nat. Ecol. Evol. 2020, 4, 210–220. [Google Scholar] [CrossRef]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.C.C.; Charles, T.; Chen, X.Y.L.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar]
- Kumawat, K.C.; Nagpal, S.; Sharma, P. Potential of plant growth-promoting rhizobacteria-plant interactions in mitigating salt stress for sustainable agriculture: A review. Pedosphere 2022, 32, 223–245. [Google Scholar] [CrossRef]
- Zhu, H.-X.; Hu, L.-F.; Hu, H.-Y.; Zhou, F.; Wu, L.-L.; Wang, S.-W.; Rozhkova, T.; Li, C.-W. Identification of a novel Streptomyces sp. Strain HU2014 showing growth promotion and biocontrol effect against Rhizoctonia spp. in wheat. Plant Dis. 2023, 107, 1139–1150. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.W.; Gan, Y.T.; Xu, B.L. Mechanisms of the IAA and ACC-deaminase producing strain of Trichoderma longibrachiatum T6 in enhancing wheat seedling tolerance to NaCl stress. BMC Plant Biol. 2019, 19, 18. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.-H.; Liu, J.; He, X.-J.; Mu, R.-L.; Zhou, H.-L.; Chen, S.-Y.; Zhang, J.-S. Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 2007, 143, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.C.; Zhao, G.Y.; Xia, C.; Jia, J.Z.; Liu, X.; Kong, X.Y. Overexpression of a wheat MYB transcription factor gene, TaMYB56-B, enhances tolerances to freezing and salt stresses in transgenic Arabidopsis. Gene 2012, 505, 100–107. [Google Scholar] [CrossRef]
- Yang, L.Y.; Zhou, S.Y.D.; Lin, C.S.; Huang, X.R.; Neilson, R.; Yang, X.R. Effects of biofertilizer on soil microbial diversity and antibiotic resistance genes. Sci. Total Environ. 2022, 820, 153170. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.C.; Wang, M.K.; Cai, L.; Cang, L. Deciphering biodegradable chelant-enhanced phytoremediation through microbes and nitrogen transformation in contaminated soils. Environ. Sci. Pollut. Res. 2017, 24, 14627–14636. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.K. Methods for Agricultural Chemical Analysis of Soil; China Agricultural Science and Technology Press: Beijing, China, 2000; pp. 50–95. ISBN 7-80119-925-1. [Google Scholar]
- Karlsson, I.; Friberg, H.; Steinberg, C.; Persson, P. Fungicide effects on fungal community composition in the wheat phyllosphere. PLoS ONE 2014, 9, e111786. [Google Scholar] [CrossRef]
- Logue, J.B.; Stedmon, C.A.; Kellerman, A.M.; Nielsen, N.J.; Andersson, A.F.; Laudon, H.; Lindström, E.S.; Kritzberg, E.S. Experimental insights into the importance of aquatic bacterial community composition to the degradation of dissolved organic matter. Isme J. 2016, 10, 533–545. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.; Kindt, R.; Legendre, P.; Wagner, H. Vegan: Community Ecology Package, R Package Version 2.3-5; R Foundation: Vienna, Austria, 2016. [Google Scholar]
- Salwan, R.; Sharma, A.; Sharma, V. Microbes mediated plant stress tolerance in saline agricultural ecosystem. Plant Soil 2019, 442, 1–22. [Google Scholar] [CrossRef]
- Akbari, A.; Gharanjik, S.; Koobaz, P.; Sadeghi, A. Plant growth promoting Streptomyces strains are selectively interacting with the wheat cultivars especially in saline conditions. Heliyon 2020, 6, e03445. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, M.; Chandra, P.; Dixit, B.; Nehra, V.; Choudhary, U.; Choudhary, S. Plant growth-promoting microbes: Role and prospective in amelioration of salt stress. Commun. Soil Sci. Plant Anal. 2022, 53, 1692–1711. [Google Scholar] [CrossRef]
- Liu, H.Q.; Lu, X.B.; Li, Z.H.; Tian, C.Y.; Song, J. The role of root-associated microbes in growth stimulation of plants under saline conditions. Land Degrad. Dev. 2021, 32, 3471–3486. [Google Scholar] [CrossRef]
- Boamah, S.; Zhang, S.W.; Xu, B.L.; Li, T.; Calderón-Urrea, A. Trichoderma longibrachiatum (TG1) enhances wheat seedlings tolerance to salt stress and resistance to Fusarium pseudograminearum. Front. Plant Sci. 2021, 12, 741231. [Google Scholar] [CrossRef] [PubMed]
- Wafaa, M.H.; Tawfik, M.M.; Abouziena, H.F.; Abd El Wahed, M.S.; Ali, R.R. Enhancing wheat production under arid climate stresses using bio-elicitors. Gesunde Pflanz. 2017, 69, 149–158. [Google Scholar] [CrossRef]
- Li, H.Y.; Guo, Q.; Jing, Y.X.; Liu, Z.; Zheng, Z.H.; Sun, Y.F.; Xue, Q.H.; Lai, H.X. Application of Streptomyces pactum Act12 enhances drought resistance in wheat. J. Plant Growth Regul. 2020, 39, 122–132. [Google Scholar] [CrossRef]
- Abbasi, S.; Sadeghi, A.; Safaie, N. Streptomyces alleviate drought stress in tomato plants and modulate the expression of transcription factors ERF1 and WRKY70 genes. Sci. Hortic. 2020, 265, 109206. [Google Scholar] [CrossRef]
- Bharti, N.; Barnawal, D.; Maji, D.; Kalra, A. Halotolerant PGPRs prevent major shifts in indigenous microbial community structure under salinity stress. Microb. Ecol. 2015, 70, 196–208. [Google Scholar] [CrossRef]
- Liang, X.L.; Wang, X.Y.; Zhang, N.; Li, B.X. Biogeographical patterns and assembly of bacterial communities in saline soils of Northeast China. Microorganisms 2022, 10, 1787. [Google Scholar] [CrossRef]
- Luo, J.Q.; Zhang, Z.C.; Hou, Y.Z.; Diao, F.W.; Hao, B.H.; Bao, Z.H.; Wang, L.X.; Guo, W. Exploring microbial resource of different rhizocompartments of dominant plants along the salinity gradient around the hypersaline lake Ejinur. Front. Microbiol. 2021, 12, 698479. [Google Scholar] [CrossRef]
- Barnawal, D.; Bharti, N.; Pandey, S.S.; Pandey, A.; Chanotiya, C.S.; Kalra, A. Plant growth-promoting rhizobacteria enhance wheat salt and drought stress tolerance by altering endogenous phytohormone levels and TaCTR1/TaDREB2 expression. Physiol. Plant. 2017, 161, 502–514. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Lv, C.Q.; Fernández-García, V.; Huang, B.L.; Yao, J.M.; Ding, W. Biochar and PGPR amendments influence soil enzyme activities and nutrient concentrations in a eucalyptus seedling plantation. Biomass Convers. Biorefinery 2021, 11, 1865–1874. [Google Scholar] [CrossRef]
- Li, M.Y.; Wang, J.L.; Yao, T.; Zhang, T.; Zhou, Q.; Mutallip, M. Bacterial diversity and community structure in the rhizosphere of four halophytes. Curr. Microbiol. 2021, 78, 2720–2732. [Google Scholar] [CrossRef] [PubMed]
- Soothar, M.K.; Hamani, A.K.M.; Sardar, M.F.; Sootahar, M.K.; Fu, Y.Y.; Rahim, R.; Soothar, J.K.; Bhatti, S.M.; Abubakar, S.A.; Gao, Y.; et al. Maize (Zea mays L.) seedlings rhizosphere microbial community as responded to acidic biochar amendment under saline conditions. Front. Microbiol. 2021, 12, 789235. [Google Scholar] [CrossRef]
- Shi, X.L.; Zhao, X.H.; Ren, J.Y.; Dong, J.L.; Zhang, H.; Dong, Q.Q.; Jiang, C.J.; Zhong, C.; Zhou, Y.F.; Yu, H.Q. Influence of peanut, sorghum, and soil salinity on microbial community composition in interspecific interaction zone. Front. Microbiol. 2021, 12, 678250. [Google Scholar] [CrossRef]
- Green, L.E.; Porras-Alfaro, A.; Sinsabaugh, R.L. Translocation of nitrogen and carbon integrates biotic crust and grass production in desert grassland. J. Ecol. 2008, 96, 1076–1085. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, H.; Hu, L.; Rozhkova, T.; Li, C. Enhancing Resistance to Salinity in Wheat by Using Streptomyces sp. HU2014. Agronomy 2024, 14, 39. https://doi.org/10.3390/agronomy14010039
Zhu H, Hu L, Rozhkova T, Li C. Enhancing Resistance to Salinity in Wheat by Using Streptomyces sp. HU2014. Agronomy. 2024; 14(1):39. https://doi.org/10.3390/agronomy14010039
Chicago/Turabian StyleZhu, Hongxia, Linfeng Hu, Tetiana Rozhkova, and Chengwei Li. 2024. "Enhancing Resistance to Salinity in Wheat by Using Streptomyces sp. HU2014" Agronomy 14, no. 1: 39. https://doi.org/10.3390/agronomy14010039
APA StyleZhu, H., Hu, L., Rozhkova, T., & Li, C. (2024). Enhancing Resistance to Salinity in Wheat by Using Streptomyces sp. HU2014. Agronomy, 14(1), 39. https://doi.org/10.3390/agronomy14010039





