The Genetic Diversity of 69 Widely Used Chinese Sorghum Hybrids Released between the 1970s and 2010s
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Genotyping
2.3. Statistical Analysis
3. Result
3.1. Diversity of SSR Markers in Hybrid Sorghum Varieties
3.2. Analysis of Genetic Diversity and Genetic Similarity in Hybrid Varieties Released in Different Breeding Development Stages
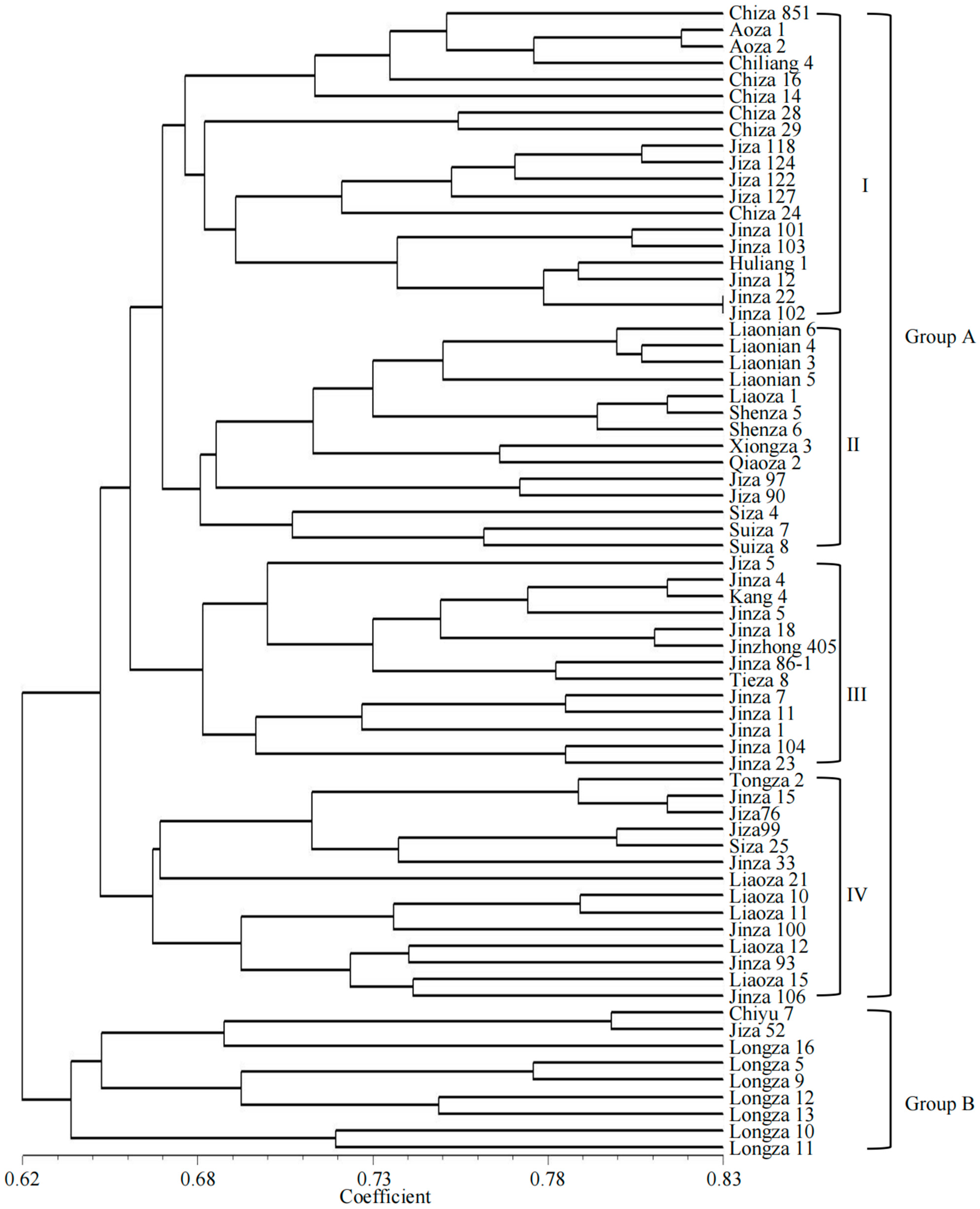
3.3. Cluster Analysis of Hybrid Sorghum Varieties
4. Discussion
4.1. SSR Marker Performed High Polymorphism in Hybrid Variety Population
4.2. The Overall Genetic Diversity of Sorghum Varieties in China Was Gradually Increasing
4.3. Hybrid Varieties Could Be Divided into Two Major Groups with No Significant Correlation between Group Differentiation and Regional Origin of Varieties
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- FAOSTAT. Food and Agriculture Organization of the United Nations. FAO, 2023. Available online: https://www.fao.org/faostat/en/#data/QCL/visualize (accessed on 28 April 2024).
- Baloch, F.S.; Altaf, M.T.; Liaqat, W.; Bedir, M.; Nadeem, M.A.; Cömertpay, G.; Çoban, N.; Habyarimana, E.; Barutçular, C.; Cerit, I.; et al. Recent advancements in the breeding of sorghum crop: Current status and future strategies for marker-assisted breeding. Front. Genet. 2023, 14, 1150616. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.P.; Ramu, P.; Deshpande, S.P.; Hash, C.T.; Shah, T.; Upadhyaya, H.D.; Riera-Lizarazu, O.; Brown, P.J.; Acharya, C.B.; Mitchell, S.E.; et al. Population genomic and genome-wide association studies of agronomic traits in sorghum. Proc. Natl. Acad. Sci. USA 2012, 110, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Paterson, A.H.; Bowers, J.E.; Feltus, F.A. Genomics of Sorghum, a Semi-Arid Cereal and Emerging Model for Tropical Grass Genomics. In Genomics of Tropical Crop Plants; Moore, P.H., Ming, R., Eds.; Springer: New York, NY, USA, 2008; pp. 469–482. [Google Scholar]
- Sweta, S.; Kumaravadivel, N. Understanding genetic diversity of sorghum using quantitative traits. Int. J. Bio-Resour. Stress Manag. 2015, 6, 552–557. [Google Scholar] [CrossRef]
- Behera, P.P.; Saharia, N.; Borah, N.; Devi, S.H.; Sarma, R.N. Sorghum physiology and adaptation to abiotic stresses. Int. J. Environ. Clim. Chang. 2022, 12, 1005–1022. [Google Scholar] [CrossRef]
- Popescu, A.; Dinu, T.A.; Stoian, E. Sorghum—An important cereal in the world, in the European Union and Romania. Sci. Pap.-Ser. Manag. Econ. Eng. Agric. Rural Dev. 2018, 18, 271–284. [Google Scholar]
- United States Department of Agricultural: Economics, Statistics and Market Information System. 2023. Available online: https://usda.library.cornell.edu/concern/publications/5q47rn72z?locale=en (accessed on 3 June 2024).
- Lu, Q.-S.; Jeffery, A.D. Chinese sorghum genetic resources. Econ. Bot. 2001, 55, 401–425. [Google Scholar] [CrossRef]
- Gao, S.-J.; Liu, X.-H.; Guo, Z.-X.; Li, J.-H. Chinese hybrid sorghum idioplasm foundation and superiority use pattern. Chin. Agric. Sci. Bull. 2005, 21, 106. [Google Scholar] [CrossRef]
- Lv, F.-T.; Han, A.-Q.; Du, X.-L.; Zhang, F.-Y.; Li, T.-Y. Development and tendency of Chinese sorghum since the funding of P.R.China. J. Shanxi Agric. Sci. 2002, 30, 20–24. [Google Scholar]
- Hutchison, P.D. Grain Sorghum in the United States; Food and Fiber National Institute of Achievement: Washington, DC, USA, 1977; pp. 44–52.
- Li, T.-Y.; Zhang, F.-Y.; Li, S.-M.; Wei, Y.-M.; Liu, Q.-S. Development of a new type cytoplasm male sterile line A2V4A and its application in sorghum. Chin. Agric. Sci. Bull. 1995, 11, 10–13. [Google Scholar]
- Liu, Q.-S. Research course and prospects of Shanxi hybrid sorghum breeding. J. Shanxi Agric. Sci. 2023, 51, 1115–1120. [Google Scholar]
- Lobell, D.B.; Burke, M.B.; Tebaldi, C.; Mastrandrea, M.D.; Falcon, W.P.; Naylor, R.L. Prioritizing climate change adaptation needs for food security in 2030. Science 2008, 319, 607–610. [Google Scholar] [CrossRef] [PubMed]
- Gepts, P. Plant genetic resources conservation and utilization: The accomplishments and future of a societal insurance policy. Crop Sci. 2006, 46, 2278–2292. [Google Scholar] [CrossRef]
- Cuevas, H.E.; Knoll, J.E.; Prom, L.K.; Stutts, L.R.; Vermerris, W. Genetic diversity, population, structure and anthracnose resistance response in a novel sorghum diversity panel. Front. Plant Sci. 2023, 14, 1249555. [Google Scholar] [CrossRef] [PubMed]
- Dickson, N.; Mulatu, G.; Tomas, B. Genetic diversity in sorghum (Sorghum bicolor (L.) Moench) accessions of Zambia as revealed by simple sequence repeats (SSR). Hereditas 2011, 148, 52–62. [Google Scholar] [CrossRef]
- Ge, F.-Y.; Xie, P.; Wu, Y.-R.; Xie, Q. Genetic architecture and molecular regulation of sorghum domestication. aBIOTECH 2023, 4, 57–71. [Google Scholar] [CrossRef]
- Smith, S.; Primomo, V.; Monk, R.; Nelson, B.; Jones, E.; Porter, K. Genetic diversity of widely used U.S. sorghum hybrids 1980–2008. Crop Sci. 2010, 50, 1664–1673. [Google Scholar] [CrossRef]
- Folkertsma, R.T.; Rattunde, H.F.W.; Chandra, S.; Raju, G.S.; Hash, C.T. The pattern of genetic diversity of Guinea-race Sorghum bicolor (L.) Moench landraces as revealed with SSR markers. Theor. Appl. Genet. 2005, 111, 399–409. [Google Scholar] [CrossRef]
- Mamo, W.; Enyew, M.; Mekonnen, T.; Tesfaye, K.; Feyissa, T. Genetic diversity and population structure of sorghum (Sorghum bicolor (L.) Moench) genotypes in Ethiopia as revealed by microsatellite markers. Heliyon 2023, 9, e12830. [Google Scholar] [CrossRef]
- Kale, S.S.; Borde, G.; Ansari, M.A.; Ahmad, S.F.; Malik, A.; Shahid, M. Genetic diversity analysis in sorghum [Sorghum bicolor (L.)] by using SSR markers. Res. Sq. 2024. [Google Scholar] [CrossRef]
- Lu, Q.-S.; Zou, J.-Q. Theory of Sorghum, 2nd ed.; China Agriculture Press: Beijing, China, 2023; pp. 215–216. [Google Scholar]
- Vieira, M.B.; Faustino, M.V.; Lourenco, T.F.; Oliveira, M.M. DNA-based tools to certify authenticity of rice varieties—An overview. Foods 2022, 11, 258. [Google Scholar] [CrossRef]
- Fang, W.; Cheng, H.; Duan, Y.; Jiang, X.; Li, X. Genetic diversity and relationship of clonal tea (Camellia sinensis) cultivars in China as revealed by SSR markers. Plant Syst. Evol. 2011, 298, 469–483. [Google Scholar] [CrossRef]
- Ma, J.-Q.; Jin, J.-Q.; Yao, M.-Z.; Ma, C.-L.; Xu, Y.-X.; Hao, W.-J.; Chen, L. Quantitative trait loci mapping for theobromine and caffeine contents in tea plant (Camellia sinensis). J. Agric. Food Chem. 2018, 66, 13321–13327. [Google Scholar] [CrossRef] [PubMed]
- Roussel, V.; Leisova, L.; Exbrayat, F.; Stehno, Z.; Balfourier, F. SSR allelic diversity changes in 480 European bread wheat varieties released from 1840 to 2000. Theor. Appl. Genet. 2005, 111, 162–170. [Google Scholar] [CrossRef]
- An, Y.-L.; Xia, X.-B.; Zheng, H.-Y.; Yu, S.-R.; Jing, T.-T.; Zhang, F. Multi-genome comprehensive identification of SSR/SV and development of molecular markers database to serve Sroghum bicolor (L.) breeding. BMC Genom. Data 2023, 24, 62. [Google Scholar] [CrossRef] [PubMed]
- Menz, M.A.; Klein, R.B.; Unruh, N.C.; Rooney, W.L.; Klein, P.E.; Mullet, J.E. Genetic diversity of public inbreds of sorghum determined by mapped AFLP and SSR markers. Crop Sci. 2004, 44, 1236–1244. [Google Scholar] [CrossRef]
- Perumal, P.; Krishnaramanujam, P.; Menz, M.A. Genetic diversity among sorghum races and working groups based on AFLPs and SSRs. Crop Sci. 2007, 47, 1375–1383. [Google Scholar] [CrossRef]
- Ghebru, B.; Schmidt, R.J.; Bennetzen, J.L. Genetic diversity of Eritrean sorghum landraces assessed with simple sequence repeat (SSR) markers. Theor. Appl. Genet. 2002, 105, 229–236. [Google Scholar] [CrossRef]
- Mudaki, P.; Wamalwa, L.N.; Muui, C.W.; Nzuve, F.; Muasya, R.M.; Nguluu, S.; Kimani, W. Genetic diversity and population structure of sorghum (Sorghum bicolor (L.) Moench) landraces using DArTseq-derived single-nucleotide polymorphism (SNP) markers. J. Mol. Evol. 2023, 91, 552–561. [Google Scholar] [CrossRef]
- Mindaye, T.T.; Mace, E.S.; Godwin, I.D.; Jordan, D.R. Genetic differentiation analysis for the identification of complementary parental pools for sorghum hybrid breeding in Ethiopia. Theor. Appl. Genet. 2015, 128, 1765–1775. [Google Scholar] [CrossRef]
- Clark, R.D.; Pinsky, M.L. Global patterns of nuclear and mitochondrial genetic diversity in marine fishes. Ecol. Evol. 2024, 14, e11365. [Google Scholar] [CrossRef]
- Mabunda, R.S.; Nephawe, K.A.; Mtileni, B.; Makgahlela, M.L. Pedigree-based genetic diversity in the South African Boerboel dog breed. Animals 2024, 14, 975. [Google Scholar] [CrossRef] [PubMed]
- van de Wouw, M.; van Hintum, T.; Kik, C.; van Treuren, R.; Visser, B. Genetic diversity trends in twentieth century crop cultivars: A meta analysis. Theor. Appl. Genet. 2010, 120, 1241–1252. [Google Scholar] [CrossRef] [PubMed]
- Schertz, K.F.; Sotomayor-Rios, A.; Torraco-Cardona, S. Cytoplasmic-nuclear male-sterility opportunities in breeding and genetics. Proc. Grain Sorghum Res. Util. Conf. 1989, 16, 175–186. [Google Scholar]
- Jordan, D.R.; Klein, R.R.; Sakrewski, K.G.; Henzell, R.G.; Klein, P.E.; Mace, E.S. Mapping and characterization of Rf5 a new gene conditioning pollen fertility restoration in A1 and A2 cytoplasm in sorghum (Sorghum bicolor (L.) Moench). Theor. Appl. Genet. 2011, 123, 383–396. [Google Scholar] [CrossRef]
- Stephens, J.C.; Miller, F.R.; Rosenow, D.T. Conversion of alien sorghums to early combine genotypes. Crop Sci. 1967, 7, 396. [Google Scholar] [CrossRef]
- Klein, R.P.; Mullet, J.E.; Jordan, D.R.; Miller, F.R.; Rooney, W.L.; Menz, M.A.; Franks, C.D.; Klein, P.E. The effect of tropical sorghum conversion and inbred development on genome diversity as revealed by high-resolution genotyping. Crop Sci. 2008, 48, S12–S26. [Google Scholar] [CrossRef]
- Klein, R.R.; Miller, F.R.; Bean, S.; Klein, P.E. Registration of 40 converted germplasm sources from the reinstated sorghum conversion program. J. Plant Regist. 2015, 10, 57–61. [Google Scholar] [CrossRef]
- Horne, D.W.; Patil, N.Y.; Klein, R.R.; Miller, F.R.; Hoffmann, L.; Klein, P.E.; Rooney, W.L. Registration of 11 diverse sorghum germplasm lines for grain and silage hybrid production. J. Plant Regist. 2020, 14, 179–188. [Google Scholar] [CrossRef]
- Crozier, D.; Winans, N.D.; Hoffmann, L., Jr.; Patil, N.Y.; Klein, P.E.; Klein, R.R.; Rooney, W.L. Evaluation and prediction the performance of sorghum lines in an elite by exotic backcross-nested association mapping population. Plants 2024, 13, 879. [Google Scholar] [CrossRef]
- Silva, K.J.; Pastina, M.M.; Guimarães, C.T.; Magalkães, J.V.; Pimentel, L.D.; Schaffert, R.E.; Pinto, M.D.O.; Souza, V.F.D.; Bernardino, K.D.C.; Silva, M.J.D.; et al. Genetic diversity and heterotic grouping of sorghum lines using SNP markers. Sci. Agric. 2021, 78, e20200039. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, J.-C.; Wang, D.-J.; Yao, F.-X.; Xu, J.-F.; Song, G.-A.; Guan, Y.A.; Li, R.Y. Assessment of genetic diversity in Chinese sorghum lanraces using SSR markers as compared with foreign accessions. Acta Agron. Sin. 2011, 37, 224–234. [Google Scholar] [CrossRef]
- Adugna, A. Analysis of in situ diversity and population structure in Ethiopian cultivated Sorghum bicolor (L.) landraces using phenotypic traits and SSR markers. SpringerPlus 2014, 3, 212. [Google Scholar] [CrossRef]
| Stages | Sorghum Hybrid | Parental Combination | Release Year and Province |
|---|---|---|---|
| 1 (1973–1982) | Jinza 1 | Tx3197A×Jinfu 1 | 1973, Shanxi |
| Jinza 4 | Tx3197A×Jinliang 5 | 1973, Shanxi | |
| Jinza 5 | Tx3197A×Sanchisan | 1973, Shanxi | |
| Jinza 7 | Tx3197A×Xin 7 | 1973, Shanxi | |
| Tongza 2 | Heilong 11A×Jihui 7384 | 1973, Shanxi | |
| Jiza 52 | Heilong 30A×Jihui 13 | 1979, Jilin | |
| Chiyu 7 | Hei 30A×Yin 71 | 1980, Inner Mongolia | |
| 2 (1983–1992) | Liaoza 1 | Tx622A×Jinfu 1 | 1983, Liaoning |
| Jiza 5 | Cheng 3A×7501 | 1983, Hebei | |
| Aoza 1 | 314A×5933 | 1984, Inner Mongolia | |
| Chiza 851 | 314A×7657 | 1987, Inner Mongolia | |
| Jinzhong 405 | 7501A×Jinliang 5 | 1987, Shanxi | |
| Jinza 11 | Tx623A×Xin 7 | 1987, Shanxi | |
| Kang 4 | Tx623A×Jinliang 5 | 1988, Shanxi | |
| Shenza 5 | Tx622A×0-30 | 1988, Liaoning | |
| Qiaoza 2 | Tx622A×L654 | 1988, Liaoning | |
| Tieza 8 | Tx623A×Tieling 157 | 1989, Liaoning | |
| Shenza 6 | Tx622A×5-27 | 1989, Liaoning | |
| Siza 4 | 2731A×140R | 1990, Jilin | |
| Jinza 86-1 | Tx623A×HM65 | 1991, Shanxi | |
| Chiza 14 | Chi 12A×7663 | 1991, Inner Mongolia | |
| Aoza 2 | 314A×7788 | 1992, Inner Mongolia | |
| 3 (1993–2002) | Jinza 93 | 232EA/2036A×Shen 5-27 | 1993, Liaoning |
| Jinza 12 | A2V4A×1383-2 | 1994, Shanxi | |
| Huliang 1 | F14A×0-30 | 1995, Liaoning | |
| Xiongza 3 | Tx622A×4930 | 1994, Liaoning | |
| Jiza 76 | Heilong 11A×7431-24 | 1995, Jilin | |
| Liaoza 10 | 7050A×LR9198 | 1997, Liaoning | |
| Siza 25 | TAM428×Nan 133 | 1998, Jilin | |
| Jinza 15 | Heilong 11A×Qikangqi | 1998, Shanxi | |
| Longza 5 | 301A×Hahui 118 | 1999, Heilongjiang | |
| Jinza 18 | 7501A×R111 | 1999, Shanxi | |
| Liaoza 12 | 7050A×654 | 2001, Liaoning | |
| Jinza 100 | 7050A×9544 | 2001, Liaoning | |
| Jiza 90 | 4190A×9060 | 2001, Jilin | |
| Liaoza 11 | 7050A×148 | 2001, Liaoning | |
| Chiza 16 | Fan 8A×7654 | 2002, Inner Mongolia | |
| 4 (2003–2014) | Liaoza 15 | LA-17×LR9198 | 2003, Liaoning |
| Jiza 97 | 352A×133-6-8 | 2004, Jilin | |
| Liaoza 21 | 363A×0-01 | 2005, Liaoning | |
| Jiza 99 | TAM428A×Ji R107 | 2005, Jilin | |
| Jinza 101 | F44A×363/2691 | 2005, Shanxi | |
| Longza 9 | 325A×Hahui 118 | 2006, Heilongjiang | |
| Jiza 118 | Ji 2055A×R8063 | 2007, Jilin | |
| Liaonian 3 | Liaonian A-2×R-2 | 2008, Liaoning | |
| Loangza 10 | 454A×Hahui 591 | 2008, Heilongjiang | |
| Loangza 11 | 403A×Hahui 576 | 2008, Heilongjiang | |
| Jinza 22 | SX44A×SXR-30 | 2008, Shanxi | |
| Jinza 23 | SX45A×SXR-30-1 | 2008, Shanxi | |
| Chiza 24 | 0253A×0282 | 2008, Inner Mongolia | |
| Loangza 12 | Heilong 429A×Non 68 | 2009, Heilongjiang | |
| Jiza 122 | Ji 2055A×R105 | 2009, Jilin | |
| Jiza 124 | Ji 2055A×Ji R107 | 2009, Jilin | |
| Jinza 102 | F44A×0-30 Hong | 2009, Shanxi | |
| Jinza 103 | F44A×LR233 | 2009, Shanxi | |
| Liaonian 4 | LA-25×7037 | 2010, Liaoning | |
| Jinza 106 | 081A×580 | 2010, Liaoning | |
| Longza 13 | Heilong 423A×Nong 68 | 2010, Heilongjiang | |
| Jiza 127 | Ji 2055A×R117 | 2010, Jilin | |
| Chiza 28 | Chi A7×7654 | 2010, Inner Mongolia | |
| Chiza 29 | Chi A6×7654 | 2010, Inner Mongolia | |
| Jinza 104 | Lu 45A×Z233 | 2011, Shanxi | |
| Liaonian 5 | Fu A-1×Liaonian R-4 | 2012, Liaoning | |
| Liaonian 6 | LA-34×0-01 Xuan | 2012, Liaoning | |
| Suiza 7 | Suibuyu 30A×Suihui 25 | 2012, Heilongjiang | |
| Chiliang 4 | 314A×R185 | 2013, Inner Mongolia | |
| Jinza 33 | SX605A×Nan 133 | 2013, Shanxi | |
| Longza 16 | Heilong 433A×Hahui 591 | 2014, Heilongjiang | |
| Suiza 8 | Suibuyu 26A×Suihui 27 | 2014, Heilongjiang |
| Marker | Number of Alleles (Na) | Number of Effective Alleles (Ne) | Polymorphism Information Content (PIC) | Shannon’s Genetic Diversity Index (H) |
|---|---|---|---|---|
| Xtxp46 | 5 | 4.13 | 0.76 | 1.46 |
| Xtxp58 | 4 | 3.52 | 0.72 | 1.3 |
| Xtxp75 | 6 | 4.02 | 0.75 | 1.54 |
| Xtxp78 | 7 | 6.22 | 0.84 | 1.86 |
| Xtxp248 | 5 | 4.11 | 0.76 | 1.47 |
| Xtxp279 | 6 | 4.78 | 0.79 | 1.6 |
| Xtxp302 | 9 | 6.58 | 0.85 | 1.99 |
| Xtxp8 | 4 | 3.06 | 0.67 | 1.25 |
| Xtxp84 | 5 | 3.64 | 0.73 | 1.44 |
| Xtxp96 | 5 | 4.83 | 0.79 | 1.59 |
| Xtxp100 | 7 | 6.58 | 0.85 | 1.91 |
| Xtxp286 | 9 | 8.15 | 0.88 | 2.15 |
| Xtxp298 | 9 | 7.76 | 0.87 | 2.12 |
| Xtxp315 | 8 | 6.97 | 0.86 | 2.01 |
| Xtxp31 | 9 | 7.96 | 0.87 | 2.13 |
| Xtxp34 | 3 | 2.94 | 0.66 | 1.09 |
| Xtxp69 | 6 | 4.46 | 0.78 | 1.62 |
| Xtxp120 | 8 | 5.22 | 0.81 | 1.82 |
| Xtxp266 | 2 | 1.88 | 0.47 | 0.66 |
| Xtxp285 | 7 | 5.59 | 0.82 | 1.80 |
| Xtxp60 | 3 | 2.51 | 0.60 | 1.00 |
| Xtxp177 | 4 | 2.40 | 0.58 | 1.01 |
| Xtxp212 | 3 | 1.90 | 0.47 | 0.83 |
| Xtxp15 | 5 | 4.64 | 0.78 | 1.57 |
| Xtxp94 | 4 | 3.04 | 0.67 | 1.23 |
| Xtxp262 | 4 | 3.33 | 0.70 | 1.29 |
| Xtxp17 | 6 | 5.59 | 0.82 | 1.76 |
| Xtxp95 | 6 | 3.92 | 0.74 | 1.55 |
| Xtxp145 | 4 | 3.45 | 0.71 | 1.29 |
| Xtxp176 | 4 | 2.38 | 0.58 | 1.05 |
| Xcup-37 | 2 | 1.69 | 0.41 | 0.60 |
| Xtxp36 | 2 | 1.44 | 0.30 | 0.48 |
| Xtxp40 | 4 | 2.43 | 0.59 | 1.11 |
| Xtxp159 | 6 | 5.76 | 0.83 | 1.77 |
| Xtxp168 | 5 | 4.12 | 0.76 | 1.46 |
| Xtxp278 | 3 | 2.89 | 0.65 | 1.08 |
| Xtxp312 | 5 | 3.23 | 0.69 | 1.38 |
| Xtxp18 | 7 | 5.50 | 0.82 | 1.80 |
| Xtxp47 | 2 | 1.78 | 0.44 | 0.63 |
| Xtxp105 | 5 | 2.31 | 0.57 | 0.99 |
| Xtxp210 | 4 | 2.65 | 0.62 | 1.16 |
| Xtxp250 | 5 | 4.28 | 0.77 | 1.51 |
| Xtxp354 | 3 | 2.98 | 0.66 | 1.09 |
| Xcup-47 | 5 | 3.32 | 0.70 | 1.31 |
| Xtxp10 | 6 | 4.13 | 0.76 | 1.55 |
| Xtxp67 | 4 | 3.68 | 0.73 | 1.34 |
| Xtxp287 | 2 | 1.94 | 0.48 | 0.68 |
| Xtxp289 | 2 | 1.95 | 0.49 | 0.68 |
| Xtxp23 | 8 | 6.18 | 0.84 | 1.90 |
| Xtxp141 | 5 | 4.61 | 0.78 | 1.57 |
| Xtxp217 | 5 | 4.83 | 0.79 | 1.59 |
| Average | 5.04 | 4.06 | 0.70 | 1.39 |
| Stages | Number of Hybrids | Number of Alleles | Number of Introduced Alleles | Number of Lost Alleles * | Average Number of Alleles | Nei’s Genetic Diversity Index |
|---|---|---|---|---|---|---|
| 1 (1973–1982) | 7 | 232 | / | / | 4.55 | 0.66 |
| 2 (1983–1992) | 15 | 247 | 20 | 5 | 4.84 | 0.67 |
| 3 (1993–2002) | 15 | 242 | 6 | 11 | 4.75 | 0.68 |
| 4 (2003–2014) | 32 | 254 | 12 | 0 | 4.98 | 0.69 |
| First Stage (1973–1982) | Second Stage (1983–1992) | Third Stage (1993–2002) | Fourth Stage (2003–2014) | |
|---|---|---|---|---|
| Nationwide | 0.65 | 0.68 | 0.66 | 0.66 |
| Early maturing area | 0.68 | 0.70 | 0.64 | 0.66 |
| Late maturing area | 0.71 | 0.72 | 0.67 | 0.68 |
| Shanxi Province | 0.71 | 0.76 | 0.63 | 0.69 |
| Liaoning Province | 0.76 | 0.69 | 0.70 |
| Group | Sorghum Hybrids | Number of Varieties |
|---|---|---|
| A | Subgroup I: Chiza 851, Aoza 1, Aoza 2, Chiliang 4, Chiza 16, Chiza 14, Chiza 28, Chiza 29, Jiza 118, Jiza 124, Jiza 122, Jiza 127, Chiza 24, Jinza 101, Jinza 103, Huliang 1, Jinza 12, Jinza 22, Jinza 102 | 19 |
| Subgroup II: Liaonian 6, Liaonian 4, Liaonian 3, Liaonian 5, Liaoza 1, Shenza 5, Shenza 6, Xiongza 3, Qiaoza 2, Jiza 97, Jiza 90, Siza 4, Suiza 7, Suiza 8 | 14 | |
| Subgroup III: Jiza 5, Jinza 4, Kang 4, Jinza 5, Jinza 18, Jinzhong 405, Jinza 86-1, Tieza 8, Jinza 7, Jinza 11, Jinza 1, Jinza 104, Jinza 23 | 13 | |
| Subgroup IV: Tongza 2, Jinza 15, Jiza 76, Jiza 99, Siza 25, Jinza 33, Liaoza 21, Liaoza 10, Liaoza 11, Jinza 100, Liaoza 12, Jinza 93, Liaoza 15, Jinza 106 | 14 | |
| B | Chiyu 7, Jiza 52, Longza 16, Longza 5, Longza 9, Longza 12, Longza 13, Longza 10, Longza 11 | 9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, H.; Lv, N.; Yin, F.; Wang, Y.; Niu, H.; Lv, X.; Chu, J.; Fan, F.; Ju, L.; Yu, J.; et al. The Genetic Diversity of 69 Widely Used Chinese Sorghum Hybrids Released between the 1970s and 2010s. Agronomy 2024, 14, 2180. https://doi.org/10.3390/agronomy14102180
Yan H, Lv N, Yin F, Wang Y, Niu H, Lv X, Chu J, Fan F, Ju L, Yu J, et al. The Genetic Diversity of 69 Widely Used Chinese Sorghum Hybrids Released between the 1970s and 2010s. Agronomy. 2024; 14(10):2180. https://doi.org/10.3390/agronomy14102180
Chicago/Turabian StyleYan, Haisheng, Na Lv, Feng Yin, Yubin Wang, Hao Niu, Xin Lv, Jianqiang Chu, Fangfang Fan, Lan Ju, Jizhen Yu, and et al. 2024. "The Genetic Diversity of 69 Widely Used Chinese Sorghum Hybrids Released between the 1970s and 2010s" Agronomy 14, no. 10: 2180. https://doi.org/10.3390/agronomy14102180
APA StyleYan, H., Lv, N., Yin, F., Wang, Y., Niu, H., Lv, X., Chu, J., Fan, F., Ju, L., Yu, J., Zhang, F., & Ping, J. (2024). The Genetic Diversity of 69 Widely Used Chinese Sorghum Hybrids Released between the 1970s and 2010s. Agronomy, 14(10), 2180. https://doi.org/10.3390/agronomy14102180





