Identification and Functional Analysis of Key Genes Regulating Organic Acid Metabolism in Jujube Fruit
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Growth Conditions
2.2. Determination of Organic Acid Components
2.3. Transcriptome Analysis
2.4. Transient Overexpression and Virus-Induced Gene Silencing (VIGS) Assays
2.5. qRT-PCR
2.6. Statistical Analysis
3. Results
3.1. Changes in Organic Acid Components in Jujube Fruit During Fruit Development
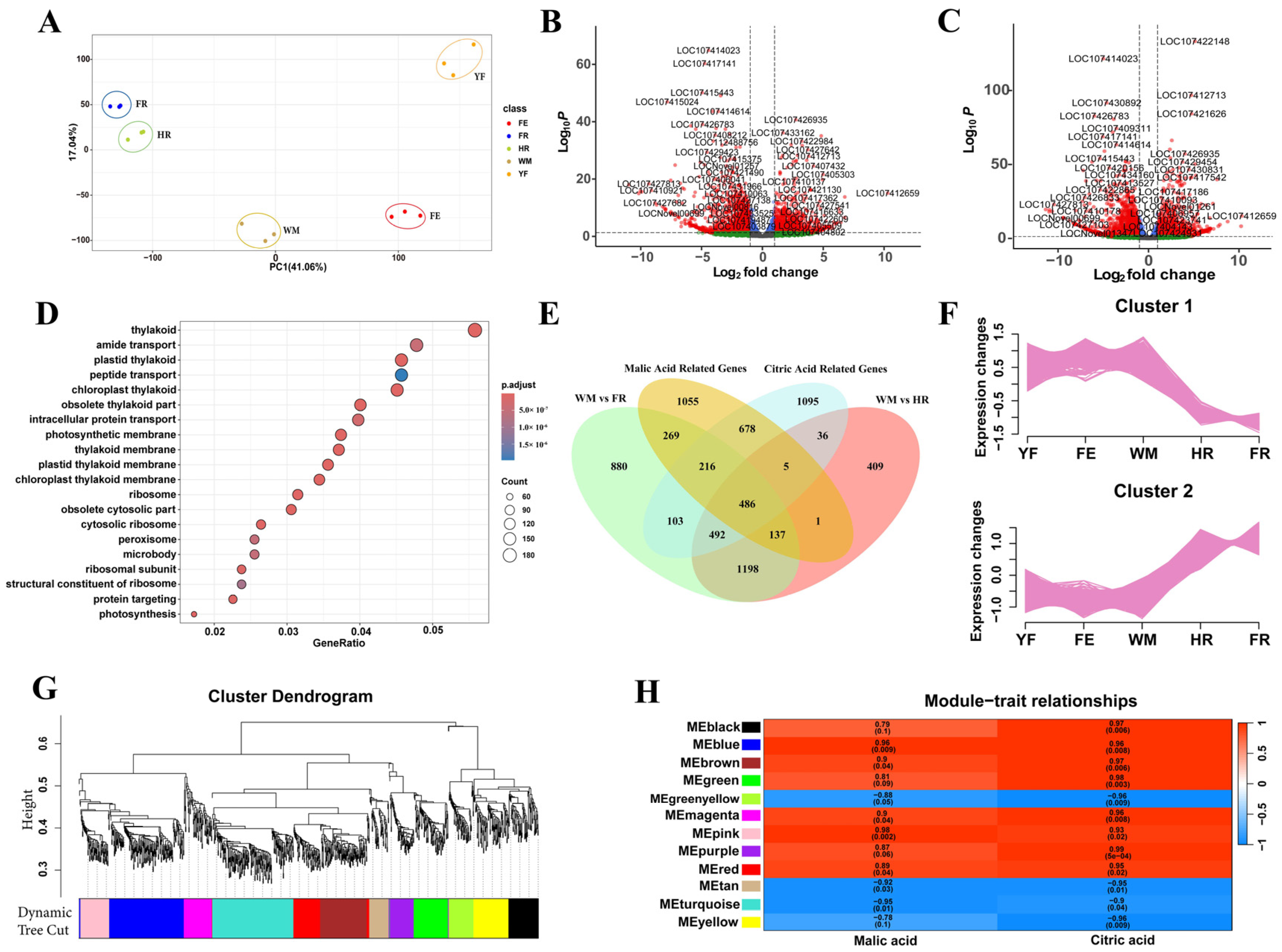
3.2. Identification of Key Genes Regulating Malic Acid and Citric Acid by Transcriptome Analysis
3.3. Real-Time Fluorescence Quantitative (qRT-PCR) Analysis
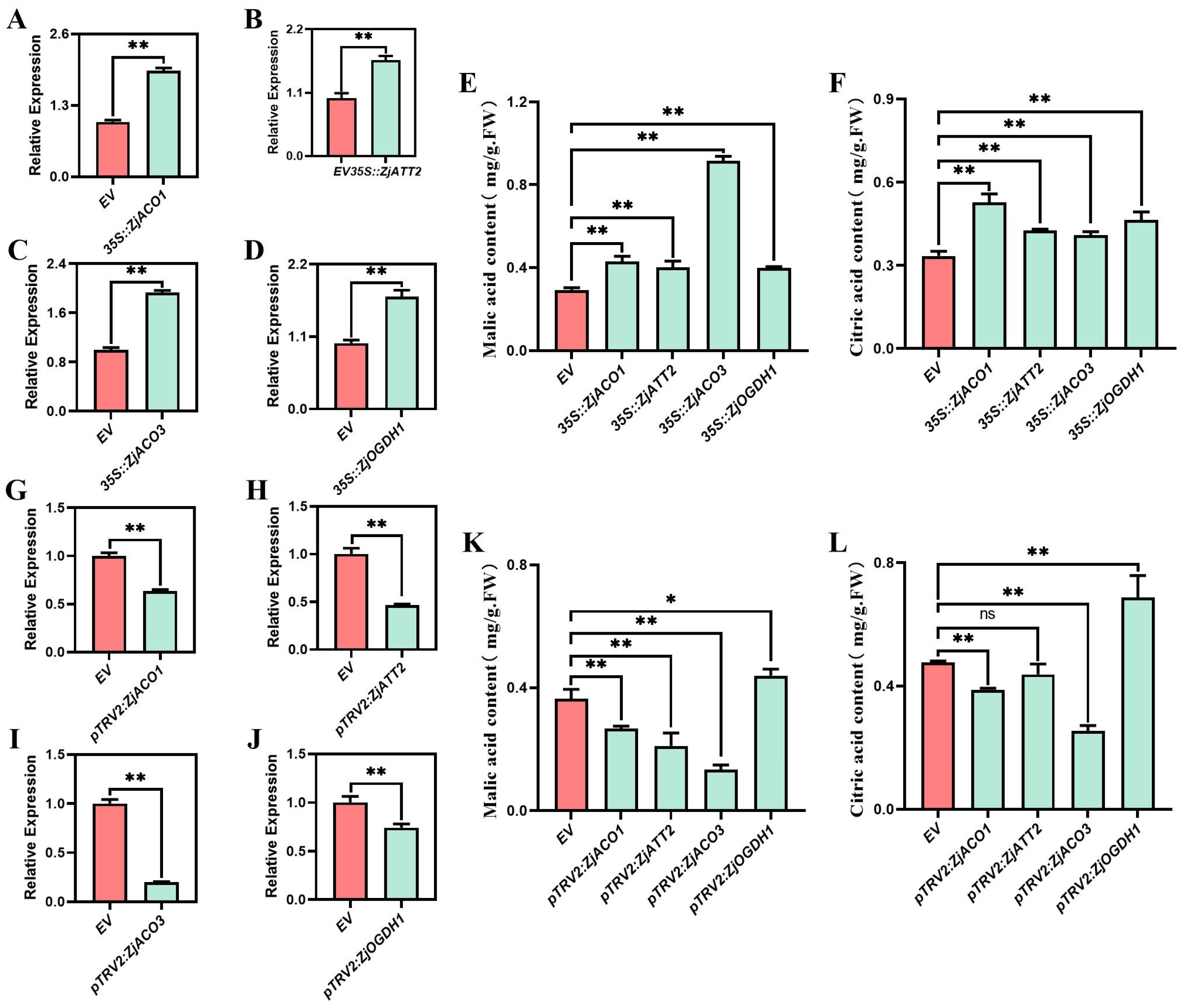
3.4. Transient Overexpression and Silencing Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, Z.; Shi, Q.Q.; Wang, B.; Ma, A.M.; Wang, Y.K.; Xue, Q.T.; Shen, B.Q.; Hamaila, H.; Tang, T.; Qi, X.Q.; et al. Jujube metabolome selection determined the edible properties acquired during domestication. Plant J. 2022, 109, 1116–1133. [Google Scholar] [CrossRef] [PubMed]
- Rashwan, A.K.; Karim, N.; Shishir, M.R.I.; Bao, T.; Lu, Y.; Chen, W. Jujube fruit: A potential nutritious fruit for the development of functional food products. J. Funct. Foods 2020, 75, 104205. [Google Scholar] [CrossRef]
- Choi, S.-H.; Ahn, J.-B.; Kim, H.-J.; Im, N.-K.; Kozukue, N.; Levin, C.E.; Friedman, M. Changes in free amino acid, protein, and flavonoid content in jujube (Ziziphus jujube) fruit during eight stages of growth and antioxidative and cancer cell inhibitory effects by extracts. J. Agric. Food Chem. 2012, 60, 10245–10255. [Google Scholar] [CrossRef] [PubMed]
- Klee, H.J. Improving the flavor of fresh fruits: Genomics, biochemistry, and biotechnology. New Phytol. 2010, 187, 44–56. [Google Scholar] [CrossRef]
- Panchal, P.; Miller, A.J.; Giri, J. Organic acids: Versatile stress-response roles in plants. J. Exp. Bot. 2021, 72, 4038–4052. [Google Scholar] [CrossRef]
- Igamberdiev, A.U.; Eprintsev, A.T. Organic acids: The pools of fixed carbon involved in redox regulation and energy balance in higher plants. Front. Plant Sci. 2016, 7, 1042. [Google Scholar] [CrossRef]
- Xue, X.F.; Zhao, A.L.; Wang, Y.K.; Sui, C.L.; Ren, H.Y.; Li, D.K.; Liang, Q. Analysis and comprehensive evaluation of fruit quality of different jujube varieties. China Fruits 2016, 3, 11–15. [Google Scholar] [CrossRef]
- Zhao, A.L.; Xue, X.F.; Ren, H.Y.; Wang, Y.K.; Li, D.K.; Li, Y. Analysis of composition and content characteristics of organic acids in jujube germplasm. Acta Agric. Boreali-Occident. Sin. 2021, 30, 1185–1198. [Google Scholar]
- Martinoia, E.; Neuhaus, E. A complex network regulating malate contents during fruit ripening in climacteric fruits. New Phytol. 2023, 239, 821–823. [Google Scholar] [CrossRef]
- He, J.; Sun, J.; Huang, Y.; Wang, L.; Liu, S. Transcriptome analysis reveals the common and specific pathways of citric acid accumulation in different citrus species. Hortic. Plant J. 2024. [Google Scholar] [CrossRef]
- Shi, Q.Q.; Du, J.T.; Zhu, D.J.; Li, X.; Li, X.G. Metabolomic and transcriptomic analyses of anthocyanin biosynthesis mechanisms in the color mutant Ziziphus jujuba cv. Tailihong. J. Agric. Food Chem. 2020, 68, 15186–15198. [Google Scholar] [CrossRef]
- Xu, J.D.; Yan, J.J.; Li, W.J.; Wang, Q.Y.; Wang, C.X.; Guo, J.X.; Geng, D.L.; Guan, Q.M.; Ma, F.W. Integrative analyses of widely targeted metabolic profiling and transcriptome data reveals molecular insight into metabolomic variations during apple (Malus domestica) fruit development and ripening. Int. J. Mol. Sci. 2020, 21, 4797. [Google Scholar] [CrossRef] [PubMed]
- Li, S.P.; Deng, B.L.; Tian, S.; Guo, M.X.; Liu, H.X.; Zhao, X.S. Metabolic and transcriptomic analyses reveal different metabolite biosynthesis profiles between leaf buds and mature leaves in Ziziphus jujuba mill. Food Chem. 2021, 15, 129005. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.Z.; Lin, X.K.; Du, J.J.; Peng, J.J.; Zhou, K.B. Transcriptomic analysis reveals key genes regulating organic acid synthesis and accumulation in the pulp of Litchi chinensis Sonn. cv. Feizixiao. Sci. Hortic. 2022, 303, 111220. [Google Scholar] [CrossRef]
- Gong, C.S.; Zhu, H.J.; Lu, X.Q.; Yang, D.D.; Zhao, S.J.; Umer, M.J.; He, N.; Yuan, P.L.; Anees, M.; Diao, W.; et al. An integrated transcriptome and metabolome approach reveals the accumulation of taste-related metabolites and gene regulatory networks during watermelon fruit development. Planta 2021, 254, 35. [Google Scholar] [CrossRef]
- Lu, D.Y.; Zhang, L.; Wu, Y.; Pan, Q.H.; Zhang, Y.P.; Liu, P. An integrated metabolome and transcriptome approach reveals the fruit flavor and regulatory network during jujube fruit development. Front. Plant Sci. 2022, 13, 952698. [Google Scholar] [CrossRef]
- Sarker, U.; Yang, H.Y.; Tian, C.P.; Ji, S.J.; Ni, F.Z. Integrative analyses of metabolome and transcriptome reveals metabolomic variations and candidate genes involved in sweet cherry (Prunus avium L.) fruit quality during development and ripening. PLoS ONE 2021, 16, e0260004. [Google Scholar] [CrossRef]
- Gawronska, K.; Niewiadomska, E. Participation of citric acid and isocitric acid in the diurnal cycle of carboxylation and decarboxylation in the common ice plant. Acta Physiol. Plant. 2015, 37, 61. [Google Scholar] [CrossRef]
- Zhang, C.M.; Geng, Y.Q.; Liu, H.X.; Wu, M.J.; Bi, J.X.; Wang, Z.T.; Dong, X.C.; Li, X.G. Low-acidity ALUMINUM-DEPENDENT MALATE TRANSPORTER4 genotype determines malate content in cultivated jujube. Plant Physiol. 2023, 191, 414–427. [Google Scholar] [CrossRef]
- Tong, P.P. Acid Accumulation Patterns and Related Gene Mining in Jujube Fruit. Master’s Thesis, Tarim University, Alar, China, 2021. [Google Scholar]
- Tong, P.P.; Liao, G.L.; Lu, D.Y.; Zhou, X.F.; Zhang, W.; Xu, Q.; Wu, C.Y.; Wang, J.B. ZjHXK5 and ZjHXK6 negatively regulate the sugar metabolism of Ziziphus jujuba Mill. Front. Plant Sci. 2024, 15, 1335120. [Google Scholar] [CrossRef]
- Morvai, M.; Molnár-Perl, I. Simultaneous gas chromatographic quantitation of sugars and acids in citrus fruits, pears, bananas, grapes, apples and tomatoes. Chromatographia 1992, 34, 502–504. [Google Scholar] [CrossRef]
- Mamat, S.F.; Azizan, K.A.; Baharum, S.N.; Noor, N.M.; Aizat, W.M. GC-MS and LC-MS analyses reveal the distribution of primary and secondary metabolites in mangosteen (Garcinia mangostana Linn.) fruit during ripening. Sci. Hortic. 2020, 262, 109004. [Google Scholar] [CrossRef]
- Liu, M.J.; Zhao, J.; Cai, Q.L.; Liu, G.C.; Wang, J.R.; Zhao, Z.H.; Liu, P.; Dai, L.; Yan, G.; Wang, W.J.; et al. The complex jujube genome provides insights into fruit tree biology. Nat. Commun. 2014, 5, 5315. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.M.; Huang, J.; Li, X.A. Identification of appropriate reference genes for RT-qPCR analysis in Ziziphus jujuba Mill. Sci. Hortic. 2015, 197, 166–169. [Google Scholar] [CrossRef]
- Goldenberg, L.; Yaniv, Y.; Porat, R.; Carmi, N. Mandarin fruit quality: A review. J. Sci. Food Agric. 2018, 98, 18–26. [Google Scholar] [CrossRef]
- Barrett, D.M.; Beaulieu, J.C.; Shewfelt, R. Color, flavor, texture, and nutritional quality of fresh-cut fruits and vegetables: Desirable levels, instrumental and sensory measurement, and the effects of processing. Crit. Rev. Food Sci. Nutr. 2010, 50, 369–389. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, L.l.; Wang, Z.T.; Liu, Z.G.; Zhao, Z.H.; Zhou, G.F.; Liu, M.G.; Liu, P. Variations of the nutritional composition of jujube fruit (Ziziphus jujuba Mill.) during maturation stages. Int. J. Food Prop. 2020, 23, 1066–1081. [Google Scholar] [CrossRef]
- Zhen, H.W.; Zhang, Q.Y.; Li, W.H.; Zhang, S.K.; Xi, W.P. Changes in soluble sugars and organic acids of Xinjiang apricot during fruit development and ripening. Sci. Agric. Sin. 2016, 49, 3981–3992. [Google Scholar] [CrossRef]
- Mao, J.P.; Gao, Z.; Wang, X.L.; Yao, D.L.; Lin, M.F.; Chen, L. Integrated transcriptome and targeted metabolome analyses provide insights into flavonoid biosynthesis in kiwifruit (Actinidia chinensis). Sci. Rep. 2024, 14, 19417. [Google Scholar] [CrossRef]
- Hanson, J.; Hanssen, M.; Wiese, A.; Hendriks, M.M.W.B.; Smeekens, S. The sucrose regulated transcription factor bZIP11 affects amino acid metabolism by regulating the expression of ASPARAGINE SYNTHETASE1 and PROLINE DEHYDROGENASE2. Plant J. 2008, 53, 935–949. [Google Scholar] [CrossRef]
- Bastías, A.; López-Climent, M.; Valcárcel, M.; Rosello, S.; Gómez-Cadenas, A.; Casaretto, J.A. Modulation of organic acids and sugar content in tomato fruits by an abscisic acid-regulated transcription factor. Physiol. Plant. 2011, 14, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.Q.; Gu, K.D.; Sun, C.H.; Zhang, Q.Y.; Wang, J.H.; Ma, F.F.; You, C.X.; Hu, D.G.; Hao, Y.J. The apple bHLH transcription factor MdbHLH3 functions in determining the fruit carbohydrates and malate. Plant Biotechnol. J. 2021, 19, 285–299. [Google Scholar] [CrossRef] [PubMed]
- Sweetman, C.; Deluc, L.G.; Cramer, G.R.; Ford, C.M.; Soole, K.L. Regulation of malate metabolism in grape berry and other developing fruits. Phytochemistry 2009, 70, 1329–1344. [Google Scholar] [CrossRef] [PubMed]
- Li, S.J.; Yin, X.R.; Wang, W.L.; Liu, X.F.; Zhang, B.; Chen, K.S. Citrus CitNAC62 cooperates with CitWRKY1 to participate in citric acid degradation via up-regulation of CitAco3. J. Exp. Bot. 2017, 68, 3419–3426. [Google Scholar] [CrossRef]
- Huang, X.Y.; Wang, C.K.; Zhao, Y.W.; Sun, C.H.; Hu, D.G. Mechanisms and regulation of organic acid accumulation in plant vacuoles. Hortic. Res. 2021, 8, 227. [Google Scholar] [CrossRef]
- Peng, J. Gene redundancy and gene compensation: An updated view. J. Genet. Genom. 2019, 46, 329–333. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tong, P.; Lu, D.; Liao, G.; Wu, C.; Wang, J. Identification and Functional Analysis of Key Genes Regulating Organic Acid Metabolism in Jujube Fruit. Agronomy 2024, 14, 2515. https://doi.org/10.3390/agronomy14112515
Tong P, Lu D, Liao G, Wu C, Wang J. Identification and Functional Analysis of Key Genes Regulating Organic Acid Metabolism in Jujube Fruit. Agronomy. 2024; 14(11):2515. https://doi.org/10.3390/agronomy14112515
Chicago/Turabian StyleTong, Panpan, Dengyang Lu, Guanglian Liao, Cuiyun Wu, and Jiangbo Wang. 2024. "Identification and Functional Analysis of Key Genes Regulating Organic Acid Metabolism in Jujube Fruit" Agronomy 14, no. 11: 2515. https://doi.org/10.3390/agronomy14112515
APA StyleTong, P., Lu, D., Liao, G., Wu, C., & Wang, J. (2024). Identification and Functional Analysis of Key Genes Regulating Organic Acid Metabolism in Jujube Fruit. Agronomy, 14(11), 2515. https://doi.org/10.3390/agronomy14112515






