Validation of KASP Markers Associated with Hydrogen Cyanide in Fresh Cassava Roots in Uganda Cassava Germplasm
Abstract
1. Introduction
2. Materials and Methods
2.1. Description of the Validation Population
2.2. Phenotyping
2.3. HCN Marker Discovery and Genotyping
2.4. Genotyping
2.5. Marker Robustness, Segregation Ability and Marker Effects
3. Results
3.1. Phenotypic Variation for HCN
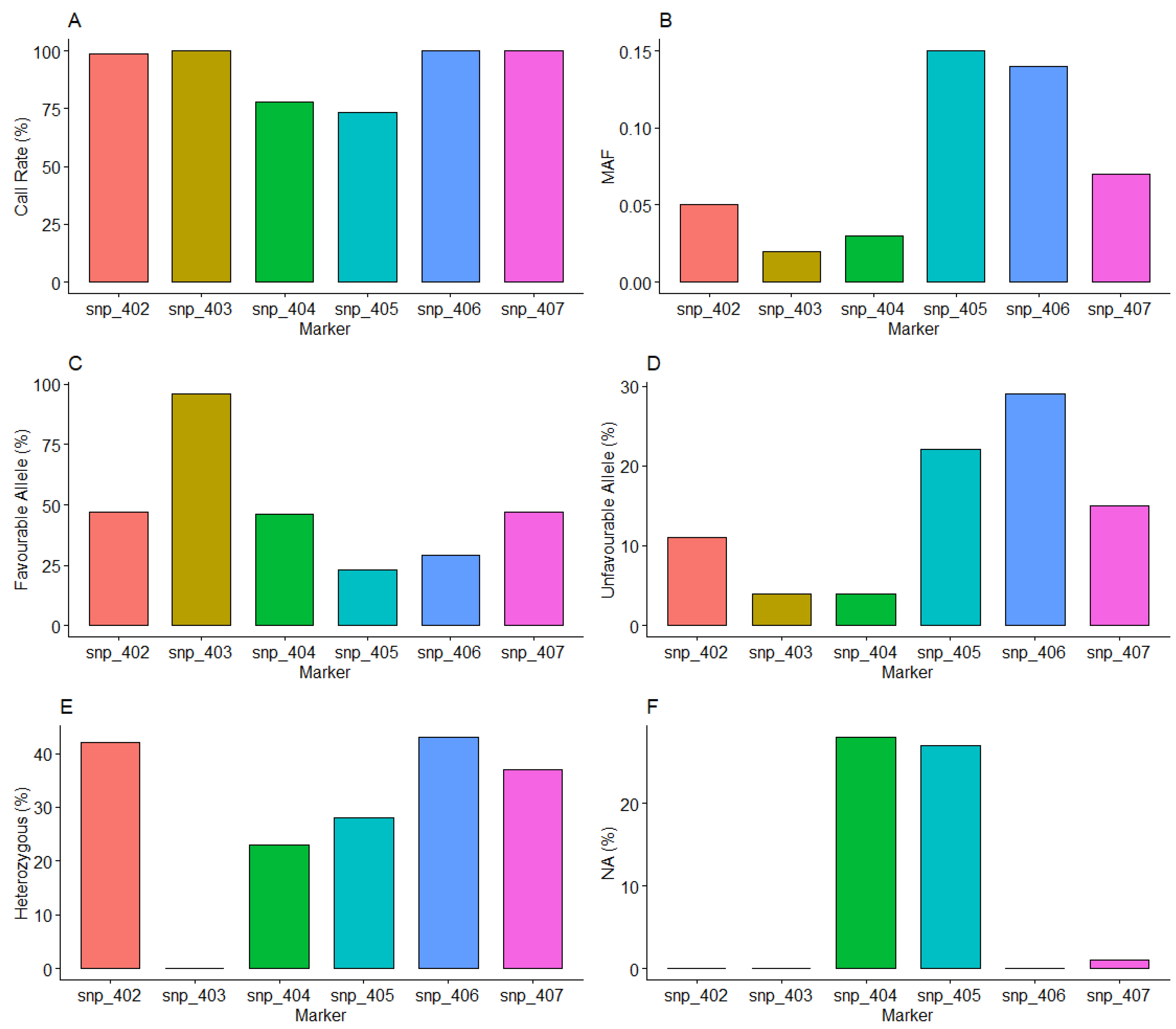
3.2. Performance of HCN KASP Markers
SNP Call Rate and Frequency of Marker Genotypes
3.3. Biological Validation
3.4. Marker Effects on Fresh Cassava Root HCN Levels
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Olsen, K.M.; Schaal, B.A. Evidence on the origin of cassava: Phylogeography of Manihot esculenta. Proc. Natl. Acad. Sci. USA 1999, 96, 5586–5591. [Google Scholar] [CrossRef] [PubMed]
- Olsen, K.M. SNPs, SSRs and inferences on cassava’s origin. Plant Mol. Biol. 2004, 56, 517–526. [Google Scholar] [CrossRef] [PubMed]
- El-sharkawy, M.A. Stress-Tolerant Cassava: The Role of Integrative Ecophysiology-Breeding Research in Crop Improvement. Open J. Soil Sci. 2012, 2, 162–186. [Google Scholar] [CrossRef]
- Bechoff, A.; Tomlins, K.; Fliedel, G.; Becerra Lopez-lavalle, L.A.; Westby, A.; Hershey, C.; Dufour, D. Cassava traits and end-user preference: Relating traits to consumer liking, sensory perception, and genetics. Crit. Rev. Food Sci. Nutr. 2018, 58, 547–567. [Google Scholar] [CrossRef] [PubMed]
- Atwijukire, E.; Hawumba, J.F.; Baguma, Y.; Wembabazi, E.; Esuma, W.; Kawuki, R.S.; Nuwamanya, E. Starch quality traits of improved provitamin A cassava (Manihot esculenta Crantz). Heliyon 2019, 5, e01215. [Google Scholar] [CrossRef]
- Iragaba, P.; Hamba, S.; Nuwamanya, E.; Kanaabi, M.; Nanyonjo, R.A.; Mpamire, D.; Muhumuza, N.; Khakasa, E.; Tufan, H.A.; Kawuki, R.S. Identification of cassava quality attributes preferred by Ugandan users along the food chain. Int. J. Food Sci. Technol. 2021, 56, 1184–1192. [Google Scholar] [CrossRef]
- Gleadow, R.M.; Møller, B.L. Cyanogenic glycosides: Synthesis, physiology, and phenotypic plasticity. Annu. Rev. Plant Biol. 2014, 65, 155–185. [Google Scholar] [CrossRef]
- Alves, A.A.C. Cassava Botany and Physiology. In Cassava Biology, Production and Utilization; CABI Publishing: Wallingford, UK, 2002; pp. 67–90. [Google Scholar] [CrossRef]
- Wheatley, C.C.; Ghuzel, G.; Zakhia, N. The Nature of the Tuber. In Encyclopedia of Food Sciences and Nutrition; Academic Press: Cambridge, MA, USA, 2003; pp. 964–969. [Google Scholar]
- Teles, F.F.F. Chronic poisoning by hydrogen cyanide in cassava and its prevention in Africa and Latin America. Food Nutr. Bull. 2002, 23, 407–412. [Google Scholar] [CrossRef]
- Alitubeera, P.H.; Eyu, P.; Kwesiga, B.; Ario, A.R.; Zhu, B.-P. Outbreak of Cyanide Poisoning Caused by Consumption of Cassava Flour—Kasese District, Uganda, September 2017. MMWR Morb. Mortal. Wkly. Rep. 2019, 68, 308–311. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6611475/ (accessed on 18 March 2024). [CrossRef]
- Cliff, J.; Muquingue, H.; Nhassico, D.; Nzwalo, H.; Bradbury, J.H. Konzo and continuing cyanide intoxication from cassava in Mozambique. Food Chem. Toxicol. 2011, 49, 631–635. [Google Scholar] [CrossRef]
- Nhassico, D.; Muquingue, H.; Cliff, J.; Cumbana, A.; Bradbury, J.H. Rising African cassava production, diseases due to high cyanide intake and control measures. J. Sci. Food Agric. 2008, 2049, 2043–2049. [Google Scholar] [CrossRef]
- McKey, D.; Cavagnaro, T.R.; Cliff, J.; Gleadow, R. Chemical ecology in coupled human and natural systems: People, manioc, multitrophic interactions and global change. Chemoecology 2010, 20, 109–133. [Google Scholar] [CrossRef]
- World Health Organization. FAO/WHO Book Review: Safety Evaluation of Certain Food Additives and Contaminants. Nutr. Health 2001, 15, 74. [Google Scholar] [CrossRef]
- Cardoso, A.P.; Mirione, E.; Ernesto, M.; Massaza, F.; Cliff, J.; Rezaul Haque, M.; Massaza, F.; Cliff, J.; Haque, M.R.; Bradbury, J. Processing of cassava roots to remove cyanogens. J. Food Compos. Anal. 2005, 18, 451–460. [Google Scholar] [CrossRef]
- Teeken, B.; Olaosebikan, O.; Haleegoah, J.; Oladejo, E.; Madu, T.; Bello, A.; Parkes, E.; Egesi, C.; Kulakow, P.; Kirscht, H.; et al. Cassava Trait Preferences of Men and Women Farmers in Nigeria: Implications for Breeding. Econ. Bot. 2018, 72, 263–277. [Google Scholar] [CrossRef]
- Dufour, D.; Hershey, C.; Hamaker, B.R.; Lorenzen, J. Integrating end-user preferences into breeding programmes for roots tubers and bananas. Int. J. Food Sci. Technol. 2021, 56, 1071–1075. [Google Scholar] [CrossRef]
- Polar, V.; Ashby, J.A.; Thiele, G.; Tufan, H. When is choice empowering? Examining gender differences in varietal adoption through case studies from sub-saharan africa. Sustainability 2021, 13, 3678. [Google Scholar] [CrossRef]
- Kawuki, R.; Nuwamanya, E.; Kanaabi, M.; Iragaba, P.; Williams, E.; Nanyonjo, A.R. NaCRRI Activities & Achievements for RTBfoods Project in Period 3; RTBfoods: Kampala, Uganda, 2020; p. 3. [Google Scholar]
- Ceballos, H.; Kulakow, P.; Hershey, C. Cassava Breeding: Current Status, Bottlenecks and the Potential of Biotechnology Tools. Trop. Plant Biol. 2012, 5, 73–87. [Google Scholar] [CrossRef]
- Ceballos, H.; Rojanaridpiched, C.; Phumichai, C. Excellence in Cassava Breeding: Perspectives for the Future. Crop Breed. Genet. Genom. 2020, 2, e200008. [Google Scholar] [CrossRef][Green Version]
- Baguma, J.K.; Mukasa, S.B.; Nuwamanya, E.; Alicai, T.; Omongo, C.; Hyde, P.T.; Setter, T.L.; Ochwo-Ssemakula, M.; Esuma, W.; Kanaabi, M.; et al. Flowering and fruit-set in cassava under extended red-light photoperiod supplemented with plant-growth regulators and pruning. BMC Plant Biol. 2023, 23, 335. [Google Scholar] [CrossRef]
- Njoku, D.N.; Ano, C.U.C. A review on plant genomic development; its importance, constraints and prospects. Niger. Agric. J. 2018, 49, 161–174. [Google Scholar]
- Xu, Y.; Crouch, J.H. Genomics of Tropical Maize, a Staple Food and Feed across the World. In Genomics of Tropical Crop Plants; Plant Genetics and Genomics: Crops and Models; Moore, P.H., Ming, R., Eds.; Springer: New York, NY, USA, 2008; Volume 1. [Google Scholar] [CrossRef]
- Ige, A.D.; Olasanmi, B.; Mbanjo, E.G.N.; Kayondo, I.S.; Parkes, E.Y.; Kulakow, P.; Egesi, C.; Bauchet, G.J.; Ng, E.; Lopez-Lavalle, L.A.B.; et al. Conversion and validation of uniplex snp markers for selection of resistance to cassava mosaic disease in cassava breeding programs. Agronomy 2021, 11, 420. [Google Scholar] [CrossRef]
- Wolfe, M.D.; Rabbi, I.Y.; Egesi, C.; Hamblin, M.; Kawuki, R.; Kulakow, P.; Lozano, R.; Del Carpio, D.P.; Ramu, P.; Jannink, J.-L. Genome-Wide Association and Prediction Reveals Genetic Architecture of Cassava Mosaic Disease Resistance and Prospects for Rapid Genetic Improvement. Plant Genome 2016, 9, 1–13. [Google Scholar] [CrossRef]
- Codjia, E.D.; Olasanmi, B.; Agre, P.A.; Uwugiaren, R.; Ige, A.D.; Rabbi, I.Y. Selection for resistance to cassava mosaic disease in African cassava germplasm using single nucleotide polymorphism markers. S. Afr. J. Sci. 2022, 118, 73–79. [Google Scholar] [CrossRef]
- Esuma, W.; Eyoo, O.; Gwandu, F.; Mukasa, S.; Alicai, T.; Ozimati, A.; Nuwamanya, E.; Rabbi, I.; Kawuki, R. Validation of KASP markers associated with cassava mosaic disease resistance, storage root dry matter and provitamin A carotenoid contents in Ugandan cassava germplasm. Front. Plant Sci. 2022, 23, 13. [Google Scholar] [CrossRef]
- Balyejusa Kizito, E.; Chiwona-Karltun, L.; Egwang, T.; Fregene, M.; Westerbergh, A. Genetic diversity and variety composition of cassava on small-scale farms in Uganda: An interdisciplinary study using genetic markers and farmer interviews. Genetica 2007, 130, 301–318. [Google Scholar] [CrossRef]
- Whankaew, S.; Poopear, S.; Kanjanawattanawong, S.; Tangphatsornruang, S.; Boonseng, O.; Lightfoot, D.A.; Triwitayakorn, K. A genome scan for quantitative trait loci affecting cyanogenic potential of cassava root in an outbred population. BMC Genom. 2011, 12, 266. [Google Scholar] [CrossRef]
- Ogbonna, A.C.; Braatz de Andrade, L.R.; Rabbi, I.Y.; Mueller, L.A.; Jorge de Oliveira, E.; Bauchet, G.J. Large-scale genome-wide association study, using historical data, identifies conserved genetic architecture of cyanogenic glucoside content in cassava (Manihot esculenta Crantz) root. Plant J. 2021, 105, 754–770. [Google Scholar] [CrossRef]
- Platten, J.D.; Cobb, J.N.; Zantua, R.E. Criteria for evaluating molecular markers: Comprehensive quality metrics to improve marker-assisted selection. PLoS ONE 2019, 14, e0210529. [Google Scholar] [CrossRef]
- Atim, B.P.; Kabami, Z.; Gidudu, S. Cyanide Poisoning Investigation, Terego District, Uganda, February 2023. Uganda Natl. Inst. Public Health Q. Epidemiol. Bull. 2023, 8, e5030-41. Available online: https://uniph.go.ug/cyanide-poisoning-investigation-terego-district-uganda-february-2023/ (accessed on 9 November 2024).
- Ozimati, A.; Kawuki, R.; Esuma, W.; Kayondo, I.S.; Wolfe, M. Training Population Optimization for Prediction of Cassava Brown Streak Disease Resistance in West African Clones. G3 Genes Genomes Genet. 2018, 12, 3903–3913. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, M.G.; Egan, S.V.; Bradbury, J.H. Picrate paper kits for determination of total cyanogens in cassava roots and all forms of cyanogens in cassava products. J. Sci. Food Agric. 1999, 79, 593–601. [Google Scholar] [CrossRef]
- Fukuda, W.M.G.; Guevara, C.L.; Kawuki, R.; Ferguson, M.E. Selected Morphological and Agronomic Descriptors for the Characterization of Cassava; IITA: Ibadan, Nigeria, 2010; Available online: https://books.google.co.ug/books?hl=en&lr=&id=-SnckHhBlEYC&oi=fnd&pg=PA1&dq=FUKUDA+2010+hydrogen+cyanide+cassava+scale&ots=_tpYNzi4sg&sig=QbPKY4T0MSnmZK6biD6Z7XzTjUQ&redir_esc=y#v=onepage&q&f=false (accessed on 29 March 2022).
- Rahman, M.Z.; Hasan, M.T.; Rahman, J. Kompetitive Allele-Specific PCR (KASP): An Efficient High-Throughput Genotyping Platform and Its Applications in Crop Variety Development. Mol. Marker Technol. A Potential. Approach Crop Improv. 2023, 29, 25–54. Available online: https://link.springer.com/chapter/10.1007/978-981-99-1612-2_2 (accessed on 10 August 2024).
- Kanaabi, M.; Namakula, F.B.; Nuwamanya, E.; Kayondo, I.S.; Muhumuza, N.; Wembabazi, E.; Iragaba, P.; Nandudu, L.; Nanyonjo, A.R.; Baguma, J.; et al. Rapid analysis of hydrogen cyanide in fresh cassava roots using NIRS and machine learning algorithms: Meeting end user demand for low cyanogenic cassava. Plant Genome 2023, 17, e20403. [Google Scholar] [CrossRef] [PubMed]
- Cobb, J.N.; Biswas, P.S.; Platten, J.D. Back to the future: Revisiting MAS as a tool for modern plant breeding. Theor. Appl. Genet. 2019, 132, 647–667. [Google Scholar] [CrossRef] [PubMed]
- Cobb, J.N.; Juma, R.U.; Biswas, P.S.; Arbelaez, J.D.; Rutkoski, J.; Atlin, G.; Hagen, T.; Quinn, M.; Ng, E.H. Enhancing the rate of genetic gain in public-sector plant breeding programs: Lessons from the breeder’s equation. Theor. Appl. Genet. 2019, 132, 627–645. [Google Scholar] [CrossRef]
- Ceballos, H.; Kawuki, R.S.; Gracen, V.E.; Yencho, G.C.; Hershey, C.H. Conventional breeding, marker-assisted selection, genomic selection and inbreeding in clonally propagated crops: A case study for cassava. Theor. Appl. Genet. 2015, 128, 1647–1667. [Google Scholar] [CrossRef]
- Parveen, R.; Kumar, M.; Swapnil Singh, D.; Shahani, M.; Imam, Z.; Sahoo, J.P. Understanding the genomic selection for crop improvement: Current progress and future prospects. Mol. Genet. Genom. 2023, 298, 813–821. Available online: https://link.springer.com/article/10.1007/s00438-023-02026-0 (accessed on 1 August 2024). [CrossRef]
- Okogbenin, E.; Porto, M.C.M.; Egesi, C.; Mba, C.; Espinosa, E.; Santos, L.G.; Ospina, C.; Marín, J.; Barrera, E.; Gutiérrez, J.; et al. Marker-Assisted Introgression of Resistance to Cassava Mosaic Disease into Latin American Germplasm for the Genetic Improvement of Cassava in Africa. Crop Sci. 2007, 47, 1895–1904. Available online: https://onlinelibrary.wiley.com/doi/full/10.2135/cropsci2006.10.0688 (accessed on 10 November 2024). [CrossRef]
- Mbanjo, E.G.N.; Ogungbesan, A.; Agbona, A.; Akpotuzor, P.; Toyinbo, S.; Iluebbey, P.; Rabbi, I.Y.; Peteti, P.; Wages, S.A.; Norton, J.; et al. Validation of SNP Markers for Diversity Analysis, Quality Control, and Trait Selection in a Biofortified Cassava Population. Plants 2024, 13, 2328. [Google Scholar] [CrossRef]
- Jiang, G.L. Molecular Markers and Marker-Assisted Breeding in Plants. In Plant Breeding from Laboratories to Fields; InTech: Vienna, Austria, 2013. [Google Scholar] [CrossRef]
- Majeed, U.; Darwish, E.; Rehman, S.U.; Zhang, X. Kompetitive Allele Specific PCR (KASP): A Singleplex Genotyping Platform and Its Application. J. Agric. Sci. 2018, 11, 11. [Google Scholar] [CrossRef]
- Suo, W.; Shi, X.; Xu, S.; Li, X.; Lin, Y. Towards low cost, multiplex clinical genotyping: 4-fluorescent Kompetitive Allele-Specific PCR and its application on pharmacogenetics. PLoS ONE 2020, 15, 2–9. [Google Scholar] [CrossRef]
- Kanaabi, M.; Settumba, M.B.; Nuwamanya, E.; Muhumuza, N.; Iragaba, P.; Ozimati, A.; Namakula, F.B.; Kayondo, I.S.; Baguma, J.K.; Nanyonjo, A.R.; et al. Genetic Variation and Heritability for Hydrogen Cyanide in Fresh Cassava Roots: Implications for Low-Cyanide Cassava Breeding. Plants 2024, 13, 1186. [Google Scholar] [CrossRef] [PubMed]
- Siritunga, D.; Sayre, R.T. Generation of cyanogen-free transgenic cassava. Planta 2003, 217, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Maeda, S.I.; Konishi, M.; Yanagisawa, S.; Omata, T. Nitrite Transport Activity of a Novel HPP Family Protein Conserved in Cyanobacteria and Chloroplasts. Plant Cell Physiol. 2014, 55, 1311–1324. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Yang, J. Genome-wide comparison reveals divergence of cassava and rubber aquaporin family genes after the recent whole-genome duplication. BMC Genom. 2019, 20, 380. [Google Scholar] [CrossRef]
- Omote, H.; Hiasa, M.; Matsumoto, T.; Otsuka, M.; Moriyama, Y. The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol. Sci. 2006, 27, 587–593. [Google Scholar] [CrossRef]
- Brown, A.L.; Cavagnaro, T.R.; Gleadow, R.; Miller, R.E. Interactive effects of temperature and drought on cassava growth and toxicity: Implications for food security? Glob. Change Biol. 2016, 22, 3461–3473. [Google Scholar] [CrossRef]
- Rabbi, I.Y. Marker-assisted selection in cassava breeding. In Achieving Cultivation of Cassava; Genetics, Breeding, Pests and Diseases; Burleigh Dodds Science Publishing Limited: Cambridge, UK, 2017; Volume 2, pp. 101–115. [Google Scholar]



| Chromosome | SNP | Position, bp | Allele | p-Value | Gene | Name | Function |
|---|---|---|---|---|---|---|---|
| 16 | S16_773999 | 773999 | G/A | 7.53 × 10−22 | Manes.16G007900 | MATE efflux family protein. | Multi-drug and Toxic Compound Extrusion or Multi-Antimicrobial Extrusion. |
| 16 | S16_795990 | 795990 | A/T | 2.41 × 10−10 | Manes.16G008000 | MATE efflux family protein. | Multi-drug and Toxic Compound Extrusion or Multi-Antimicrobial Extrusion. |
| 16 | S16_796041 | 796041 | T/A | 1.36 × 10−20 | Manes.16G008100 | ||
| 14 | S14_6050078 | 6050078 | G/A | 1.09 × 10−8 | Manes.14G074300 | HPP family protein. | Integral membrane HPP family protein involved in Nitrite Transport Activity. |
| 14 | S14_5775892 | 5775892 | G/T | 1.63 × 10−8 | Manes.14G071000 | K03355—anaphase-promoting complex subunit 8 (APC8, CDC23). | Interacting selectively and non-covalently with any protein or protein complex (a complex of two or more proteins that may include other non-protein molecules). |
| 14 | S14_6021712 | 6021712 | A/T | 7.32 × 10−8 | Manes.14G073900 | H(+)-ATPase | The plasma membrane H+-ATPase mediated H+influx may be associated with the plasma membrane gradients as well as Al-induced citrate efflux mediated by a H+-ATPase-coupled MATE co-transport system. |
| SNP | Intertek SNP ID | SNPNum | AlleleY | AlleleX | Sequence |
|---|---|---|---|---|---|
| S16_773999 | snpME00402 | 140060 | A (Low) | G | TTCACTGATGGTGAA[G/A]CTGTTTCCAAAGCA |
| S16_795990 | snpME00403 | 140061 | A (Low) | T | GGCTGCCAAATCTGG[T/A]GGACTAATGACATG |
| S16_796041 | snpME00404 | 140062 | T (Low) | A | TGGATCTCAGCAGCA[A/T]TTTAACCCACTGAT |
| S14_5775892 | snpME00405 | 140063 | G (Low) | T | TTTATCTGCCTGGAC[T/G]CTTATGGGTCATGA |
| S14_6050078 | snpME00406 | 140064 | A | G (Low) | CGGAAAGATGGACCA[G/A]TTACTTGCGCCTAA |
| S14_6021712 | snpME00407 | 140065 | G (Low) | C | TGATTTAGCGAAGAA[C/G]AAAAGCTGCGCGAG |
| SOV | DF | MS |
|---|---|---|
| Clone | 61 | 5.20 *** |
| Location | 3 | 127.55 *** |
| Block | 1 | |
| Clone:Location | 149 | 1.57 * |
| Residuals | 144 | |
| Vg | 0.49 | |
| Vge | 0.35 | |
| Ve | 1.05 | |
| H2 | 0.26 |
| Marker | Degrees of Freedom | Mean Square | R2 |
|---|---|---|---|
| snpME00402 | 2 | 1.024 ns | 0.09 |
| snpME00403 | 2 | 1.225 ns | 0.05 |
| snpME00404 | 2 | 2.39 ** | 0.27 |
| snpME00405 | 2 | 1.475 * | 0.17 |
| snpME00406 | 2 | 1.587 * | 0.14 |
| snpME00407 | 2 | 0.439 ns | 0.04 |
| Count | |||||||
|---|---|---|---|---|---|---|---|
| Marker | Low | High | Total | FPR (%) | FNR (%) | Accuracy (%) | |
| snpME00404 | Low | 17 | 9 | 26 | 47 | 6 | 73 |
| High | 1 | 10 | 11 | ||||
| Total | 18 | 19 | 37 | ||||
| snpME00405 | Low | 8 | 5 | 13 | 26 | 56 | 59 |
| High | 10 | 14 | 24 | ||||
| Total | 18 | 19 | 37 | ||||
| snpME00406 | Low | 13 | 5 | 18 | 25 | 5 | 65 |
| High | 13 | 20 | 33 | ||||
| Total | 26 | 25 | 51 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanaabi, M.; Mukasa, S.B.; Nuwamanya, E.; Iragaba, P.; Baguma, J.K.; Nanyonjo, A.R.; Wagaba, H.; Muhumuza, N.; Namakula, F.B.; Wembabazi, E.; et al. Validation of KASP Markers Associated with Hydrogen Cyanide in Fresh Cassava Roots in Uganda Cassava Germplasm. Agronomy 2024, 14, 2765. https://doi.org/10.3390/agronomy14122765
Kanaabi M, Mukasa SB, Nuwamanya E, Iragaba P, Baguma JK, Nanyonjo AR, Wagaba H, Muhumuza N, Namakula FB, Wembabazi E, et al. Validation of KASP Markers Associated with Hydrogen Cyanide in Fresh Cassava Roots in Uganda Cassava Germplasm. Agronomy. 2024; 14(12):2765. https://doi.org/10.3390/agronomy14122765
Chicago/Turabian StyleKanaabi, Michael, Settumba B. Mukasa, Ephraim Nuwamanya, Paula Iragaba, Julius Karubanga Baguma, Ann Ritah Nanyonjo, Henry Wagaba, Nicholas Muhumuza, Fatumah Babirye Namakula, Enoch Wembabazi, and et al. 2024. "Validation of KASP Markers Associated with Hydrogen Cyanide in Fresh Cassava Roots in Uganda Cassava Germplasm" Agronomy 14, no. 12: 2765. https://doi.org/10.3390/agronomy14122765
APA StyleKanaabi, M., Mukasa, S. B., Nuwamanya, E., Iragaba, P., Baguma, J. K., Nanyonjo, A. R., Wagaba, H., Muhumuza, N., Namakula, F. B., Wembabazi, E., Ozimati, A., Kayondo, I. S., Esuma, W., & Kawuki, R. S. (2024). Validation of KASP Markers Associated with Hydrogen Cyanide in Fresh Cassava Roots in Uganda Cassava Germplasm. Agronomy, 14(12), 2765. https://doi.org/10.3390/agronomy14122765









