Integrative Analysis of Metabolome and Transcriptome Profiles to Evaluate the Response Mechanisms of Carex adrienii to Shade Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatment
2.2. Measurement of Physiological Indices and Photosynthetic Parameters
2.3. RNA Extraction, Library Construction and Squencing Analysis
2.4. Metabolite Extraction and UPLC-MS/MS Analysis
2.5. qRT-PCR Validation of DEGs
3. Results
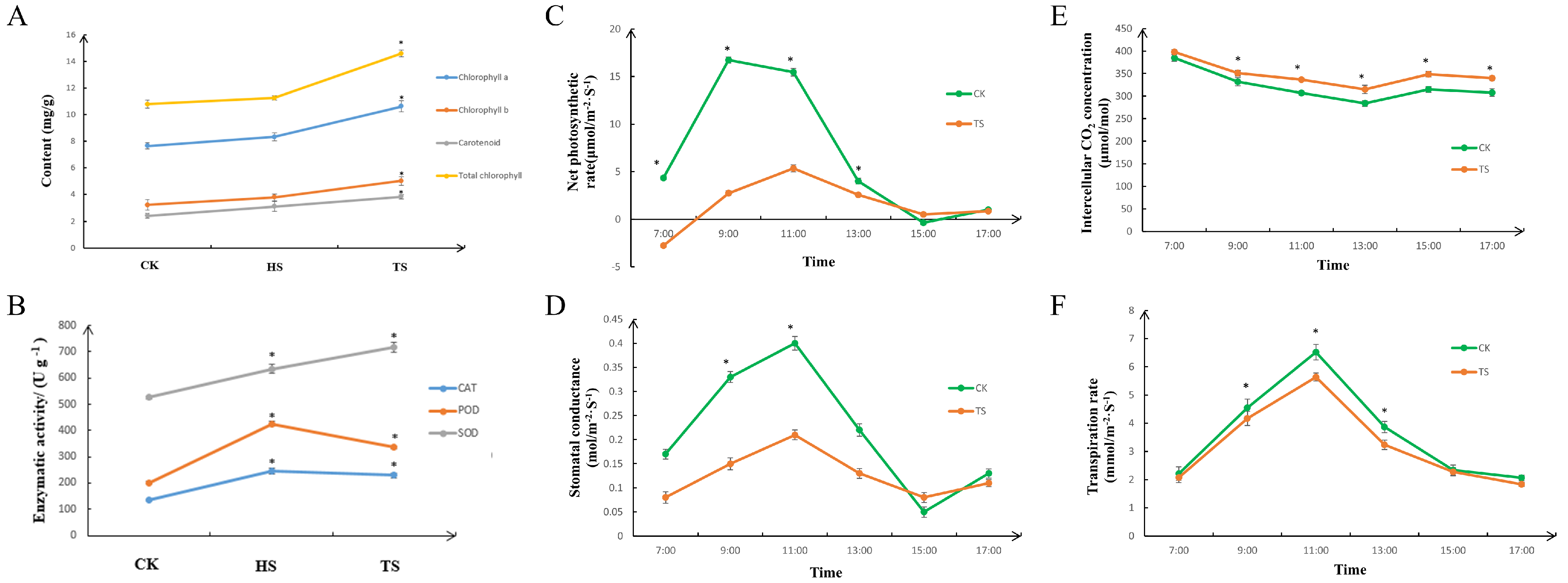
3.1. Physiological and Photosynthetic Characteristics of Carex adrienii Leaves Under Shading Treatment
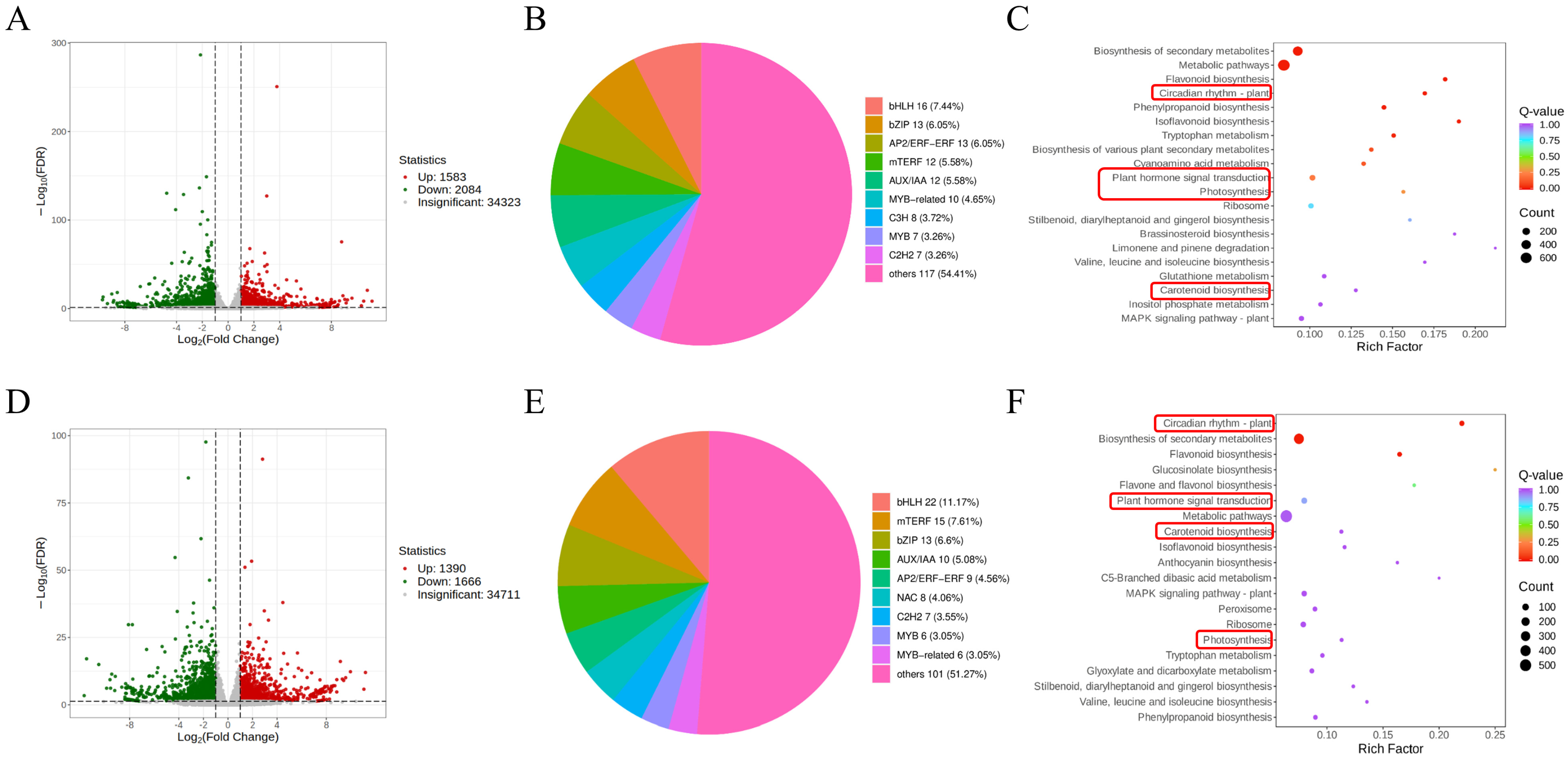
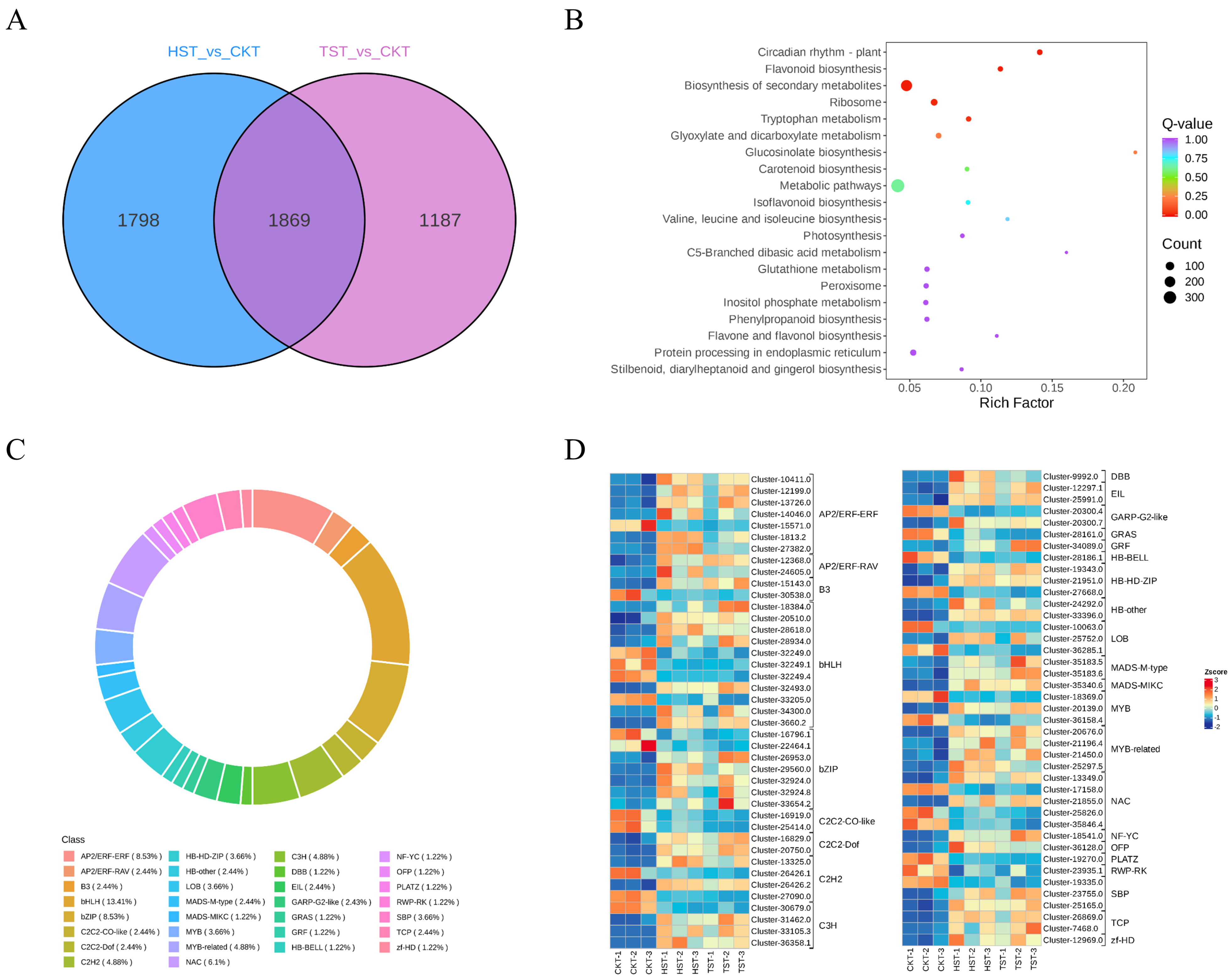
3.2. Transcriptome Sequencing and Overview of DEGs
3.3. GO Enrichment Analysis of DEGs
3.4. Analysis of Differentially Expressed Transcription Factors
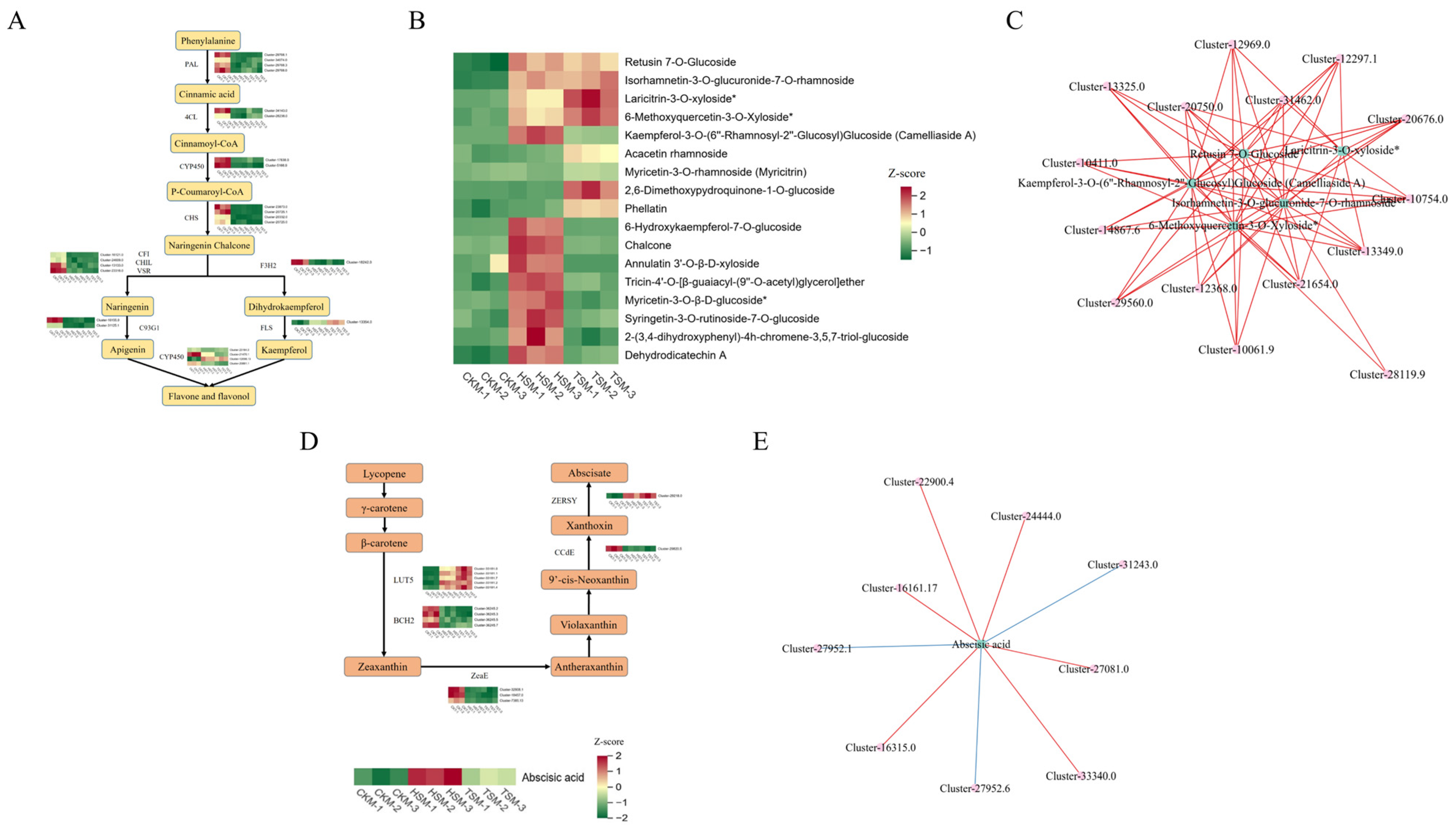
3.5. Analysis of DEGs Involved in Photosynthesis and Flavonoids Biosynthesis
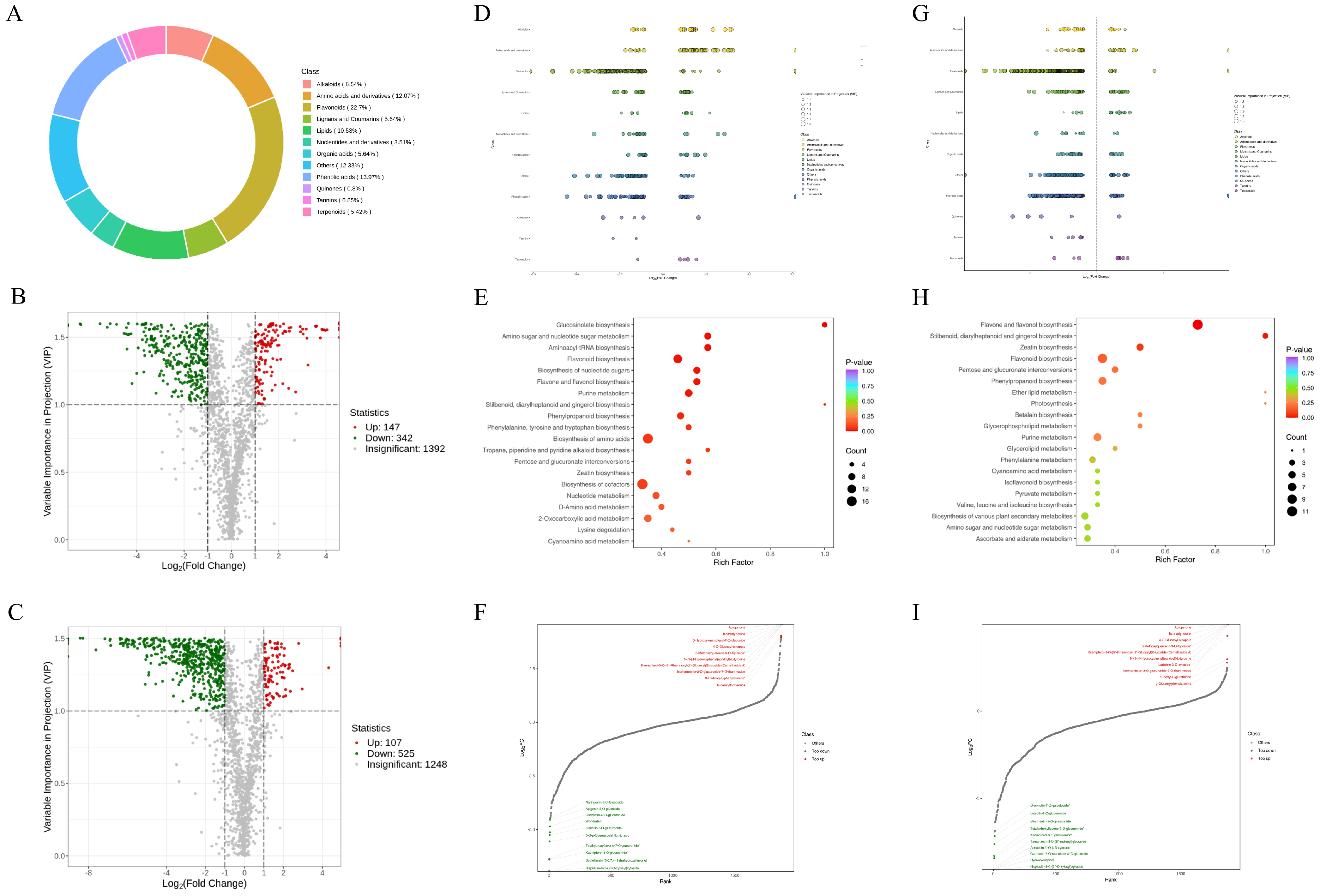
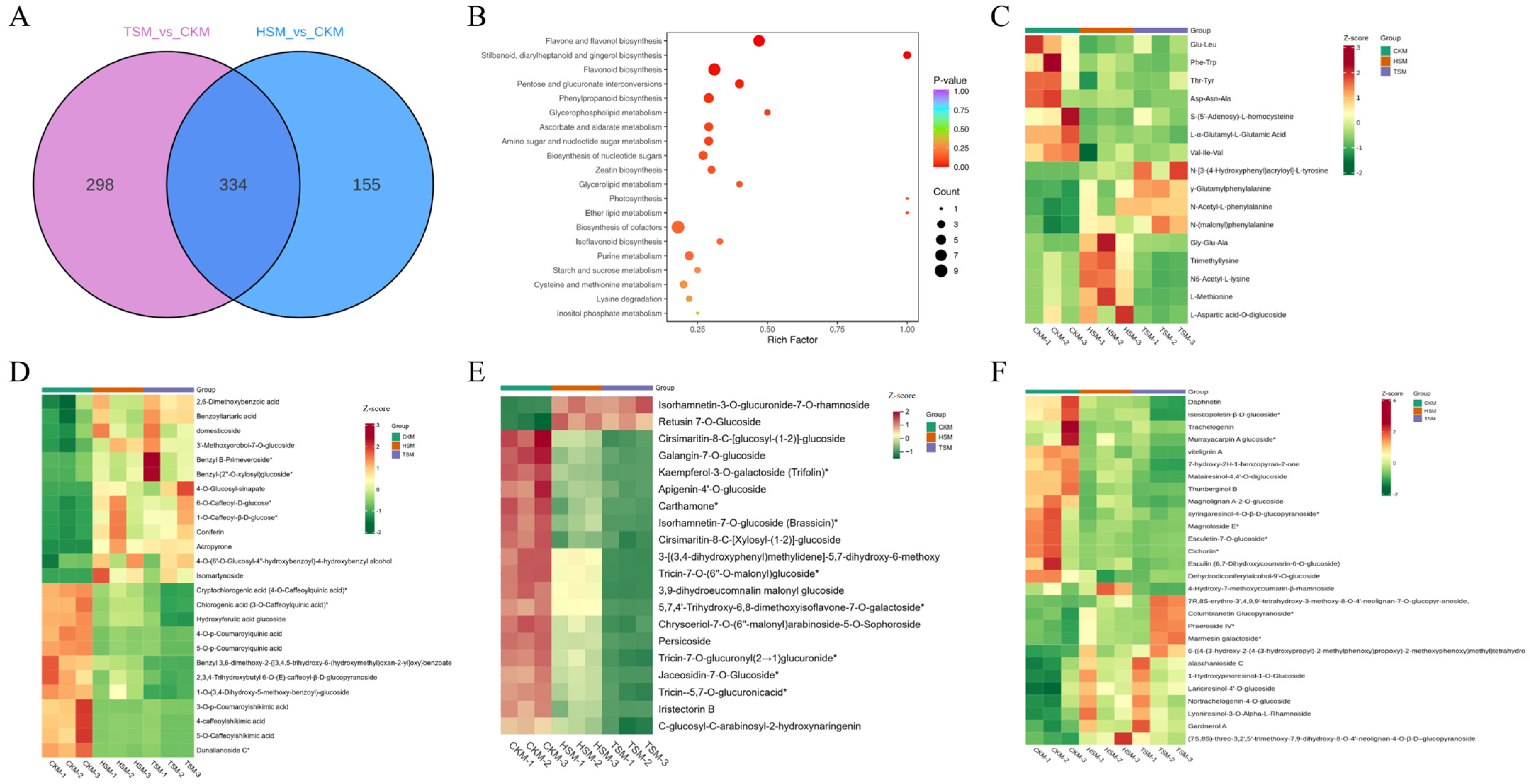
3.6. Metabolomic Analysis
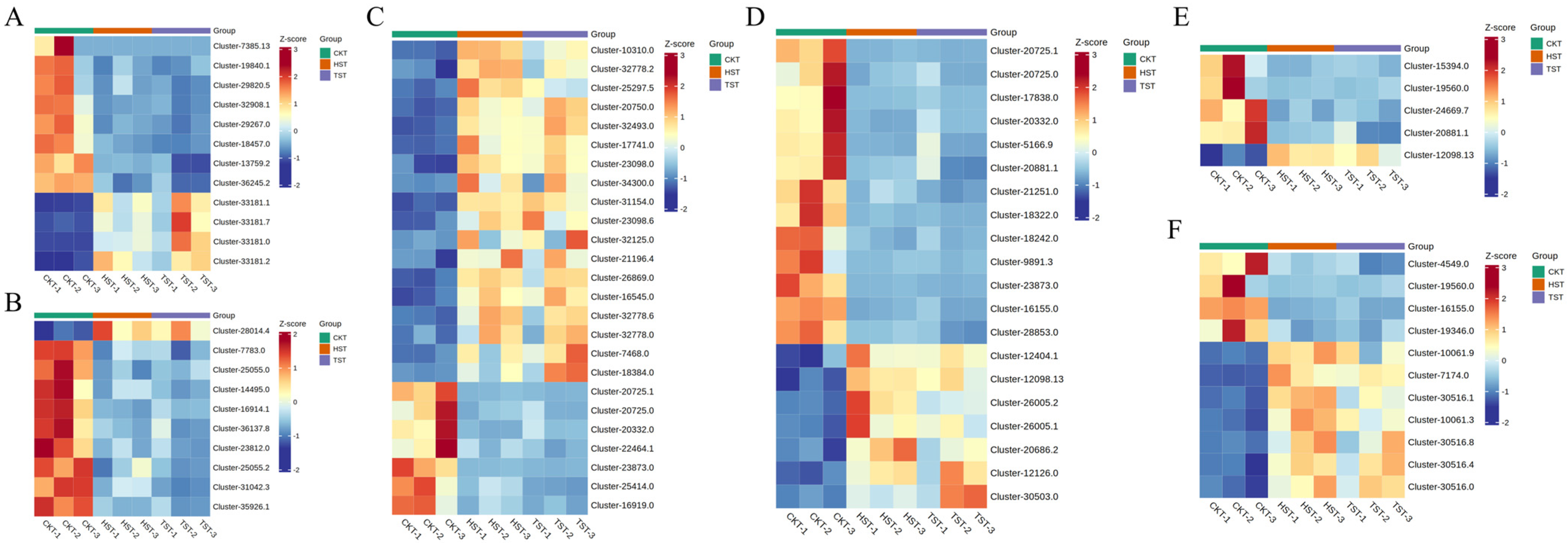
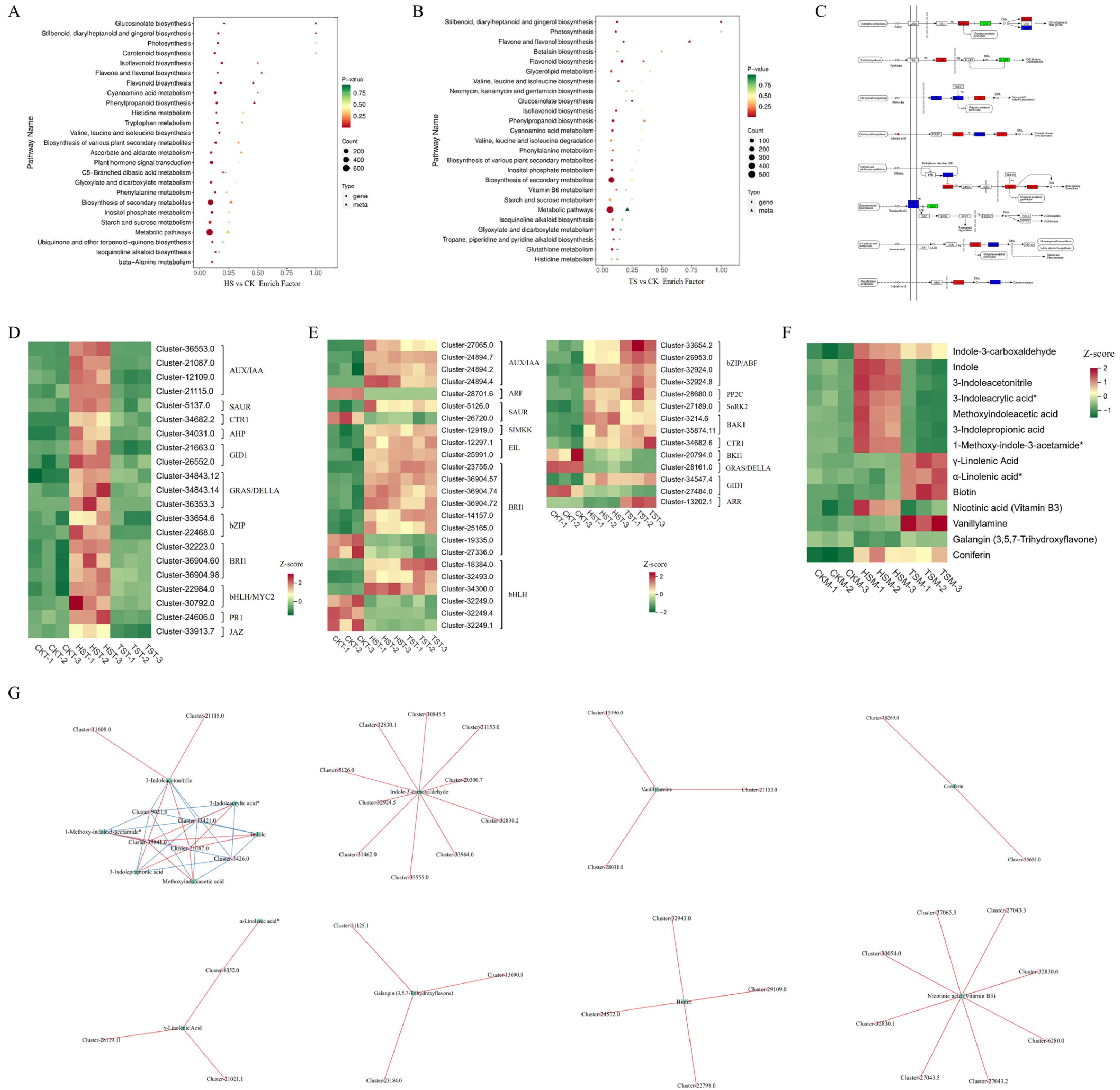
3.7. Combined Transcriptomic and Metabolomic Analyses
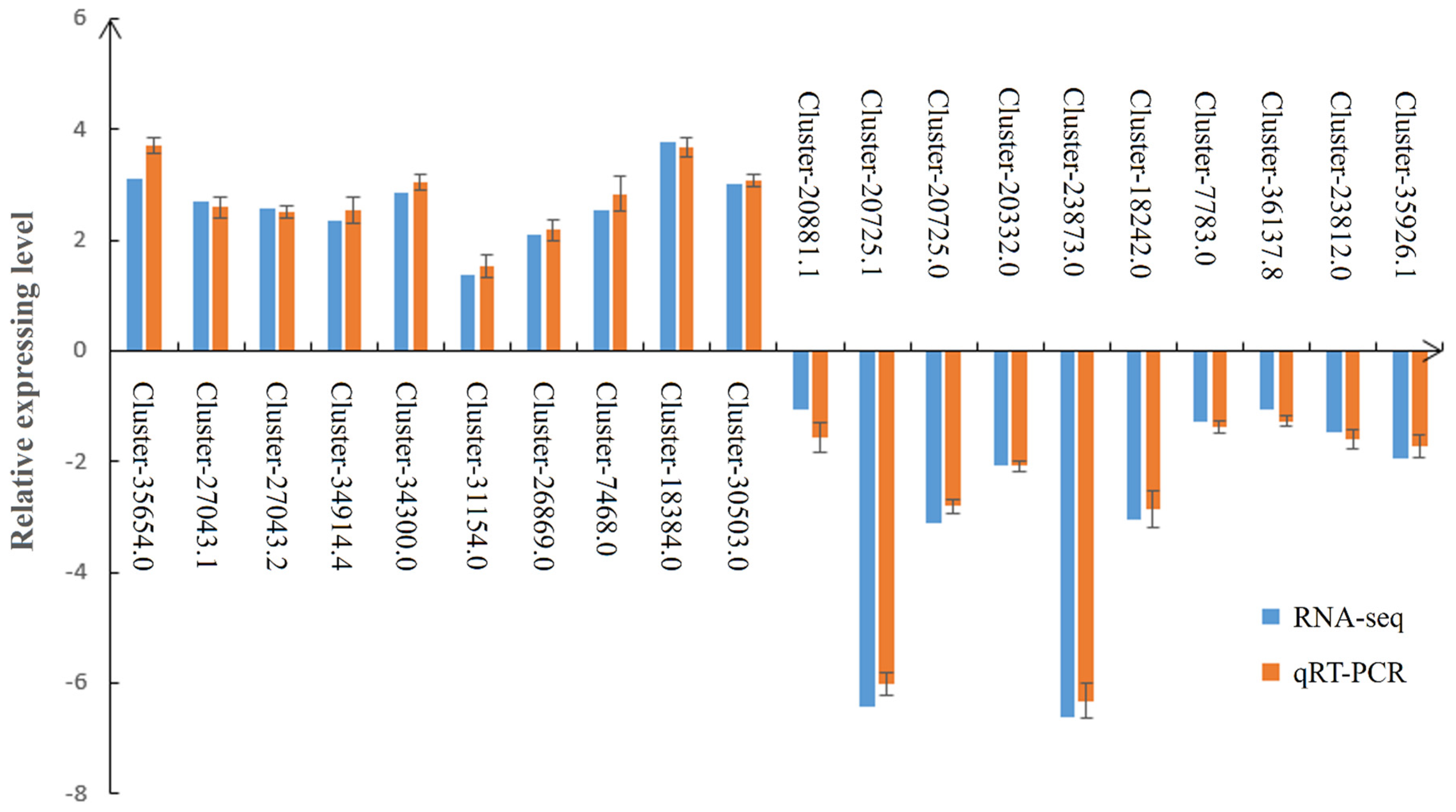
3.8. Expression Validation of RNA-Seq Data by Real-Time PCR (qRT-PCR)
4. Discussion
4.1. Effects of Shading on the Physiological Indicators and Photosynthesis of Carex adrienii
4.2. Responses of Hormone-Related Pathways to Shading Stress in Carex adrienii
4.3. Induction of Flavonoids in Carex adrienii Leaves Under Shading Conditions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wu, Z.; Man, W.; Ren, Y. Influence of tree coverage and micro-topography on the thermal environment within and beyond a green space. Agric. For. Meteorol. 2022, 316, 108846. [Google Scholar] [CrossRef]
- Cerqueira, A.F.; Rocha-Santos, L.; Benchimol, M.; Mielke, M.S. Habitat loss and canopy openness mediate leaf trait plasticity of an endangered palm in the Brazilian Atlantic Forest. Oecologia 2021, 196, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Tucker, G.F.; Maguire, D.A.; Tupinambá Simões, F. Associations between shade tolerance and wood specific gravity for conifers in contrast to angiosperm trees: Foundations of the conifer fitness-enhancing shade tolerance hypothesis. Plant-Environ. Interact. 2024, 5, e10131. [Google Scholar] [CrossRef]
- Valladares, F.; Niinemets, U. Shade tolerance, a key plant feature of complex nature and consequences. Annu. Rev. Ecol. Evol. Syst. 2008, 39, 237–257. [Google Scholar] [CrossRef]
- Lyu, X.; Mu, R.; Liu, B. Shade avoidance syndrome in soybean and ideotype toward shade tolerance. Mol. Breed. 2023, 43, 31. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, T.; Liu, J.; Li, H.; Liu, B. Shade-induced leaf senescence in plants. Plants 2023, 12, 1550. [Google Scholar] [CrossRef]
- Tang, W.; Chen, C.; Zhang, Y.; Chu, Y.; Yang, W.; Cui, Y.; Gong, R. Effect of low-light stress on sugar and acid accumulation during fruit development and ripening of sweet cherry. Horticulturae 2023, 9, 654. [Google Scholar] [CrossRef]
- Hu, Z.; Yao, X.; Chen, H.; Li, F.; Zhao, H.; Tang, H.; Lu, L. Changes and dynamics of the main quality components in tea leaves of 4 tea cultivars during the shading process. Sci. Hortic. 2024, 333, 113242. [Google Scholar] [CrossRef]
- Shokoya, G.; Fontanier, C.; Martin, D.L.; Dunn, B.L. Evaluation of sedges and nimblewill as low-input, shaded Lawns in Oklahoma, USA. HortTechnology 2022, 32, 567–577. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper Enzymes in Isolated Chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef]
- Tabatabaie, S.; Litt, J.S.; Muller, B.H.F. Sidewalks, trees and shade matter: A visual landscape assessment approach to understanding people’s preferences for walking. Urban For. Urban Green. 2023, 84, 127931. [Google Scholar] [CrossRef]
- Yu, Q.; Ji, W.; Pu, R.; Landry, S.; Acheampong, M.; O’Neil-Dunne, J.; Tanim, S.H. A preliminary exploration of the cooling effect of tree shade in urban landscapes. Int. J. Appl. Earth Obs. Geoinf. 2020, 92, 102161. [Google Scholar] [CrossRef]
- Zhang, C.T.; Zhu, X.L.; Cai, K.F.; Yu, Y.G. Evaluation of shade tolerance of Carex species available for garden-environment planting. J. Beijing For. Univ. 2010, 32, 207–212. [Google Scholar]
- Jung, C.; Arar, M. Natural vs. Artificial Light: A study on the influence of light source on chlorophyll content and photosynthetic rates on indoor Plants. Buildings 2023, 13, 1482. [Google Scholar] [CrossRef]
- Fatemeh, I.; Katja, F.; Soltani, F.; Baldermann, S. Effects of Shading on Plant Growth, Chlorophylls and carotenoids in florets of differently coloured cauliflowers (Brassica oleracea L. ssp. botrytis). Lebensmittelchemie 2021, 75, S1–S61. [Google Scholar] [CrossRef]
- Hayashi, A.; Sonobe, R.; Sano, T.; Horie, H. Estimating leaf chlorophyll content of shade grown tea based on PROSPECT model inversion. J. Jpn. Soc. Photogramm. Remote Sens. 2019, 58, 260–264. [Google Scholar] [CrossRef]
- Casal, J.J. Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 2013, 64, 403–427. [Google Scholar] [CrossRef]
- Liang, M.; Dong, Q.; Zhang, X.; Liu, Y.; Li, H.; Guo, S.; Qi, G. Metabolomics and transcriptomics analyses reveals the molecular regulatory mechanisms of Walnut (Juglans regia L.) Embryos in response to shade treatment. Int. J. Mol. Sci. 2023, 24, 10871. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Y.; Yan, H.; Cai, K.; Li, H.; Wu, Z.; Zhao, X. Integrated metabolomic and transcriptomic analyses reveal different metabolite biosynthesis profiles of Juglans mandshurica in shade. Front. Plant Sci. 2022, 13, 991874. [Google Scholar] [CrossRef]
- Bernal-Bayard, P.; Pallara, C.; Carmen Castell, M.; Molina-Heredia, F.P.; Fernández-Recio, J.; Hervás, M.; Navarro, J.A. Interaction of photosystem I from Phaeodactylum tricornutum with plastocyanins as compared with its native cytochrome c6: Reunion with a lost donor. Biochim. Biophys. Acta 2015, 1847, 1549–1559. [Google Scholar] [CrossRef]
- Alboresi, A.; Le Quiniou, C.; Yadav, S.K.N.; Scholz, M.; Meneghesso, A.; Gerotto, C.; Morosinotto, T. Conservation of core complex subunits shaped the structure and function of photosystem I in the secondary endosymbiont alga Nannochloropsis gaditana. New Phytol. 2017, 213, 714–726. [Google Scholar] [CrossRef] [PubMed]
- Bela, K.; Horváth, E.; Gallé, Á.; Szabados, L.; Tari, I.; Csiszár, J. Plant glutathione peroxidases: Emerging role of the antioxidant enzymes in plant development and stress responses. J. Plant Physiol. 2015, 176, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Møller, I.M.; Jensen, P.E.; Hansson, A. Oxidative modifications to cellular components in plants. Annu. Rev. Plant Biol. 2007, 58, 459–481. [Google Scholar] [CrossRef] [PubMed]
- Finlayson, S.A.; Krishnareddy, S.R.; Kebrom, T.H.; Casal, J.J. Phytochrome regulation of branching in Arabidopsis. Plant Physiol. 2010, 152, 1914–1927. [Google Scholar] [CrossRef]
- Stamm, P.; Kumar, P.P. Phytohormone signal network regulating elongation growth during shade avoidance. J. Exp. Bot. 2010, 61, 2889–2903. [Google Scholar] [CrossRef]
- Li, G.; Kazmi, A.; Feng, M.; Hou, H. Phytochrome-interacting factors (PIFs) regulate phytohormone-mediated plant environmental adaptation. Environ. Exp. Bot. 2024, 218, 105610. [Google Scholar] [CrossRef]
- Procko, C.; Burko, Y.; Jaillais, Y.; Ljung, K.; Long, J.A.; Chory, J.; Sveriges, L. The epidermis coordinates auxin-induced stem growth in response to shade. Genes Dev. 2016, 30, 1529–1541. [Google Scholar] [CrossRef]
- Zhao, Q.P.; Zhu, J.D.; Li, N.N.; Wang, X.N.; Zhao, X.; Zhang, X. Cryptochrome-mediated hypocotyl phototropism was regulated antagonistically by gibberellic acid and sucrose in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 614–630. [Google Scholar] [CrossRef]
- Pierik, R.; Djakovic-Petrovic, T.; Keuskamp, D.H.; de Wit, M.; Voesenek, L.A.C.J. Auxin and ethylene regulate elongation responses to neighbor proximity signals independent of gibberellin and DELLA proteins in Arabidopsis. Plant Physiol. 2009, 149, 1701–1712. [Google Scholar] [CrossRef]
- Kim, B.B.; Brownlee, S.N.; Grant, J.S.; Cannon, A.B. Gene expression characteristics in response to abscisic acid under shade. Plant Mol. Biol. Report. 2022, 40, 43–67. [Google Scholar] [CrossRef]
- Panigrahy, M.; Ranga, A.; Das, J.; Panigrahi, K.C.S. Shade tolerance in Swarnaprabha rice is associated with higher rate of panicle emergence and positively regulated by genes of ethylene and cytokinin pathway. Sci. Rep. 2019, 9, 6817. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zheng, Y.; Yao, H.; Ma, Z.; Xiao, M.; Wang, H.; Liu, Y. Light and jasmonic acid coordinately regulate the phosphate responses under shade and phosphate starvation conditions in Arabidopsis. Plant Direct 2023, 7, e504. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhang, Y.; Zhu, C.; Yao, X.; Zheng, Z.; Tian, Z.; Cai, X. EkFLS overexpression promotes flavonoid accumulation and abiotic stress tolerance in plant. Physiol. Plant. 2021, 172, 1966–1982. [Google Scholar] [CrossRef] [PubMed]
- Jaakola, L.; Hohtola, A. Effect of latitude on flavonoid biosynthesis in plants. Plant Cell Environ. 2010, 33, 1239–1247. [Google Scholar] [CrossRef]
- Zhao, X.; Zeng, X.; Lin, N.; Yu, S.; Fernie, A.R.; Zhao, J. CsbZIP1-CsMYB12 mediates the production of bitter-tasting flavonols in tea plants (Camellia sinensis) through a coordinated activator-repressor network. Hortic. Res. 2021, 8, 110. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, L.; Shan, Y.; Liu, Y.; Tian, Y.; Xia, T. Influence of shade on flavonoid biosynthesis in tea (Camellia sinensis (L.) O. Kuntze). Sci. Hortic. 2012, 141, 7–16. [Google Scholar] [CrossRef]
- Yan, H.; Pei, X.; Zhang, H.; Li, X.; Zhang, X.; Zhao, M.; Zhao, X. MYB-Mediated Regulation of Anthocyanin Biosynthesis. Int. J. Mol. Sci. 2021, 22, 3103. [Google Scholar] [CrossRef]
- Li, M.; Li, Y.; Guo, L.; Gong, N.; Pang, Y.; Jiang, W.; Xia, T. Functional characterization of tea (Camellia sinensis) MYB4a transcription factor using an integrative approach. Front. Plant Sci. 2017, 8, 943. [Google Scholar] [CrossRef]
- Li, M.; Sun, L.; Gu, H.; Cheng, D.; Guo, X.; Chen, R.; Chen, J. Genome-wide characterization and analysis of bHLH transcription factors related to anthocyanin biosynthesis in spine grapes (Vitis davidii). Sci. Rep. 2021, 11, 6863. [Google Scholar] [CrossRef]
- Luo, Y.; Yu, S.; Li, J.; Li, Q.; Wang, K.; Huang, J.; Liu, Z. Characterization of the transcriptional regulator CsbHLH62 that negatively regulates EGCG3” Me biosynthesis in Camellia sinensis. Gene 2019, 699, 8–15. [Google Scholar] [CrossRef]
- Li, P.; Xia, E.; Fu, J.; Xu, Y.; Zhao, X.; Tong, W.; Zhao, J. Diverse roles of MYB transcription factors in regulating secondary metabolite biosynthesis, shoot development, and stress responses in tea plants (Camellia sinensis). Plant J. For. Cell Mol. Biol. 2022, 110, 1144–1165. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Grain, D.; Bobet, S.; Le Gourrierec, J.; Thévenin, J.; Kelemen, Z.; Dubos, C. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol. 2014, 202, 132–144. [Google Scholar] [CrossRef]
- Schulz, E.; Tohge, T.; Winkler, J.B.; Albert, A.; Schäffner, A.R.; Fernie, A.R.; Hincha, D.K. Natural variation among Arabidopsis accessions in the regulation of flavonoid metabolism and stress gene expression by combined UV radiation and cold. Plant Cell Physiol. 2021, 62, 502–514. [Google Scholar] [CrossRef]
- Yastreb, T.O.; Kolupaev, Y.E.; Lugovaya, A.A.; Dmitriev, A.P. Content of osmolytes and flavonoids under salt stress in Arabidopsis thaliana plants defective in jasmonate signaling. Appl. Biochem. Microbiol. 2016, 52, 210–215. [Google Scholar] [CrossRef]
- Fini, A.; Brunetti, C.; Di Ferdinando, M.; Ferrini, F.; Tattini, M. Stress-induced flavonoid biosynthesis and the antioxidant machinery of plants. Plant Signal. Behav. 2011, 6, 709–711. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, T.; Wang, S.; Tian, Z.; Chen, S.; Li, X.; Zou, S.; Tan, Z.; Wang, J.; Wang, S.; Ai, L.; et al. Integrative Analysis of Metabolome and Transcriptome Profiles to Evaluate the Response Mechanisms of Carex adrienii to Shade Conditions. Agronomy 2024, 14, 2800. https://doi.org/10.3390/agronomy14122800
Guo T, Wang S, Tian Z, Chen S, Li X, Zou S, Tan Z, Wang J, Wang S, Ai L, et al. Integrative Analysis of Metabolome and Transcriptome Profiles to Evaluate the Response Mechanisms of Carex adrienii to Shade Conditions. Agronomy. 2024; 14(12):2800. https://doi.org/10.3390/agronomy14122800
Chicago/Turabian StyleGuo, Tao, Shumin Wang, Zhong Tian, Shuang Chen, Xuemei Li, Shihui Zou, Zhijian Tan, Jiao Wang, Sheng Wang, Lijiao Ai, and et al. 2024. "Integrative Analysis of Metabolome and Transcriptome Profiles to Evaluate the Response Mechanisms of Carex adrienii to Shade Conditions" Agronomy 14, no. 12: 2800. https://doi.org/10.3390/agronomy14122800
APA StyleGuo, T., Wang, S., Tian, Z., Chen, S., Li, X., Zou, S., Tan, Z., Wang, J., Wang, S., Ai, L., & Sui, S. (2024). Integrative Analysis of Metabolome and Transcriptome Profiles to Evaluate the Response Mechanisms of Carex adrienii to Shade Conditions. Agronomy, 14(12), 2800. https://doi.org/10.3390/agronomy14122800



