Prediction of Anthocyanin Content in Purple-Leaf Lettuce Based on Spectral Features and Optimized Extreme Learning Machine Algorithm
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
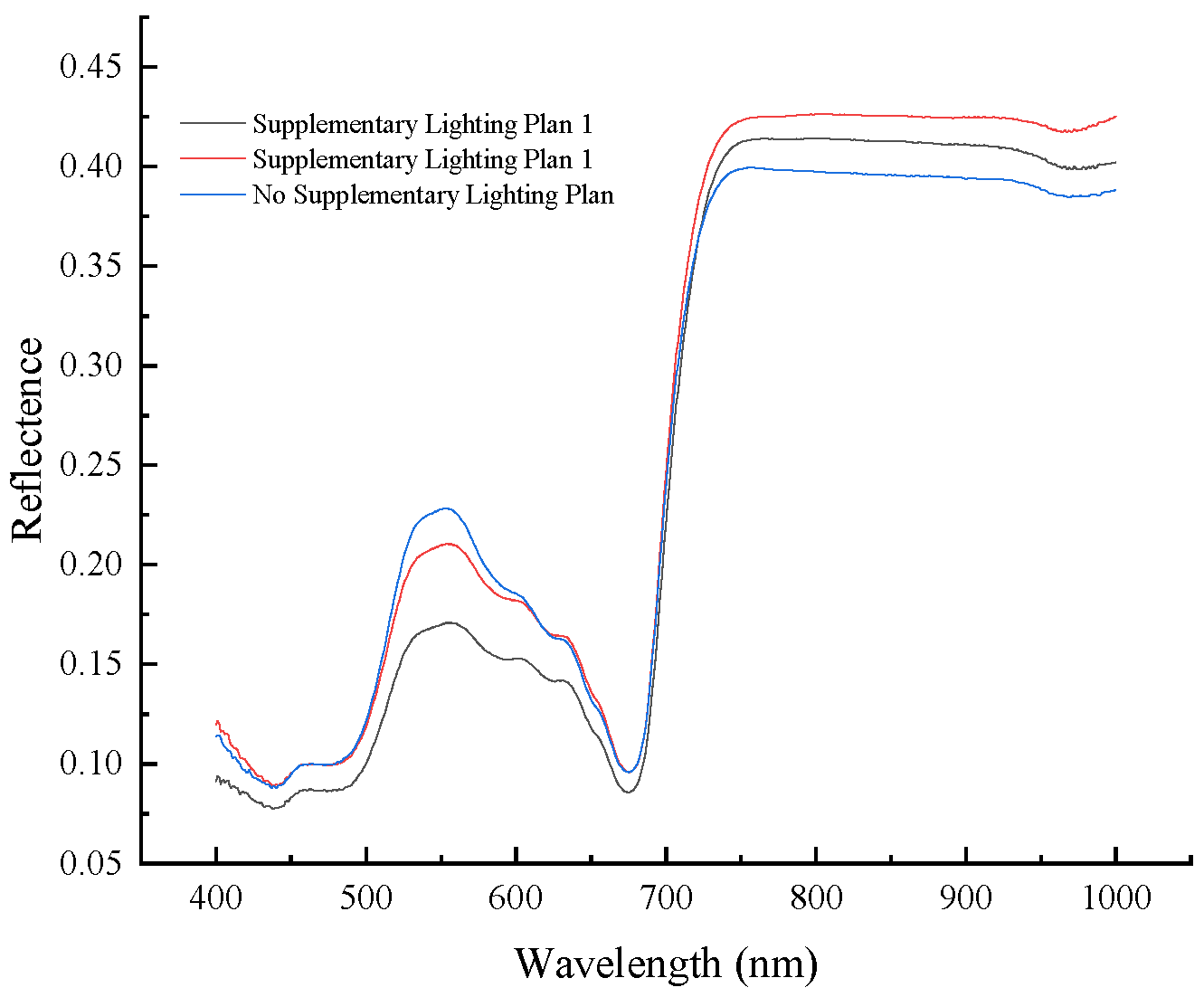
2.2. Measurement of Spectral Data
2.3. Measurement of Anthocyanins
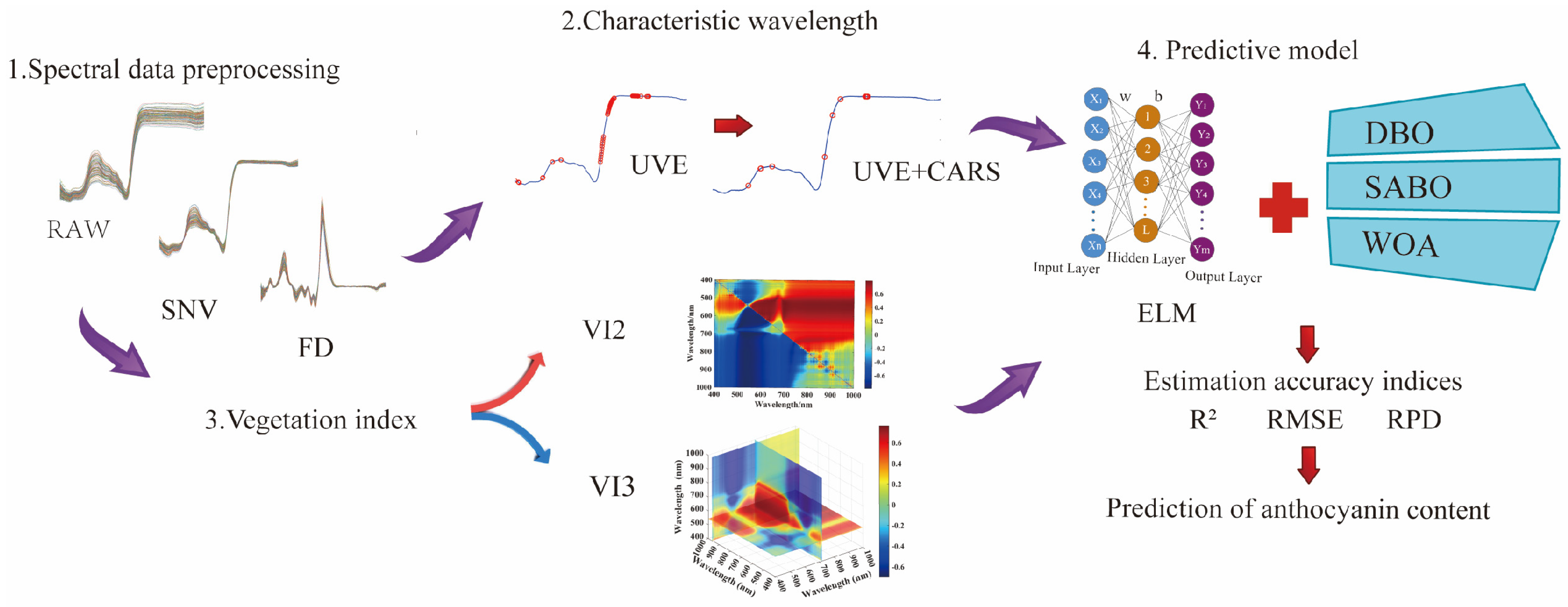
2.4. Data Processing and Modeling Methods
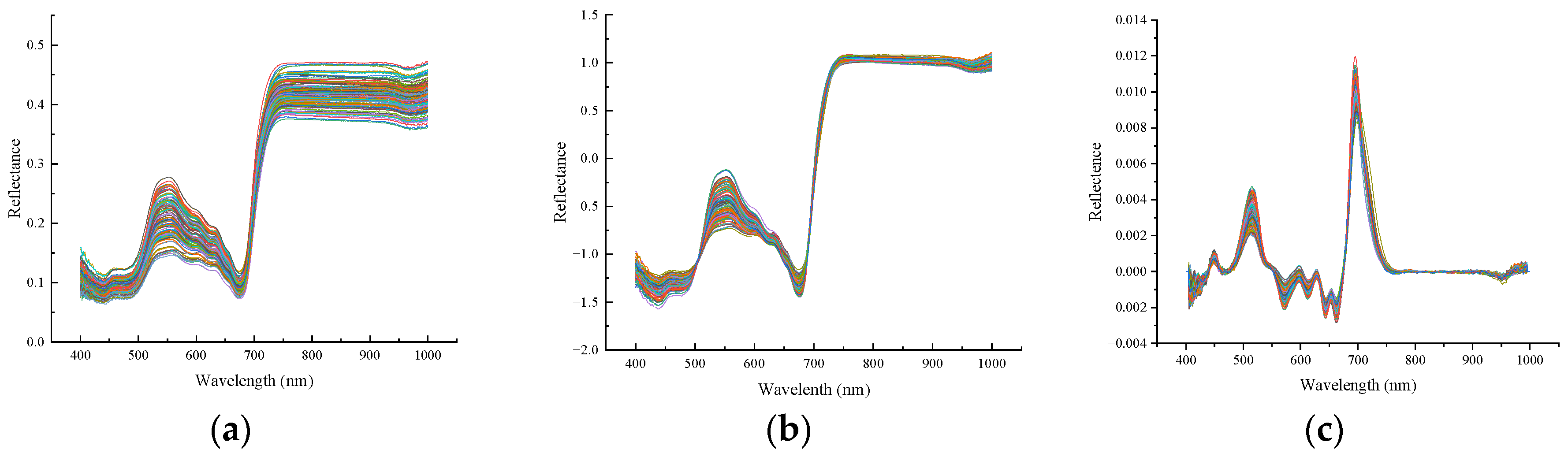
2.4.1. Data Preprocessing
2.4.2. Feature Band Extraction Method
2.4.3. Vegetation Indices
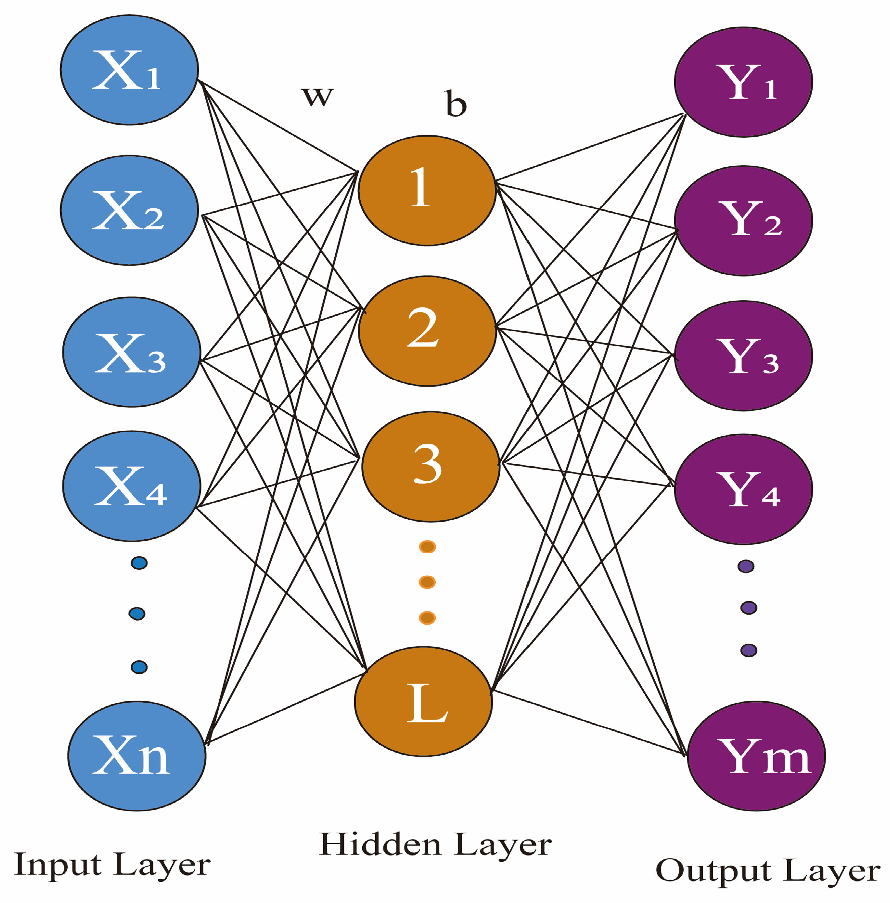
2.4.4. Construction and Evaluation of Inversion Model
- Dung Beetle Optimization (DBO)
- 2.
- Subtraction-Average-Based Optimization (SABO)
- 3.
- Whale Optimization Algorithm (WOA)
2.5. Data Analysis
3. Results
3.1. Spectra Pretreatment
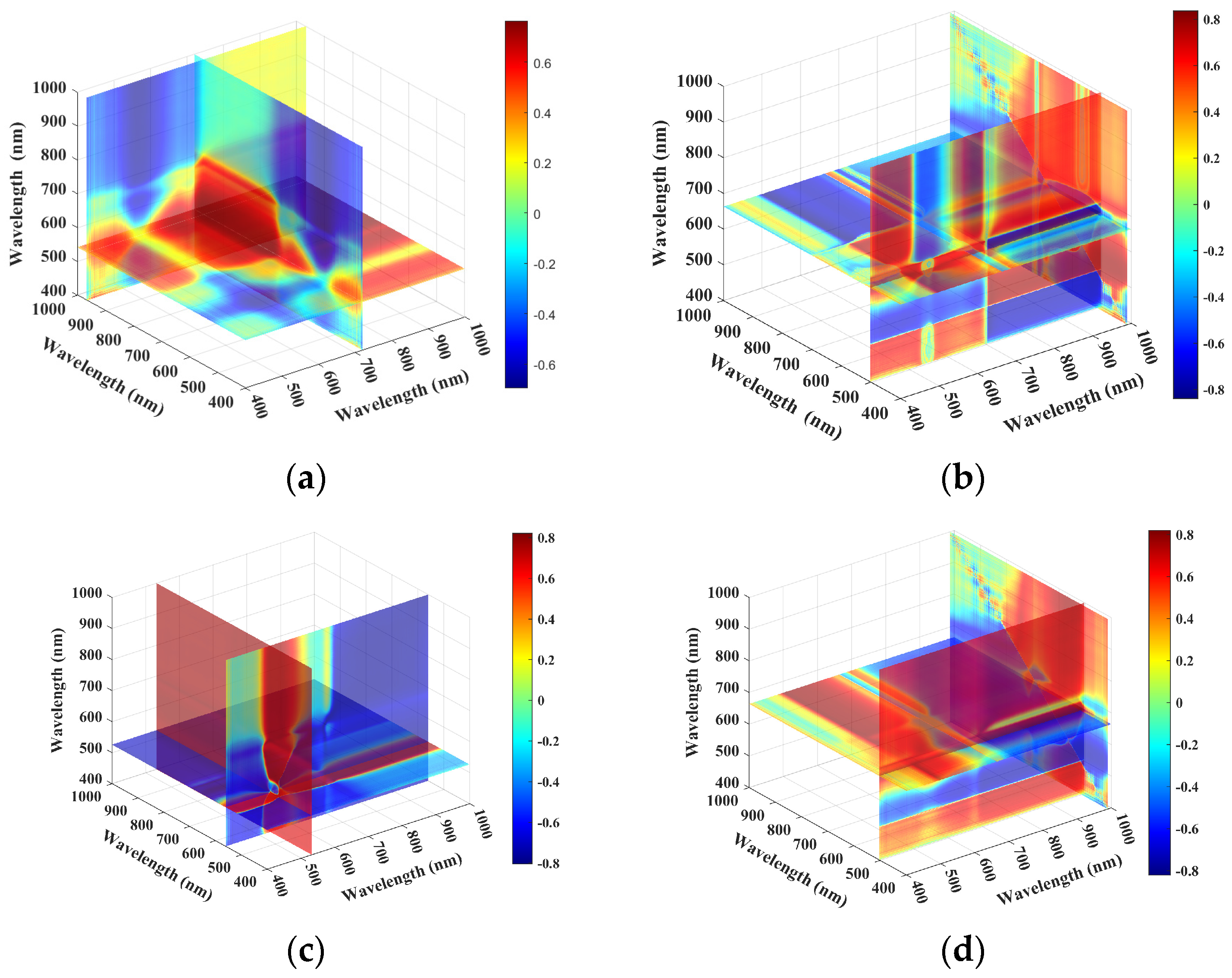
3.2. Feature Extraction
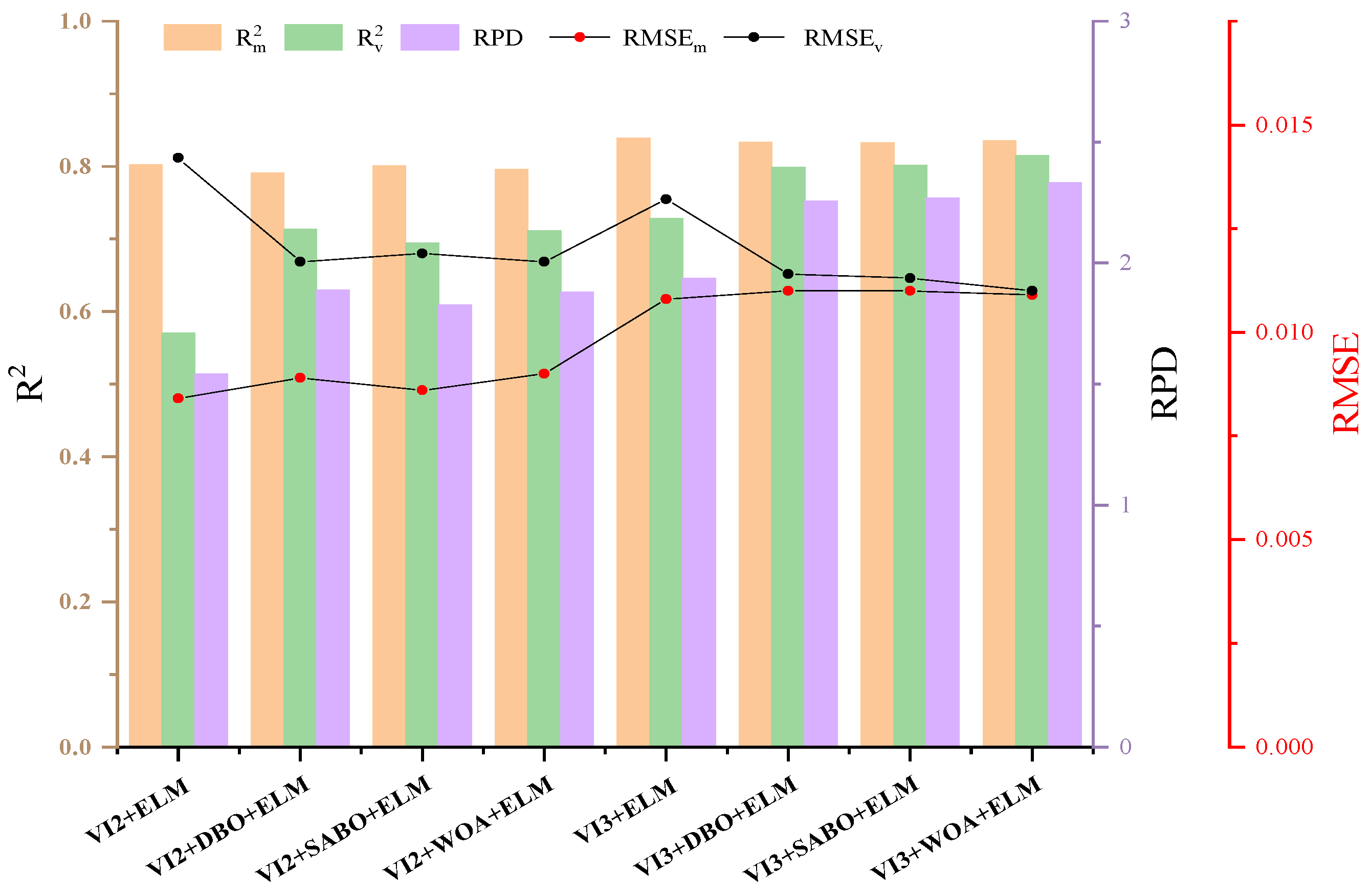
3.3. Constraction of Vegetation Indices
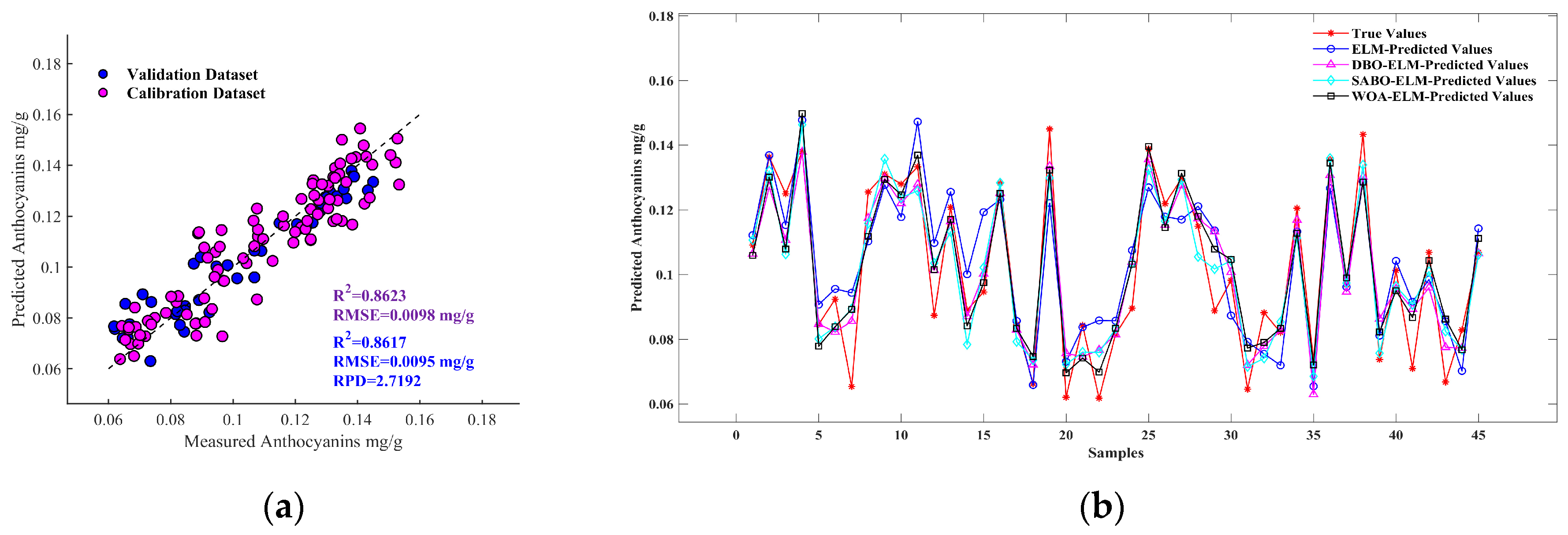
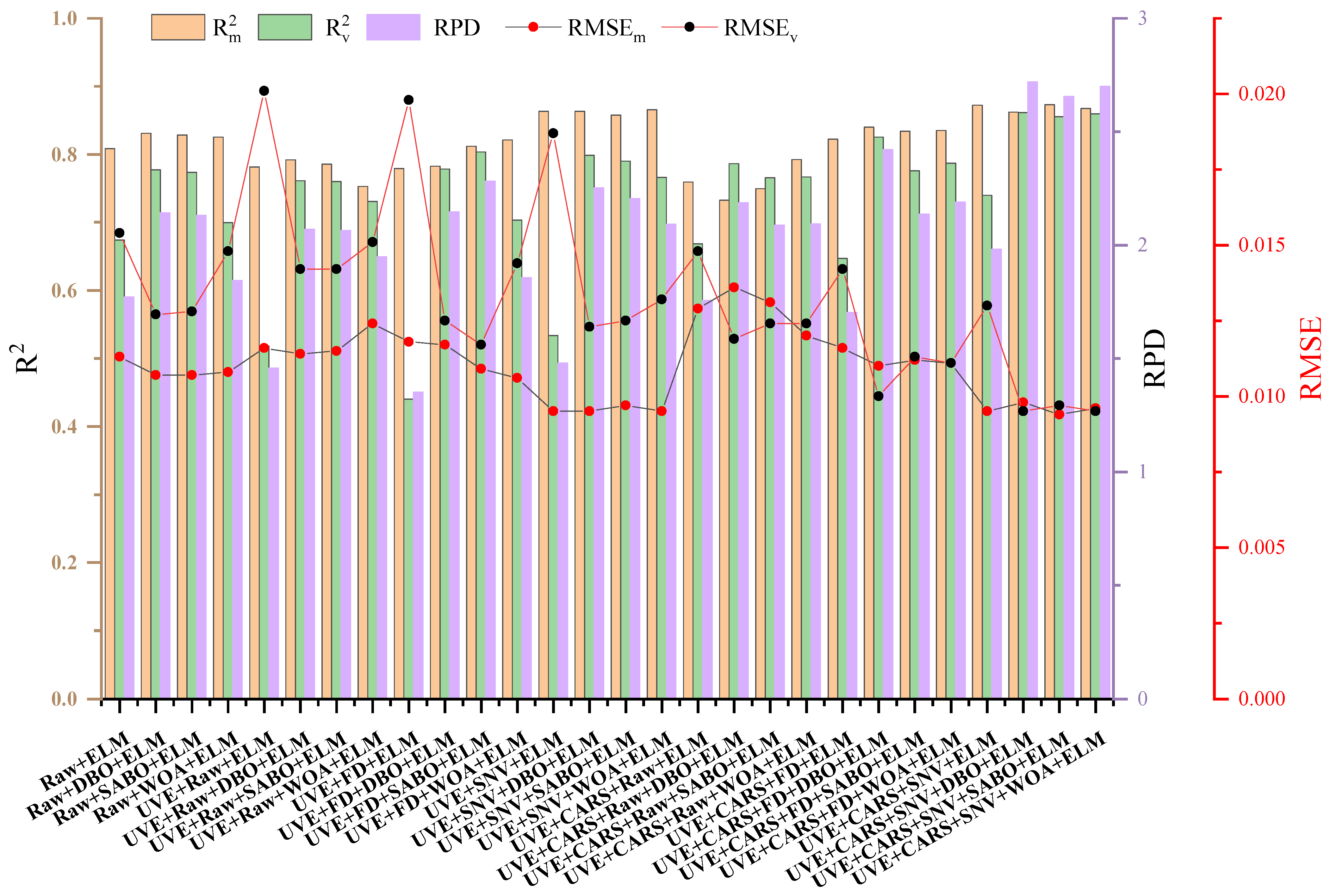
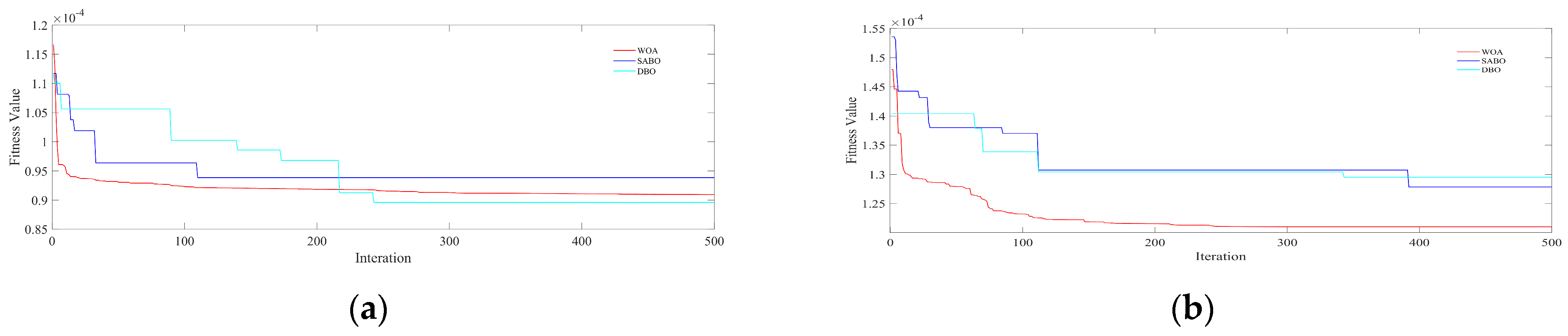
3.4. Anthocyanin Estimation Based on ELM Model and DBO-SABO-WOA Optimization
4. Discussion
4.1. Potential of Different Characteristic Wavelength Selection Methods in Estimating Anthocyanin Content of Lettuce
4.2. Potential of Vegetation Indices in Estimating Anthocyanin Content of Purple Lettuce
4.3. Influence of Model Optimization in Estimating Anthocyanin Content of Purple Lettuce
5. Conclusions
- (1)
- Two spectral preprocessing methods, the FD and SNV, were applied to reduce the impact of instrument noise, baseline drift, and other factors on the original spectra. The effects of feature wavelength selection methods, UVE and UVE-CARS, on the model performance were compared. The results indicated that UVE-CARS was the best variable selection method. The model built using the feature wavelengths selected by the UVE-CARS-SNV-DBO-ELM (Rv2 = 0.8617; RMSEv = 0.0095; RPD = 2.7192) achieved the best prediction performance for the anthocyanin content. In resource-constrained environments, UVE-CARS eliminates redundant features, thereby accelerating computation while preserving accuracy.
- (2)
- Based on the principle of the maximum correlation coefficient, two-band vegetation indices (NARI, MGRVI, ARI, OSAVI) and three-band vegetation indices (MARI, EVI, TVI, PSPR) were calculated. Compared to the prediction performance using two-band vegetation indices, the prediction performance using the three-band indices (Rv2 = 0.812; RMSEv = 0.011; RPD = 2.3323) was significantly improved.
- (3)
- The performance of the ELM model was optimized using DBO, SABO, and the WOA. The DBO algorithm achieved an improvement in the Rv2 ranging from 5.8% to 27.82%, the SABO algorithm showed an improvement from 2.92% to 26.84%, and the WOA demonstrated an improvement from 3.75% to 27.51% in the anthocyanin prediction. DBO, the WOA, and SABO optimize ELM weights and biases, balancing global and local searches to enhance performance and reduce computational costs. These techniques improve the feasibility of ELM models by maintaining high accuracy with lower resource requirements.
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Perez-Lopez, U.; Sgherri, C.; Miranda-Apodaca, J.; Lacuesta, F.M.M.; Mena-Petite, A.; Quartacci, M.; Munoz-Rueda, A. Concentration of phenolic compounds is increased in lettuce grown under high light intensity and elevated CO2. Plant Physiol. Biochem. 2018, 123, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Khanam, U.K.; Oba, S.; Yanase, E.; Murakami, Y. Phenolic acids, flavonoids and total antioxidant capacity of selected leafy vegetables. J. Func. Foods 2012, 4, 979–987. [Google Scholar] [CrossRef]
- Gould, K.S. Nature’s swiss army knife: The diverse protective roles of anthocyanins in leaves. J. Biomed. Biotechnol. 2004, 2004, 314–320. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Gu, J.; Wu, H.; Rauf, A.; Emran, T.B.; Khan, Z.; Mitra, S.; Aljohani, A.S.M.; Alhumaydhi, F.A.; Al-Awthan, Y.S.; et al. Phytochemicals, nutrition, metabolism, bioavailability, and health benefits in lettuce-a comprehensive review. Antioxidants 2022, 11, 1158. [Google Scholar] [CrossRef] [PubMed]
- Dabravolski, S.A.; Isayenkov, S.V. The role of anthocyanins in plant tolerance to drought and salt stresses. Plants 2023, 12, 2558. [Google Scholar] [CrossRef]
- Hu, Y.Z.; He, R.; Ju, J.; Zhang, S.C.; He, X.Y.; Li, Y.M.; Liu, X.J.; Liu, H.C. Effects of substituting b with fr and uva at different growth stages on the growth and quality of lettuce. Agronomy 2023, 13, 2547. [Google Scholar] [CrossRef]
- Averina, N.; Savina, S.; Dremuk, I.; Yemelyanava, H.; Pryshchepchyk, Y.; Usatov, A. Influence of 5-aminolevulinic acid on physiological and biochemical characteristics of winter wheat varieties with different levels of anthocyanins in coleoptiles. Proc. Natl. Acad. Sci. Belarus Biol. Ser. 2022, 67, 135–146. [Google Scholar] [CrossRef]
- Bhatt, V.; Sendri, N.; Swati, K.; Devidas, S.; Bhandari, P. identification and quantification of anthocyanins, flavonoids and phenolic acids in flowers of rhododendron arboreum and evaluation of their antioxidant potential. J. Sep. Sci. 2022, 45, 2555–2565. [Google Scholar] [CrossRef]
- Singh, M.C.; Price, W.; Kelso, C.; Arcot, J.; Probst, Y. Measuring the anthocyanin content of the australian fruit and vegetables for the development of a food composition database. J. Food Compos. Anal. 2022, 112, 104697. [Google Scholar] [CrossRef]
- Yu, Y.; Yu, H.Y.; Li, X.K.; Zhang, L.; Sui, Y.Y. Prediction of potassium content in rice leaves based on spectral features and random forests. Agronomy 2023, 13, 2337. [Google Scholar] [CrossRef]
- Zhou, L.; Wu, H.B.; Jing, T.T.; Li, T.H.; Li, J.S.; Kong, L.J.; Zhou, L.N. Estimation of relative chlorophyll content in lettuce (Lactuca sativa L.) Leaves under cadmium stress using visible—Near-infrared reflectance and machine-learning models. Agronomy 2024, 14, 427. [Google Scholar] [CrossRef]
- Eshkabilov, S.; Lee, A.; Sun, X.; Lee, C.W.; Simsek, H. Hyperspectral imaging techniques for rapid detection of nutrient content of hydroponically grown lettuce cultivars. Comput. Electron. Agric. 2021, 181, 105968. [Google Scholar] [CrossRef]
- Feng, H.F.; Li, Y.X.; Wu, F.; Zou, X.C. Estimating winter wheat nitrogen content using spad and hyperspectral vegetation indices with machine learning. Trans. Chin. Soc. Agric. Eng. 2024, 40, 227–237. [Google Scholar]
- Xu, T.Y.; Jin, Z.Y.; Guo, Z.H.; Yang, L.; Bai, J.C.; Feng, S.; Yu, F.H. Simultaneous inversion method of nitrogen and phosphorus contents in rice leaves using cars-run-elm algorithm. J. Agric. Eng. 2022, 38, 148–155. [Google Scholar]
- Pandey, P.; Veazie, P.; Whipker, B.; Young, S. Predicting foliar nutrient concentrations and nutrient deficiencies of hydroponic lettuce using hyperspectral imaging. Biosyst. Eng. 2023, 230, 458–469. [Google Scholar] [CrossRef]
- Jiang, S.Y.; Chang, Q.R.; Wang, X.P.; Zheng, Z.K.; Zhang, Y.; Wang, Q. Estimation of anthocyanins in whole-fertility maize leaves based on ground-based hyperspectral measurements. Remote Sens. 2023, 15, 2571. [Google Scholar] [CrossRef]
- Kim, C.; van Iersel, M.W. Image-based phenotyping to estimate anthocyanin concentrations in lettuce. Front. Plant Sci. 2023, 14, 1155722. [Google Scholar] [CrossRef]
- Liang, J.Q.; Liu, M.; Yu, K.; Liu, Z.L.; Kong, L.Q.; Hui, M.; Dong, L.Q.; Zhao, Y.J. Spectral pre-processing based on convolutional neural network. Spectrosc. Spectr. Anal. 2022, 1, 292–297. [Google Scholar]
- Wang, W.D.; Chang, Q.R.; Wang, Y.N. Hyperspectral monitoring of anthocyanins relative content in winter wheat leaves. J. Trit. Crops 2020, 40, 754–761. [Google Scholar]
- Zhang, M.L.; Chen, Y.J.; Wang, M.J.; Li, M.Z.; Zheng, L.H. A hyperspectral deep learning modelfor predicting anthocyanin contentin purple leaf lettuce. Spectrosc. Spectral Anal. 2024, 44, 865–871. [Google Scholar]
- Liu, Y.H.; Wang, Q.Q.; Shi, X.W.; Gao, X.W. Hyperspectral nondestructive detection model of chlorogenic acid content during storage of honeysuckle. J. Agric. Eng. 2019, 35, 291–299. [Google Scholar]
- Li, X.L.; Zhao, X.; Wei, F.F.; Peng, H.; Guo, H. Non-destructive prediction and visualization of anthocyanin content in mulberry fruits using hyperspectral imaging. Front. Plant Sci. 2023, 14, 1137198. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.H.; Mei, L.; Ma, M.H.; Gao, S.; Li, Q.X. Nondestructive testing and grading of preserved duck eggs based on machine vision and near-infrared spectroscopy. J. Agric. Eng. 2019, 35, 314–321. [Google Scholar]
- Zhang, H.Y.; Zhu, Q.B.; Huang, M.; Guo, Y. Automatic determination of optimal spectral peaks for classification of chinese tea varieties using laser-induced breakdown spectroscopy. Int. J. Agric. Biol. Eng. 2018, 11, 154–158. [Google Scholar]
- Li, W.; Tan, F.; Zhang, W.; Gao, L.S.; Li, J.S. Application of improved random frog algorithm in fast identification of soybean varieties. Spectrosc. Spectral Anal. 2023, 43, 3763–3769. [Google Scholar]
- Liu, Y.Y.; Wang, T.Z.; Su, R.; Hu, C.; Chen, F.; Cheng, J.H. Quantitative evaluation of color, firmness, and soluble solid content of korla fragrant pears via iriv and ls-svm. Agriculture 2021, 11, 731. [Google Scholar] [CrossRef]
- Wei, X.; Zhang, Y.C.; Wu, D.; Wei, Z.B.; Chen, K.S. Rapid and non-destructive detection of decay in peach fruit at the cold environment using a self-developed handheld electronic-nose system. Food Anal. Methods 2018, 11, 2990–3004. [Google Scholar] [CrossRef]
- Liang, K.; Liu, Q.X.; Pan, L.Q.; Shen, M.X. Detection of soluble solids content in ‘korla fragrant pear’ based on hyperspectral imaging and cars-iriv algorithm. J. Nanjing Agric. Univ. 2018, 41, 760–766. [Google Scholar]
- Tang, Z.J.; Zhen, X.Y.; Xin, W.; Wei, Z.; Li, Z.J.; Zhang, F.C.; Chen, J.Y. Nitrogen nutrition diagnosis of winter oilseed rape using spectral indexes optimized by correlation matrix method. J. Agric. Eng. 2023, 39, 97–106. [Google Scholar]
- Xiang, Y.Z.; Wang, X.; An, J.Q.; Tang, Z.J.; Li, W.Y.; Shi, H.D. Estimation of leaf area index of soybean based on fractional order differentiation and optimal spectral index. J. Agric. Mach. 2023, 54, 329–342. [Google Scholar]
- Dai, F.S.; Shi, J.; Yang, C.S.; Li, Y.; Zhao, Y.; Liu, Z.Y.; An, T.; Li, X.L.; Yan, P.; Dong, C.W. Detection of anthocyanin content in fresh zijuan tea leaves based on hyperspectral imaging. Food Control 2023, 152, 109839. [Google Scholar] [CrossRef]
- Cho, J.; Lim, J.H.; Park, K.J.; Choi, J.H.; Ok, G.S. Prediction of pelargonidin-3-glucoside in strawberries according to the postharvest distribution period of two ripening stages using vis-nir and swir hyperspectral imaging technology. LWT 2021, 141, 110875. [Google Scholar] [CrossRef]
- Chen, S.S.; Zhang, F.F.; Ning, J.F.; Liu, X.; Zhang, Z.W.; Yang, S.Q. Predicting the anthocyanin content of wine grapes by nir hyperspectral imaging. Food Chem. 2015, 172, 788–793. [Google Scholar] [CrossRef] [PubMed]
- Yue, R.; Yin, B.S.; Li, Z.F.; Lii, F.L. Monitoring of anthocyan in content of winter wheat infected by pst using uav rgb image. J. Triticeae Crops 2023, 43, 1–11. [Google Scholar]
- Liu, X.Y.; Yu, J.R.; Liu, C.X.; Den, X.F. Estimation of anthocyanin content in prunus cerasifera based on color indices and bp neural network. J. Northwest For. Univ. 2022, 37, 145–152. [Google Scholar]
- Guo, S.; Chan, Q.R.; Zhao, Z.Y.; Li, L.J.; Dong, Q.Q. Estimation of anthocyanin content in maize at different growth stages based on hyperspectral technology. Jiangsu Agric. Sci. 2024, 40, 303–311. [Google Scholar]
- Miao, H.L.; Chen, X.K.; Guo, Y.M.; Wang, Q.; Zhang, R.; Chang, Q.R. Estimation of anthocyanins in winter wheat based on band screening method and genetic algorithm optimization models. Remote Sens. 2024, 16, 2324. [Google Scholar] [CrossRef]
- Giusti, M.M.; Wrolstad, R.E. Characterization and measurement of anthocyanins by uv-visible spectroscopy. Curr. Protoc. Food Anal. Chem. 2001, F1–F2. [Google Scholar] [CrossRef]
- Li, H.H.; Lu, W.; Hong, D.L.; Dang, X.J.; Liang, K. Rapid testing method of brown rice germination rate based on characteristic spectrum and general regression neural network. Laser Optoelectron. Prog. 2015, 52, 281–287. [Google Scholar]
- Raju, C.S.; Løkke, M.M.; Sutaryo, S.; Ward, A.J.; Møller, H.B. Nir monitoring of ammonia in anaerobic digesters using a diffuse reflectance probe. Sensors 2012, 12, 2340–2350. [Google Scholar] [CrossRef]
- Liu, Y.D.; Lin, X.D.; Gao, H.G.; Gao, X.; Wang, S. Quantitative analysis of chlorophyll content in tea leaves by fluorescence spectroscopy. Laser Optoelectron. Prog. 2021, 58, 444–453. [Google Scholar]
- Wang, Q.H.; Zhou, K.; Wu, L.L.; Yun, W.C. Egg freshness detection based on hyper-spectra. Spectrosc. Spectral Anal. 2016, 36, 2596–2600. [Google Scholar]
- Jiang, J.L.; Cen, H.Y.; Zhang, C.; Lyu, X.H.; Weng, H.Y.; Xu, H.X.; He, Y. Nondestructive quality assessment of chili peppers using near-infrared hyperspectral imaging combined with multivariate analysis. Postharvest Biol. Technol. 2018, 146, 147–154. [Google Scholar] [CrossRef]
- Sun, J.; Yao, K.S.; Cheng, J.H.; Xu, M.; Zhou, X. Nondestructive detection of saponin content in panax notoginseng powder based on hyperspectral imaging. J. Pharm. Biomed. Anal. 2024, 242, 116015. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Li, C.G.; Pan, B.B.; Li, J.; Hu, J.T.; Ge, P.Z.; Liu, F.F. life prediction for vibration isolation rubber components based on dbo-elm model. Mech. Electr. Eng. Technol. 2024, 53, 13–19. [Google Scholar]
- Guo, Y.; Chen, B.; Zeng, H.Y.; Qing, G.Y.; Guo, B. Research on wear state identification of ordered grinding wheel for c/sic composites based on dbo-elm. Wear 2024, 556–557, 205529. [Google Scholar] [CrossRef]
- Zhang, Y.D.; Wang, Y.C. Study on elm power load forecasting metrics based on subtractive optimiser algorithm optimisation. Mod. Ind. Econ. Inform. 2024, 14, 124–126. [Google Scholar]
- Liu, S.L.; Gao, Y.; Lin, R.C.; Tan, W.J. Improved subtraction-average-based optimization based on hybrid strategy. Intell. Comput. Appl. 2024, 14, 70–77. [Google Scholar] [CrossRef]
- Liu, W.J.; Liu, Z.X.; Xiong, S.; Wang, M. Comparative prediction performance of the strength of a new type of ti tailings cemented backfilling body using pso-rf, ssa-rf, and woa-rf models. Case Stud. Constr. Mater. 2024, 20, e2766. [Google Scholar] [CrossRef]
- Wang, N.; Chen, C.L.; Xiang, S.; Jin, Z.Y.; Bai, J.C.; Yu, F.H. Inversing chlorophyll contents in rice using radiation transport of leaf bilayer. Agric. Eng. J. 2024, 40, 171–178. [Google Scholar]
- Osman, A.I.A.; Ahmed, A.N.; Huang, Y.F.; Kumar, P.; Birima, A.H.; Sherif, M.; Sefelnasr, A.; Ebraheemand, A.A.; El-Shafie, A. Past, present and perspective methodology for groundwater modeling-based machine learning approaches. Arch. Comput. Methods Eng. 2022, 29, 3843–3859. [Google Scholar] [CrossRef]
- Viscarra Rossel, R.A.; Mcglynn, R.N.; Mcbratney, A.B. Determining the composition of mineral-organic mixes using uv–vis–nir diffuse reflectance spectroscopy. Geoderma 2006, 137, 70–82. [Google Scholar] [CrossRef]
- Bayle, A.; Carlson, B.; Thierion, V.; Isenmann, M.; Choler, P. Improved mapping of mountain shrublands using the sentinel-2 red-edge band. Remote Sens. 2019, 11, 2807. [Google Scholar] [CrossRef]
- Zhang, J.; Sun, B.; Yang, C.H.; Wang, C.Y.; You, Y.H.; Zhou, G.S.; Liu, B.; Wang, C.F.; Kuai, J.; Xie, J. A novel composite vegetation index including solar-induced chlorophyll fluorescence for seedling rapeseed net photosynthesis rate retrieval. Comput. Electron. Agric. 2022, 198, 107031. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N.; Chivkunova, O.B. Optical properties and nondestructive estimation of anthocyanin content in plant leaves. Photochem. Photobiol. 2001, 74, 38–45. [Google Scholar] [CrossRef]
- Fang, H.; Man, W.D.; Liu, M.Y.; Zhang, Y.B.; Chen, X.T.; Li, X.; He, J.N.; Tian, D. Leaf area index inversion of spartina alterniflora using uav hyperspectral data based on multiple optimized machine learning algorithms. Remote Sens. 2023, 15, 4465. [Google Scholar] [CrossRef]
- Xing, N.C.; Huang, W.J.; Xie, Q.Y.; Shi, Y.; Ye, H.C.; Dong, Y.Y.; Wu, M.Q.; Sun, G.; Jiao, Q.J. A transformed triangular vegetation index for estimating winter wheat leaf area index. Remote Sens. 2020, 12, 16. [Google Scholar] [CrossRef]
- Ren, S.L.; Chen, X.Q.; An, S. Assessing plant senescence reflectance index-retrieved vegetation phenology and its spatiotemporal response to climate change in the inner mongolian grassland. Int. J. Biometeorol. 2017, 61, 601–612. [Google Scholar] [CrossRef]
- Ji, J.T.; Li, P.G.; Jin, X.; Ma, H.; Yong, L.M. Study on quantitative detection of tomato seedling robustness in spring seedling transplanting period based on vis-nir spectroscopy. Spectrosc. Spectral Anal. 2022, 42, 1741–1748. [Google Scholar]
- Jiang, W.; Fang, J.L.; Wang, S.W.; Wang, R.T. using cars-spa algorithm combined with hyperspectral to determine reducing sugars content in potatoes. J. Northeast Agric. Univ. 2016, 47, 88–95. [Google Scholar]
- Mu, W.Z.; Zhang, G.Y.; Zhang, W.; Yao, R.; Fu, N. Optimization of quantitative modeling of starch in huangshui based on near-infrared spectral feature extraction using competitive adaptive reweighted sampling combined with successive projections algorithm. Food Sci. 2024, 45, 8–14. [Google Scholar]
- Ma, Z.L.; Wei, C.B.; Wang, W.H.; Lin, W.Q.; Nie, H.; Duan, Z.; Liu, K.; Xiao, X.O. Non-destructive prediction of anthocyanin concentration in whole eggplant peel using hyperspectral imaging. PeerJ 2024, 12, e17379. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Wang, X.C.; Zhang, X.L.; Sun, Y.; Sun, H.Y.; Wang, D.Z.; Xu, X. Spectral quantitative analysis and research of fusarium head blight infection degree in wheat canopy visible areas. Agronomy 2023, 13, 933. [Google Scholar] [CrossRef]
- Zhu, Y.X.; Yu, L.; Hong, Y.S.; Zhang, T.; Zhu, Q.; Li, S.D.; Guo, L.; Liu, J.S. Hyperspectral features and wavelength variables selection methods of soil organic matter. Sci. Agric. Sin. 2017, 50, 4325–4337. [Google Scholar]
- Gitelson, A.A.; Gritz, U.; Merzlyak, M.N. Relationships between leaf chlorophyll content and spectral reflectance and algorithms for non-destructive chlorophyll assessment in higher plant leaves. J. Plant Physiol. 2003, 160, 271–282. [Google Scholar] [CrossRef]
- Zhang, Z.P.; Ding, J.L.; Wang, J.Z.; Ge, X.Y.; Li, Z.S. Quantitative estimation of soil organic matter content using three-dimensional spectral index: A case study of the ebinur lake basin in xinjiang. Spectrosc. Spectral Anal. 2020, 40, 1514–1522. [Google Scholar]
- Nijat, K.; Zhang, Z.C.; Umut, H.; Zinhar, Z. Estimation of leaf water content of spring wheat based on 3d spectral index. J. Triticeae Crops 2024, 44, 522–531. [Google Scholar]
- Li, X.C.; Zhang, Y.J.; Bao, Y.S.; Luo, J.H.; Jin, X.L.; Xu, X.G.; Song, X.Y.; Yang, G.J. Exploring the best hyperspectral features for lai estimation using partial least squares regression. Remote Sens. 2014, 6, 6221–6241. [Google Scholar] [CrossRef]
- Yu, D.F.; Zhou, Y.; Xing, Q.G.; Ying, G.Y.; Zhou, B.; Fan, Y.G. Retrieval of secchi disk depth using modis satellite remote sensing and in situ observations in the yellow sea and the east china sea. Mar. Environ. Sci. 2016, 35, 774–779. [Google Scholar]
- Xue, J.K.; Shen, B. Dung beetle optimizer: A new meta-heuristic algorithm for global optimization. J. Supercomput. 2023, 79, 7305–7336. [Google Scholar] [CrossRef]
- Wu, C.L.; Fu, J.C.; Huang, X.; Xu, X.F.; Meng, J.H. Lithium-ion battery health state prediction based on vmd and dbo-svr. Energies 2023, 16, 3993. [Google Scholar] [CrossRef]
- Du, X.J.; Yao, Y.P.; Qian, Q. Measurement modeling of wastewater treatment process based on idbo-elm. Ship Electr. Eng. 2024, 44, 103–107. [Google Scholar]
- Trojovský, P.; Dehghani, M. Subtraction-average-based optimizer: A new swarm-inspired metaheuristic algorithm for solving optimization problems. Biomimetics 2023, 8, 149. [Google Scholar] [CrossRef]
- Liu, X.W.; Li, J.; Liu, J.; Huang, C.; Liu, L.L. Prediction of permanent deformation of subgrade soils under f-t cycles using sabo-optimized cnn-bilstm network. Case Stud. Constr. Mater. 2024, 21, e3807. [Google Scholar] [CrossRef]
- Lv, H.; Ling, W.; Zhu, Y.Z.; Du, W.L.; Liu, N.; Yang, D.M.; Cen, B.Y. prediction of power grid based on improved sabo-bp algorithm. Guangdong Electr. Power 2024, 37, 56–65. [Google Scholar]
- Zhang, J.; Zhang, H.; Gao, X. Clustering routing algorithm for wsns based on fuzzy logic optimized by whale optimization algorithm. Inf. Control 2023, 52, 797–810. [Google Scholar]
- Chen, Z.; Guo, Y.L.; Guo, C. Prediction of ghg emissions from chengdu metro in the construction stage based on woa-delm. Tunn. Undergr. Space Technol. 2023, 139, 105235. [Google Scholar] [CrossRef]
- Dou, L.; Zheng, W.; Li, B.Q.; Li, F. Study on wine quality evaluation based on extreme learning machine improved by whale optimization algorithm. Food Mach. 2024, 40, 62–68. [Google Scholar]
- Hao, R.; Wang, H.R.; Zhu, F.G. Indirect rul prediction of lithium-ion battery based on woa-delm. Control Instrum. Chem. Ind. 2023, 50, 37–43. [Google Scholar]
- Lu, X.Q.; Li, C.A.; Wu, Z.Q. Microgrid fault diagnosis based on extreme learning machine optimized by whale algorithm. Smart Power J. 2022, 50, 15–21. [Google Scholar]
- Yuan, S.Y. Prediction of grain yield model based on empirical mode decomposition and extreme learning machine. Comput. Mod. 2024, 3, 47–53. [Google Scholar]



| Treatments | Number of Samples | Med | Min | Max | Sd | Cv% | |
|---|---|---|---|---|---|---|---|
| Training Data | Supplementary Lighting Plan 1 | 90 | 0.106 | 0.062 | 0.153 | 0.027 | 25.9 |
| Supplementary Lighting Plan 2 | |||||||
| No Supplementary Lighting Plan | |||||||
| Testing data | Supplementary Lighting Plan 1 | 45 | 0.107 | 0.064 | 0.153 | 0.025 | 22.85 |
| Supplementary Lighting Plan 2 | |||||||
| No Supplementary Lighting Plan |
| Vegetation Index Type | Vegetation Index | Maximum Correlation Coefficient | Computational Formula | Wavelength Position | References |
|---|---|---|---|---|---|
| Two-band vegetation index | NARI | 0.7902 | (Ri−1 − Rj−1)/(Ri−1 + Rj−1) | (620 nm, 634 nm) | [53] |
| MGRVI | 0.7944 | (Ri2 − Rj2)/(Ri2 + Rj2) | (703 nm, 532 nm) | [54] | |
| ARI | 0.7676 | Ri−1 − Rj−1 | (532 nm, 697 nm) | [55] | |
| OSAVI | 0.7976 | 1.16 × (Ri − Rj)/(Ri + Rj + 0.16) | (563 nm, 558 nm) | [16] | |
| There-band vegetation index | MARI | 0.7636 | (Ri−1/Rj−1) × Rk | (542 nm, 966 nm, 720 nm) | [17] |
| EVI | 0.8345 | 2.5 × (Ri − Rj)/(Ri + 6 × Rj − 7.5 × Rk + 1) | (503 nm, 663 nm, 989 nm) | [56] | |
| TVI | 0.8217 | 0.5 × (120 × (Ri − Rj) − 200 ×(Rk − Rj)) | (526 nm, 560 nm, 532 nm) | [57] | |
| PSRI | 0.8193 | (Ri − Rj)/Rk | (664 nm, 503 nm, 989 nm) | [58] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Yu, H.; Liu, Y.; Zhang, L.; Li, D.; Zhang, J.; Li, X.; Sui, Y. Prediction of Anthocyanin Content in Purple-Leaf Lettuce Based on Spectral Features and Optimized Extreme Learning Machine Algorithm. Agronomy 2024, 14, 2915. https://doi.org/10.3390/agronomy14122915
Liu C, Yu H, Liu Y, Zhang L, Li D, Zhang J, Li X, Sui Y. Prediction of Anthocyanin Content in Purple-Leaf Lettuce Based on Spectral Features and Optimized Extreme Learning Machine Algorithm. Agronomy. 2024; 14(12):2915. https://doi.org/10.3390/agronomy14122915
Chicago/Turabian StyleLiu, Chunhui, Haiye Yu, Yucheng Liu, Lei Zhang, Dawei Li, Junhe Zhang, Xiaokai Li, and Yuanyuan Sui. 2024. "Prediction of Anthocyanin Content in Purple-Leaf Lettuce Based on Spectral Features and Optimized Extreme Learning Machine Algorithm" Agronomy 14, no. 12: 2915. https://doi.org/10.3390/agronomy14122915
APA StyleLiu, C., Yu, H., Liu, Y., Zhang, L., Li, D., Zhang, J., Li, X., & Sui, Y. (2024). Prediction of Anthocyanin Content in Purple-Leaf Lettuce Based on Spectral Features and Optimized Extreme Learning Machine Algorithm. Agronomy, 14(12), 2915. https://doi.org/10.3390/agronomy14122915






