Exploring the Physiological and Molecular Mechanisms by Which Potassium Regulates Low-Temperature Tolerance of Coconut (Cocos nucifera L.) Seedlings
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Determination of Dry Weight, Soil and Plant Analyzer Development Values, and K and Chlorophyll Contents
2.3. Determination of SS, SPr, Pro, and Malondialdehyde Contents; Enzyme Activities; and Endogenous Hormones
2.4. Transcriptomics Analysis
2.5. Metabolomics Analysis
2.6. Integrated Metabolome and Transcriptome Analyses
2.7. Quantitative Real-Time PCR
2.8. Statistical Analysis
3. Results
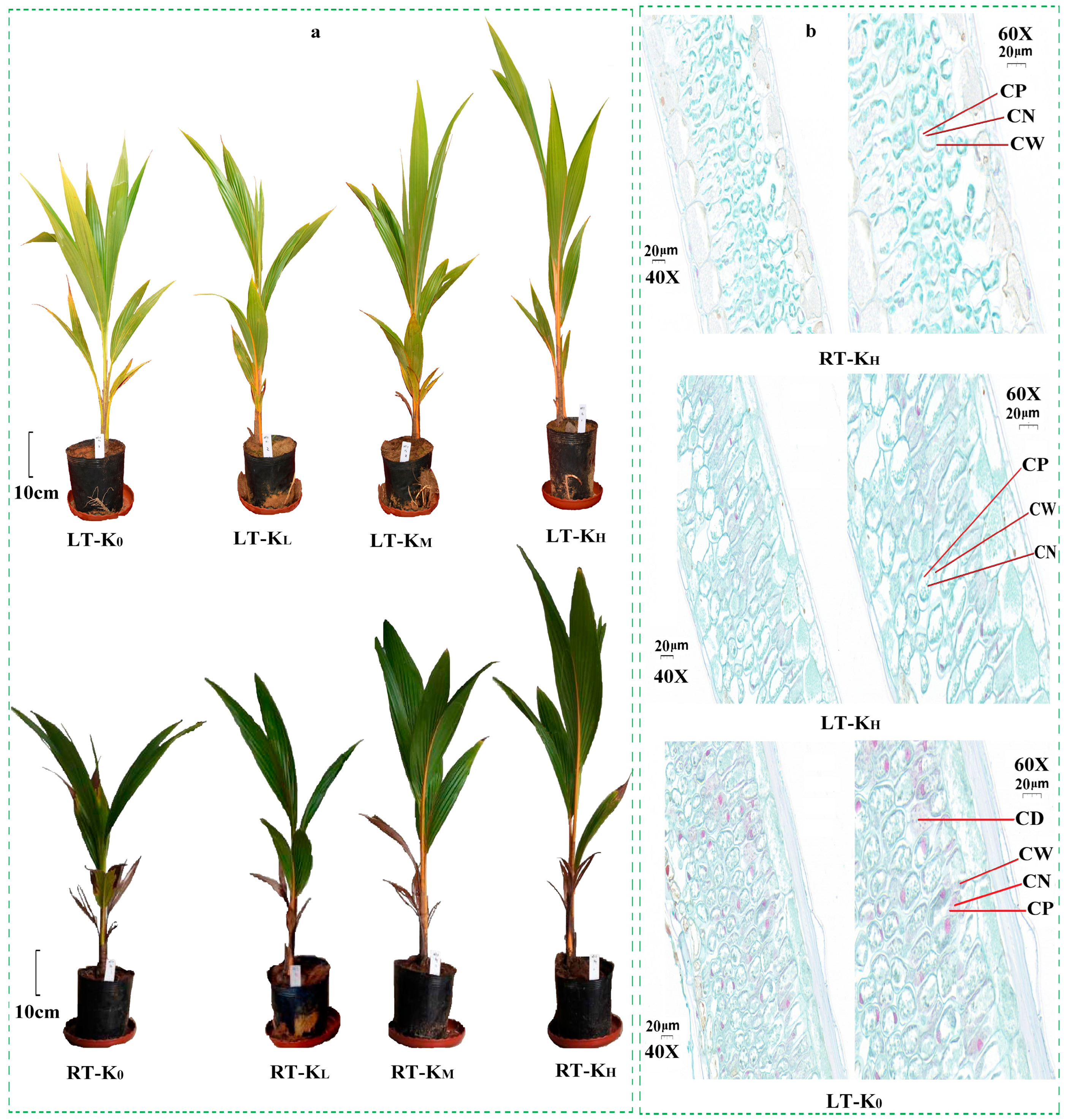
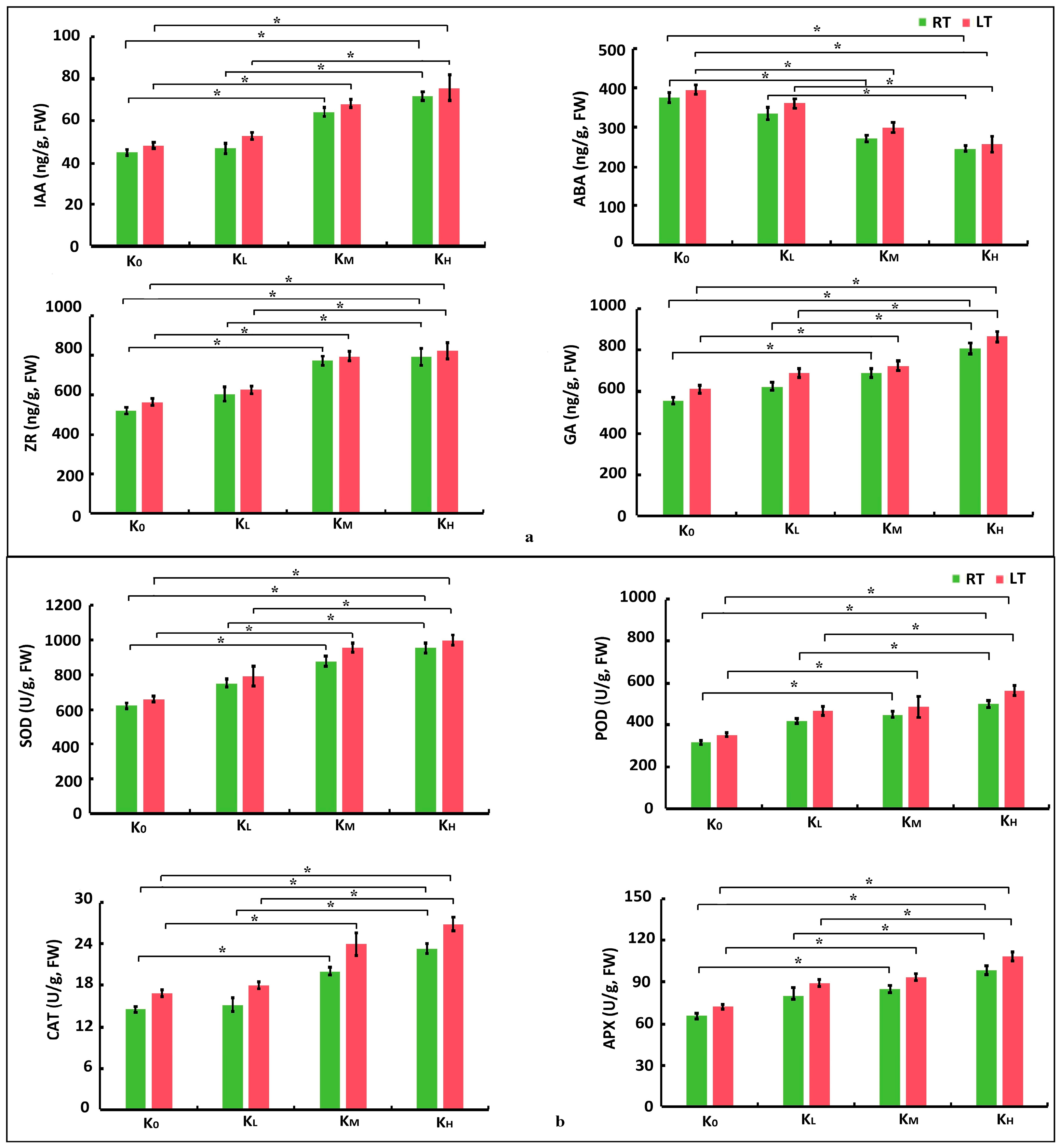
3.1. Morphological Traits and Physiological Indicators
3.2. Transcriptome Analysis
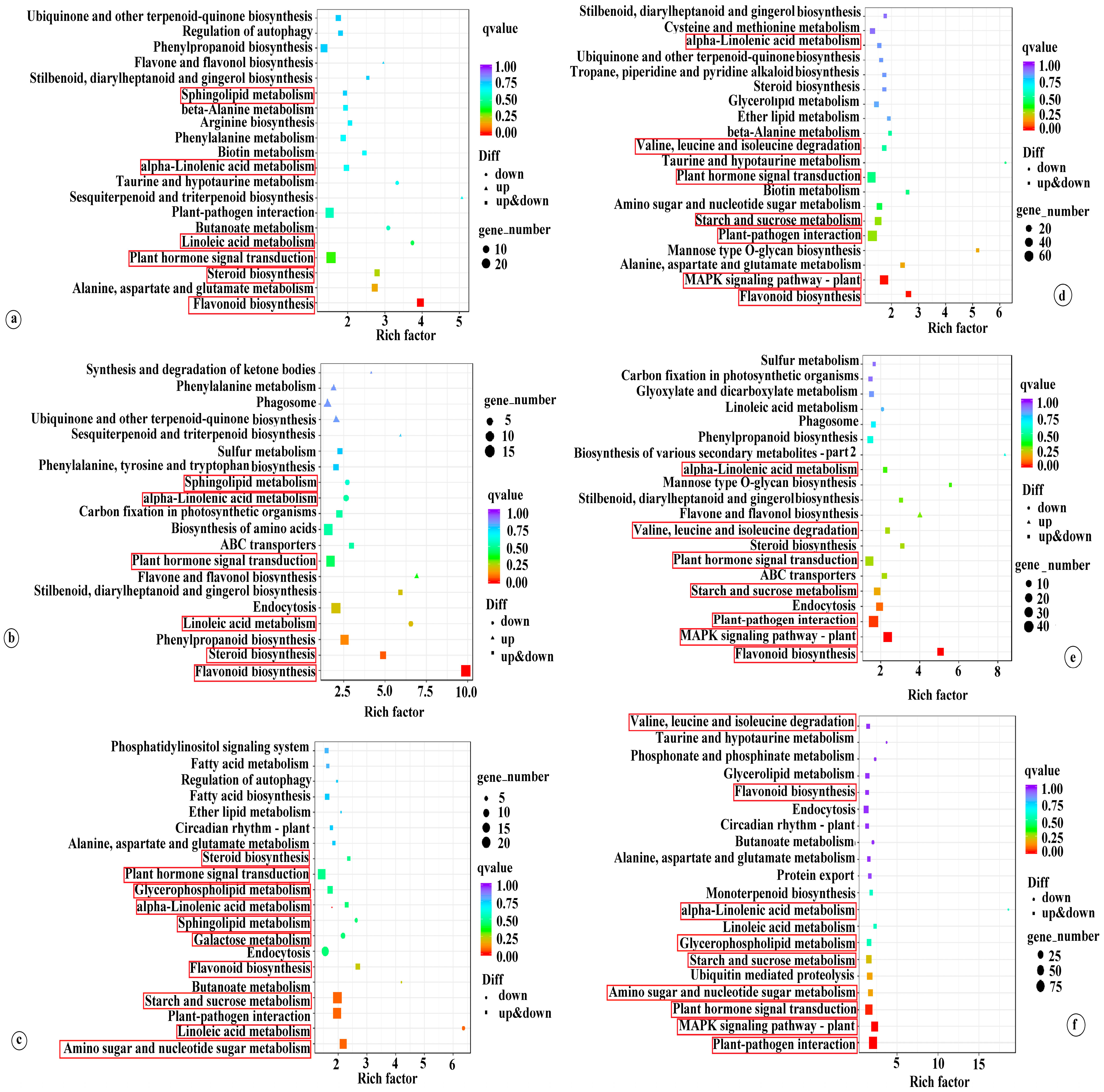
3.2.1. Plant Hormone Signal Transduction
3.2.2. Flavonoid Biosynthesis
3.2.3. Alpha-Linolenic Acid Metabolism
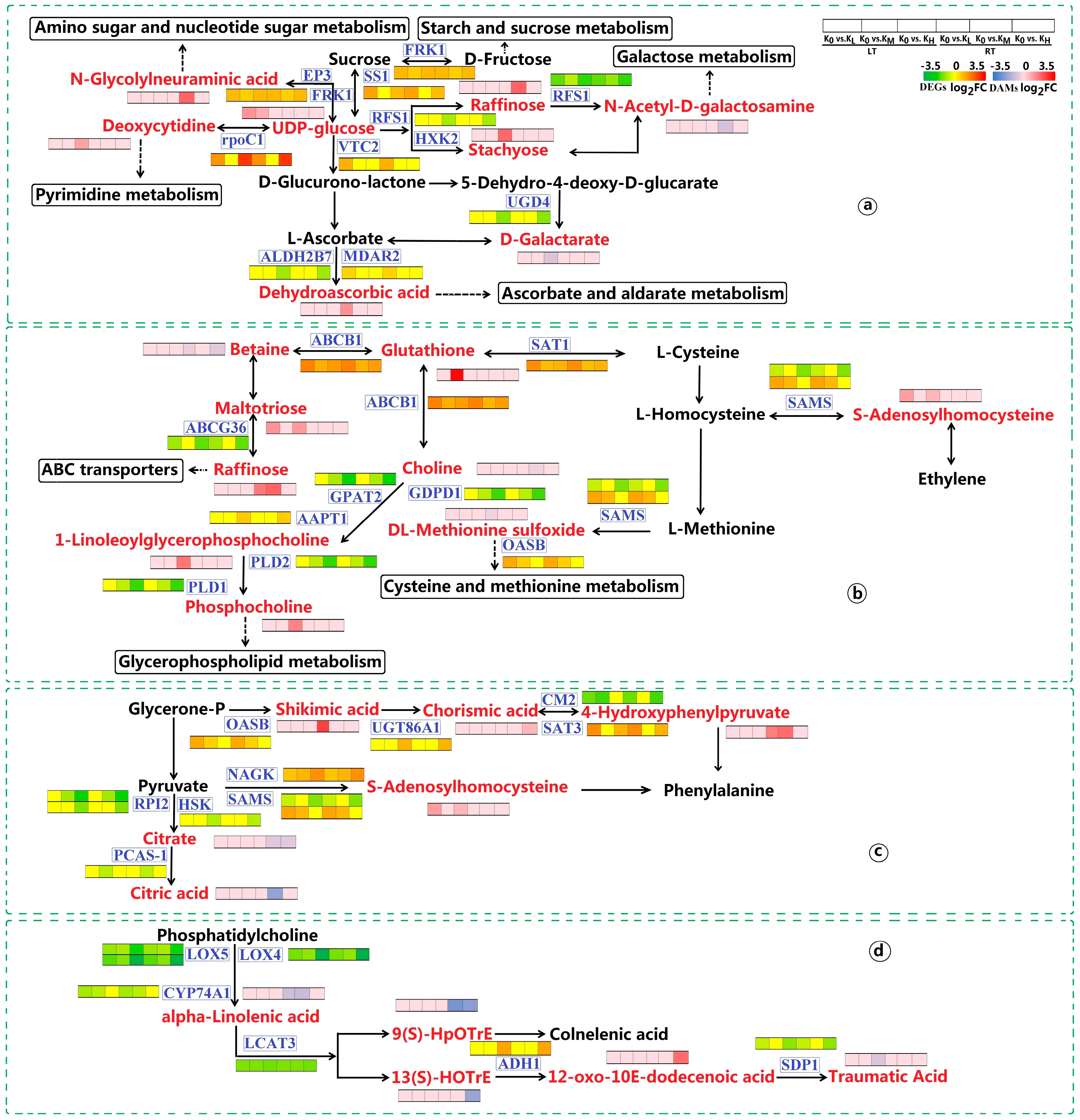
3.2.4. Starch and Sucrose Metabolism
3.2.5. Amino Sugar and Nucleotide Sugar and Glycerophospholipid Metabolism
3.2.6. Galactose Metabolism
3.2.7. Major Regulator Genes
3.2.8. TFs
3.2.9. qRT-PCR Verification
3.3. Metabolome Analysis
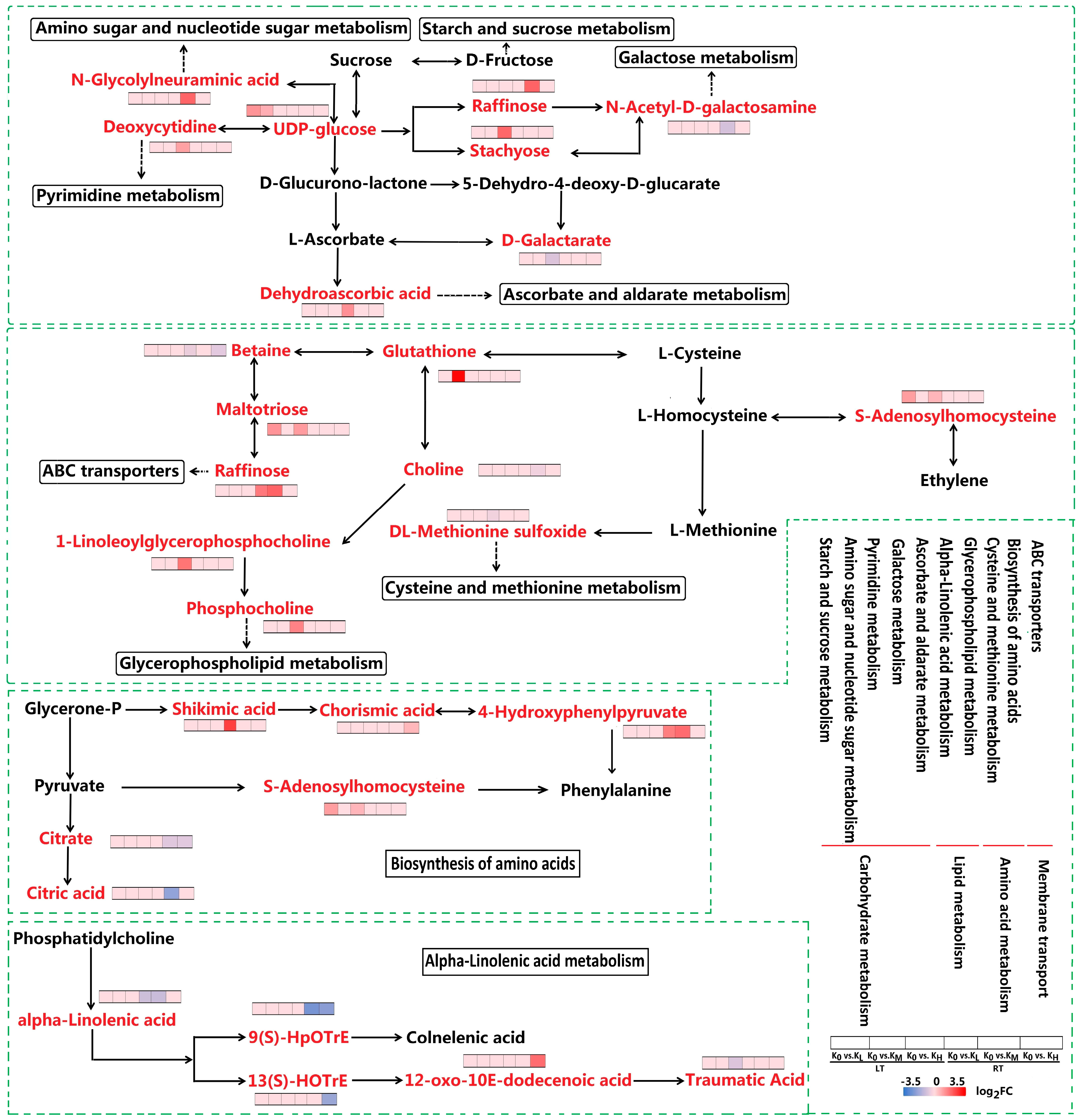
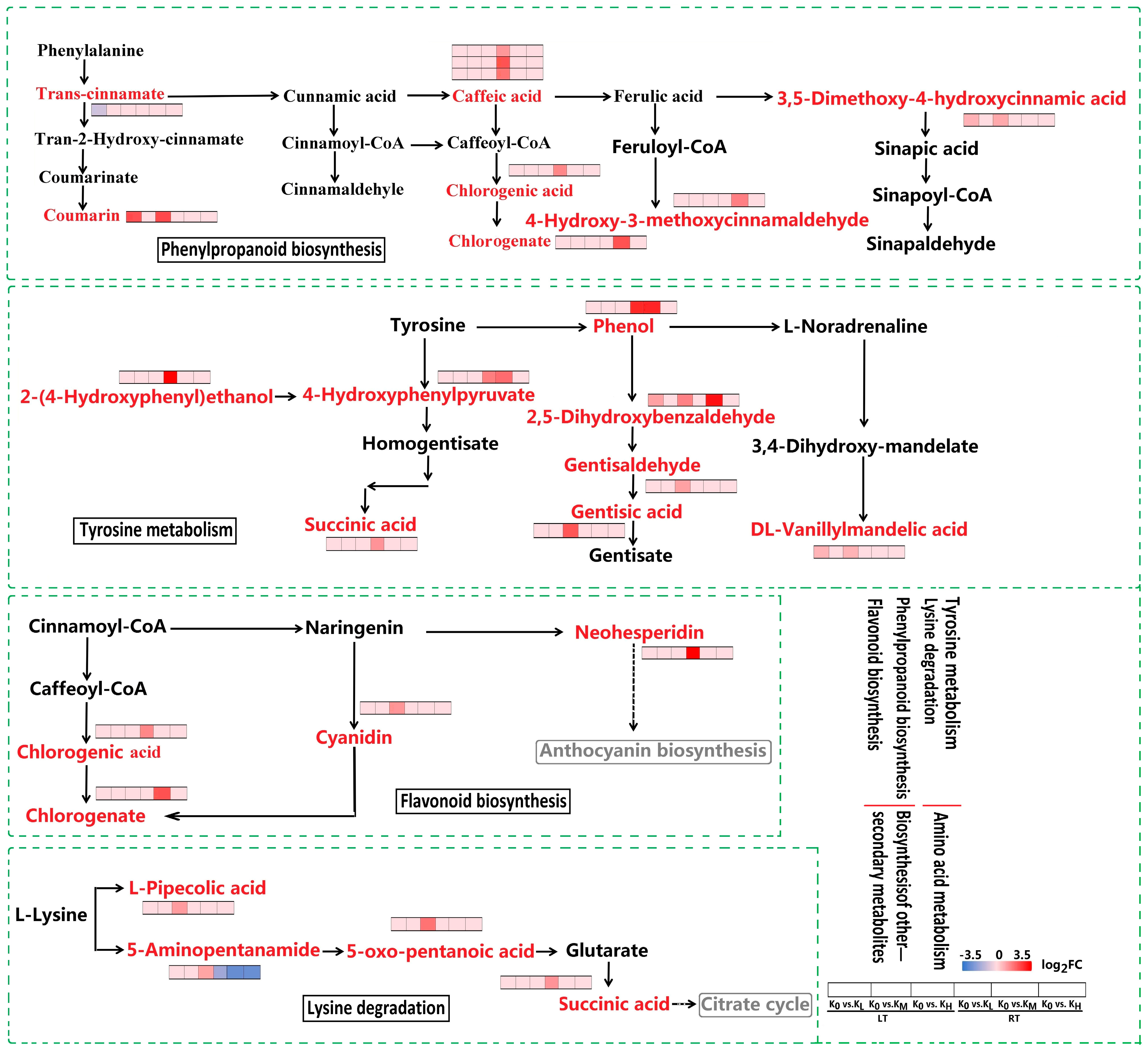
3.3.1. DAMs in Carbohydrate Metabolism
3.3.2. DAMs in Biosynthesis of Other Secondary Metabolites
3.3.3. DAMs in Amino Acid Metabolism
3.3.4. DAMs in Lipid Metabolism
3.3.5. DAMs in ABC Transporters
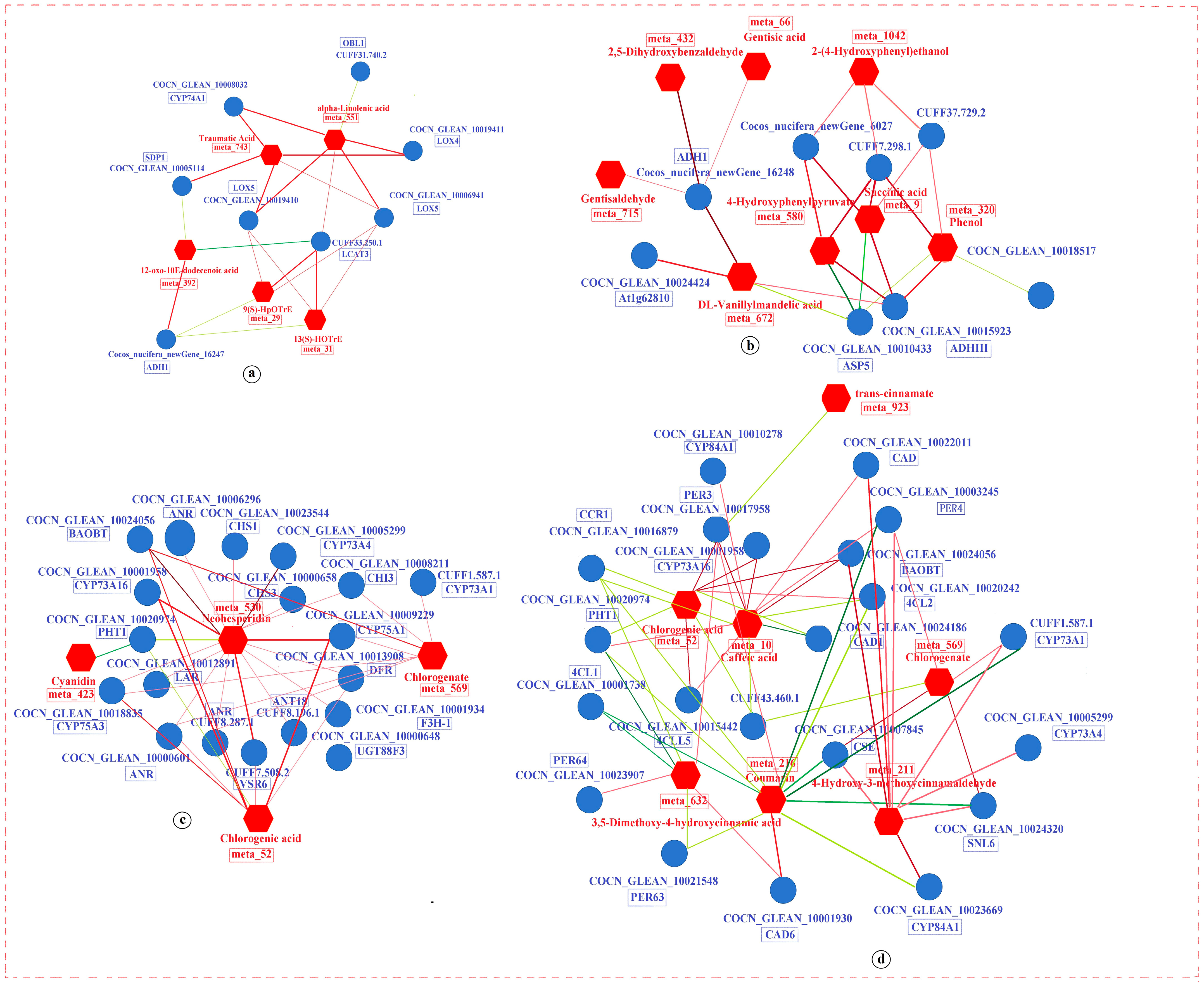
3.4. Integrative Analysis of the Transcriptome and Metabolome
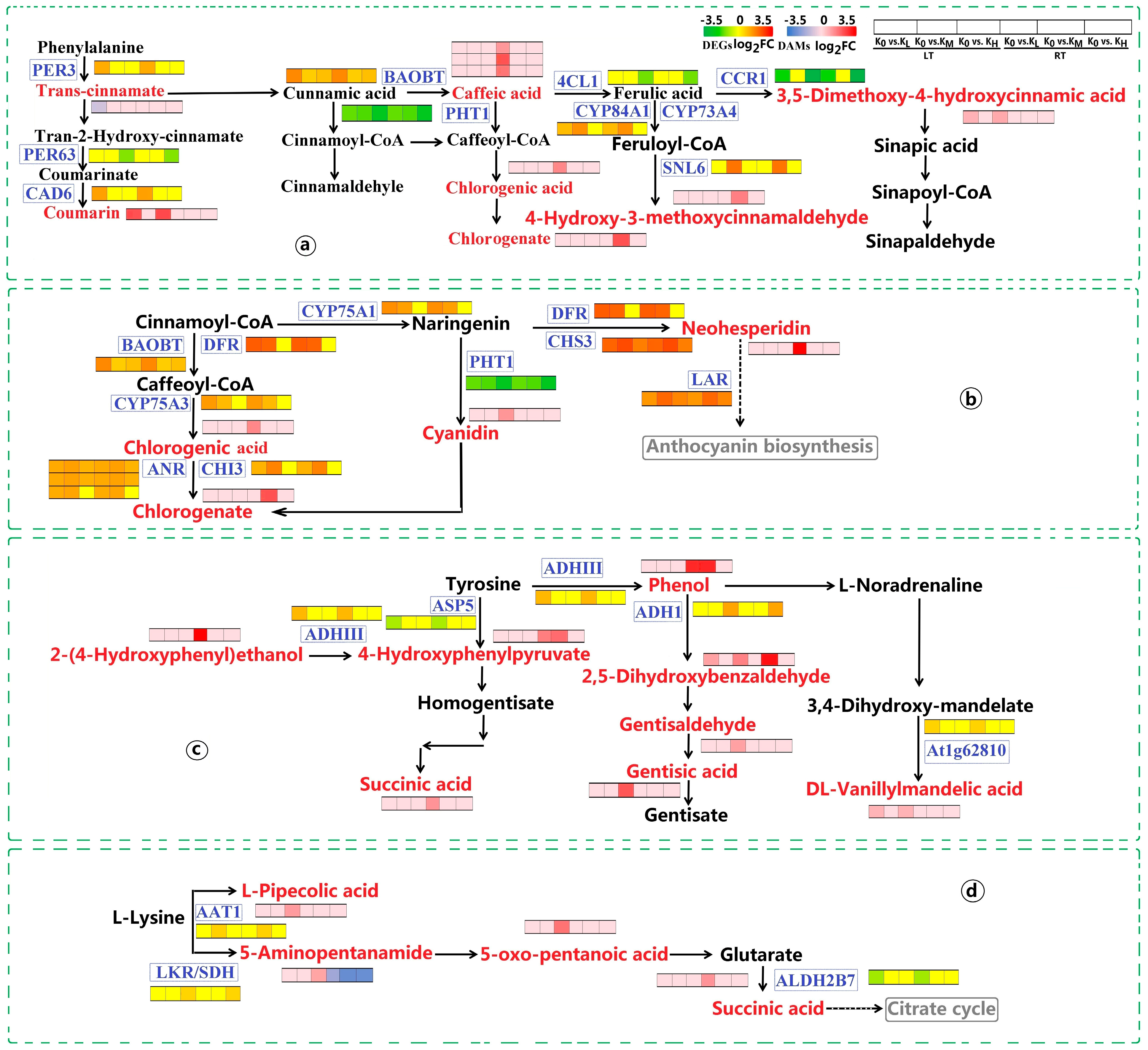
3.4.1. DEGs and DAMs in Carbohydrate Metabolism
3.4.2. DEGs and DAMs in Biosynthesis of Other Secondary Metabolites
3.4.3. DEGs and DAMs in Amino Acid Metabolism
3.4.4. DEGs and DAMs in Lipid Metabolism
3.4.5. DEGs and DAMs in ABC Transporters
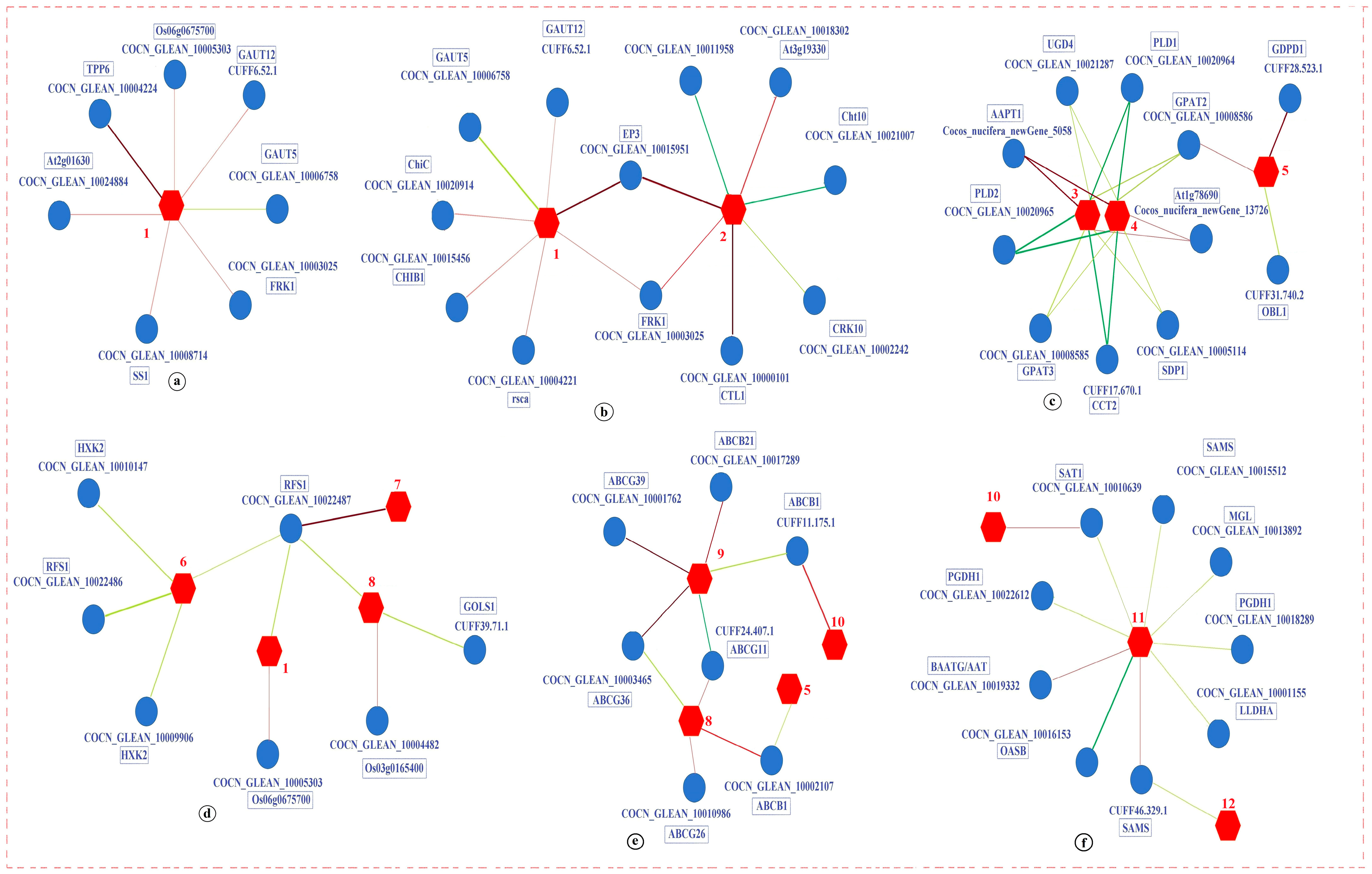
3.5. Network Regulation Diagram Based on the PCC Model
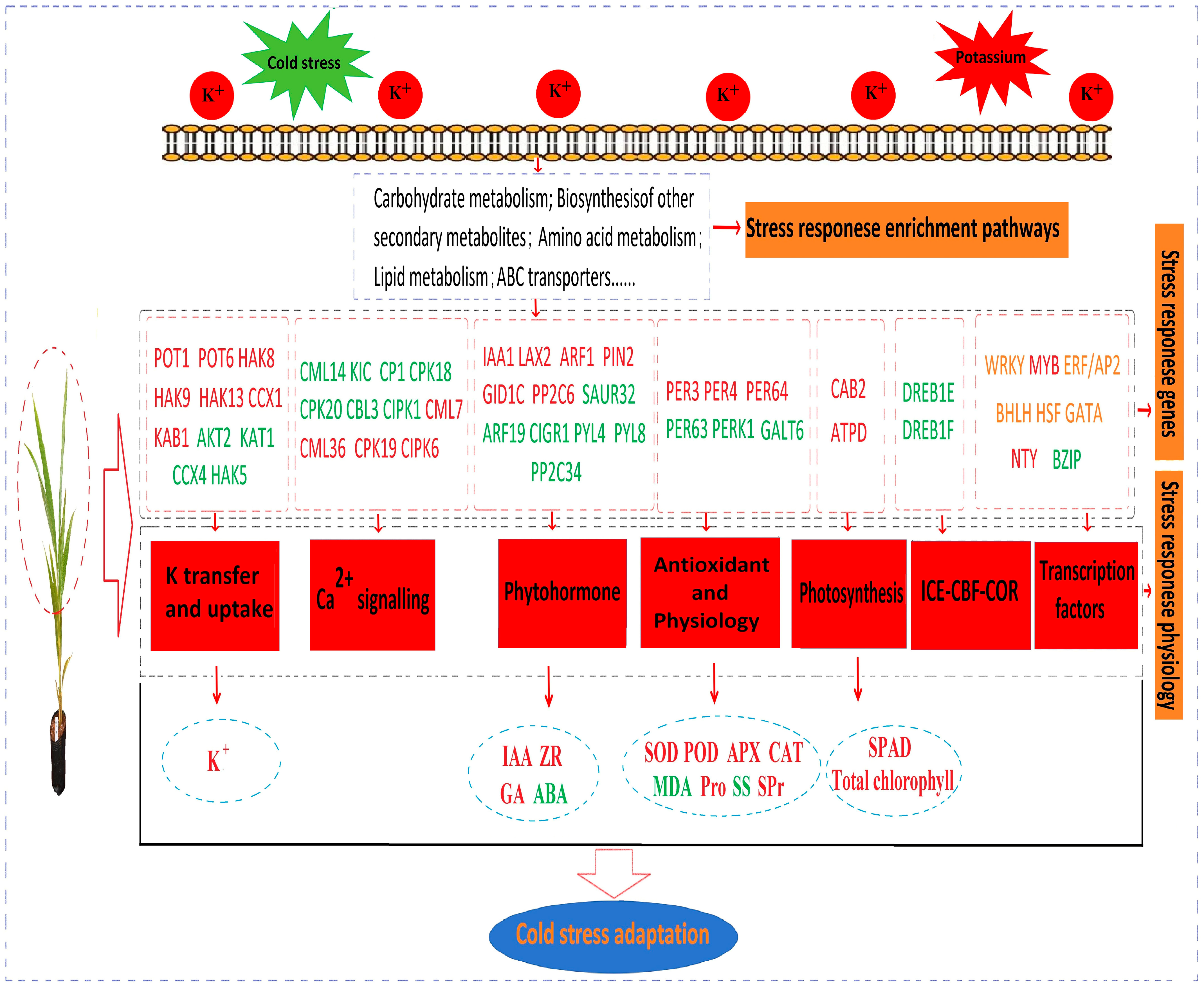
4. Discussion
4.1. K Can Accelerate the Clearance of ROS in Coconut Seedlings and Reduce LT Stress
4.2. K Can Regulate the Crucial Genes and TFs to Alleviate LT Stress Damage
4.3. K Activates Crucial Pathways Involved in Synthesis and Metabolism in Response to LT Stress
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhao, Y.; Zhou, M.; Xu, K.; Li, J.H.; Li, S.S.; Zhang, S.H.; Yang, X.J. Integrated transcriptomics and metabolomics analyses provide insights into cold stress response in wheat. Crop J. 2019, 7, 857–866. [Google Scholar] [CrossRef]
- Hu, Y.M.; Peng, X.J.; Wang, F.F.; Chen, P.L.; Zhao, M.L.; Shen, S.H. Natural population re-sequencing detects the genetic basis of local adaptation to low temperature in a woody plant. Plant Mol. Biol. 2021, 105, 585–599. [Google Scholar] [CrossRef] [PubMed]
- Sakina, A.; Wani, W.; Mushtaq, M.; Wani, S.H.; Shikari, A.B. Omics approaches for cold stress tolerance in plants. In Recent Approaches in Omics for Plant Resilience to Climate Change; Wani, S.H., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 331–356. [Google Scholar] [CrossRef]
- Mou, D.; Li, Z.R.; Liu, W.H.; Liang, G.L.; Jia, Z.F.; Yu, H.Y.; Wang, J.L.; Ou, W.Y.; Liu, K.; Yao, X.X.; et al. Integrated transcriptomic and metabolomic analyses of Caucasian clover (Trifolium ambiguum Bieb.) in response to freezing stress. Braz. J. Bot. 2022, 45, 573–585. [Google Scholar] [CrossRef]
- Ma, L.; Coulter, J.A.; Liu, L.J.; Zhao, Y.H.; Chang, Y.; Pu, Y.Y.; Zeng, X.C.; Xu, Y.Z.; Wu, J.Y.; Fang, Y.; et al. Transcriptome analysis reveals key cold-stress-responsive genes in winter rapeseed (Brassica rapa L.). Int. J. Mol. Sci. 2019, 20, 1071. [Google Scholar] [CrossRef]
- Kapoor, D.; Singh, S.; Kumar, V.; Romero, R.; Prasad, R.; Singh, J. Antioxidant enzymes regulation in plants in reference to reactive oxygen species (ROS) and reactive nitrogen species (RNS). Plant Gene 2019, 19, 100182. [Google Scholar] [CrossRef]
- Fürtauer, L.; Weiszmann, J.; Weckwerth, W.; Nägele, T. Dynamics of plant metabolism during cold acclimation. Int. J. Mol. Sci. 2019, 20, 5411. [Google Scholar] [CrossRef]
- Lu, L.; Yang, W.; Dong, Z.; Tang, L.; Liu, Y.; Xie, S.; Yang, Y. Integrated Transcriptomic and Metabolomics Analyses Reveal Molecular Responses to Cold Stress in Coconut (Cocos nucifera L.) Seedlings. Int. J. Mol. Sci. 2023, 24, 14563. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.D.; Li, W.Y.; Ye, J.L.; Sun, X.H.; Ding, Y.D.; Cheng, Y.J.; Deng, X.X. Microarray expression profiling of postharvest ponkan mandarin (Citrus reticulata) fruit under cold storage reveals regulatory gene candidates and implications on soluble sugars metabolism. J. Int. Plant Biol. 2011, 53, 358–374. [Google Scholar] [CrossRef]
- Halman, J.M.; Schaberg, P.G.; Hawley, G.J.; Eagar, C. Calcium addition at the Hubbard Brook Experimental Forest increases sugar storage, antioxidant activity and cold tolerance in native red spruce (Picea rubens). Tree Physiol. 2008, 28, 855–862. [Google Scholar] [CrossRef]
- Zhang, J.; Liang, L.; Xie, Y.; Zhao, Z.; Su, L.; Tang, Y.; Sun, B.; Lai, Y.; Li, H. Transcriptome and Metabolome Analyses Reveal Molecular Responses of Two Pepper (Capsicum annuum L.) Cultivars to Cold Stress. Front. Plant Sci. 2022, 13, 819630. [Google Scholar] [CrossRef]
- Zhang, W.J.; Wang, J.Q.; Huang, Z.L.; Mi, L.; Xu, K.F.; Wu, J.J.; Fan, Y.H.; Ma, S.Y.; Jiang, D.G. Effects of low temperature at booting stage on sucrose metabolism and endogenous hormone contents in winter wheat Spikelet. Front. Plant Sci. 2019, 10, 498. [Google Scholar] [CrossRef]
- Yang, Y.N.; Yao, Z.K.; Jia, J.; Duan, F.J.; Chen, Z.Y.; Sang, L.Y. Effect of exogenous abscisic acid on cold acclimation in two Magnolia species. Biol. Plant. 2016, 60, 555–562. [Google Scholar] [CrossRef]
- Xu, G.X.; Li, L.J.; Zhou, J.; Lyu, D.G.; Zhao, D.Y.; Qin, S.J. Comparison of transcriptome and metabolome analysis revealed differences in cold resistant metabolic pathways in different apple cultivars under low temperature stress. Hortic. Plant J. 2023, 9, 183–198. [Google Scholar] [CrossRef]
- Paterlini, A. Uncharted routes: Exploring the relevance of auxin movement via plasmodesmata. Biol. Open 2020, 9, bio055541. [Google Scholar] [CrossRef]
- Taïbi, K.; Campo, A.D.D.; Vilagrosa, A.; Bellés, J.M.; López-Gresa, M.P.; López-Nicolás, J.M.; Mulet, J.M. Distinctive physiological and molecular responses to cold stress among cold-tolerant and cold-sensitive Pinus halepensis seed sources. BMC Plant Biol. 2018, 18, 236. [Google Scholar] [CrossRef]
- Suzuki, N. Temperature stress and responses in plants. Int. J. Mol. Sci. 2019, 20, 2001. [Google Scholar] [CrossRef] [PubMed]
- Yuan, P.; Yang, T.; Poovaiah, B.W. Calcium signaling-mediated plant response to cold stress. Int. J. Mol. Sci. 2018, 19, 3896. [Google Scholar] [CrossRef]
- Kim, Y.S.; Lee, M.; Lee, J.H.; Lee, H.J.; Park, C.M. The unified ICE-CBF pathway provides a transcriptional feedback control of freezing tolerance during cold acclimation in Arabidopsis. Plant Mol. Biol. 2015, 89, 187–201. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Zhao, L.; Ren, Y.; Yang, S.; Zhu, J.K.; Zhao, C. The transcription factor ICE1 functions in cold stress response by binding to the promoters of CBF and COR genes. J. Integr. Plant Biol. 2020, 62, 258–263. [Google Scholar] [CrossRef] [PubMed]
- Dong, B.; Zheng, Z.; Zhong, S.; Ye, Y.; Wang, Y.; Yang, L.; Xiao, Z.; Fang, Q.; Zhao, H. Integrated Transcriptome and Metabolome Analysis of Color Change and Low-Temperature Response during Flowering of Prunus mume. Int. J. Mol.Sci. 2022, 23, 12831. [Google Scholar] [CrossRef]
- Sun, S.; Fang, J.; Lin, M.; Hu, C.; Qi, X.; Chen, J.; Zhong, Y.; Muhammad, A.; Li, Z.; Li, Y. Comparative Metabolomic and Transcriptomic Studies Reveal Key Metabolism Pathways Contributing to Freezing Tolerance Under Cold Stress in Kiwifruit. Front. Plant Sci. 2021, 12, 628969. [Google Scholar] [CrossRef]
- Wang, J.; Guo, J.P.; Zhang, Y.X.; Yan, X.R. Integrated transcriptomic and metabolomic analyses of yellow horn (Xanthoceras sorbifolia) in response to cold stress. PLoS ONE 2020, 15, e0236588. [Google Scholar] [CrossRef]
- Raza, A.; Su, W.; Hussain, M.A.; Mehmood, S.S.; Zhang, X.; Cheng, Y.; Zou, X.; Lv, Y. Integrated Analysis of Metabolome and Transcriptome Reveals Insights for Cold Tolerance in Rapeseed (Brassica napus L.). Front. Plant Sci. 2021, 12, 721681. [Google Scholar] [CrossRef]
- Gao, C.; Mumtaz, M.A.; Zhou, Y.; Yang, Z.; Shu, H.; Zhu, J.; Bao, W.; Cheng, S.; Yin, L.; Huang, J.; et al. Integrated Transcriptomic and Metabolomic Analyses of Cold-Tolerant and Cold-Sensitive Pepper Species Reveal Key Genes and Essential Metabolic Pathways Involved in Response to Cold Stress. Int. J. Mol. Sci. 2022, 23, 6683. [Google Scholar] [CrossRef]
- Cheng, Y.; Ban, Q.; Mao, J.; Lin, M.; Zhu, X.; Xia, Y.; Cao, X.; Zhang, X.; Li, Y. Integrated Metabolomic and Transcriptomic Analysis Reveals That Amino Acid Biosynthesis May Determine Differences in Cold-Tolerant and Cold-Sensitive Tea Cultivars. Int. J. Mol. Sci. 2023, 24, 1907. [Google Scholar] [CrossRef]
- Guo, Y.G.; Wang, C.Y.; Fu, M.K.; El-Kassaby, F.F.; Wang, Y.A.; Wang, T.L.; Gui, B. Metabolome and transcriptome analyses reveal flavonoids biosynthesis differences in ginkgo biloba associated with environmental conditions. Ind. Crops Prod. 2020, 158, 112963. [Google Scholar] [CrossRef]
- Mehmood, S.S.; Lu, G.; Luo, D.; Hussain, M.A.; Raza, A.; Zafar, Z.; Zhang, X.K.; Cheng, Y.; Zou, X.L.; Lv, Y. Integrated analysis of transcriptomics and proteomics provides insights into the molecular regulation of cold response in Brassica napus. Environ. Exp. Bot. 2021, 187, 104480. [Google Scholar] [CrossRef]
- Sanyal, S.K.; Rajasheker, G.; Kishor, P.K.; Kumar, S.A.; Kumari, P.H.; Saritha, K.V.; Rathnagiri, P.; Pandey, G.K. Role of protein phosphatases in signaling, potassium transport, and abiotic stress responses. In Protein Phosphatases and Stress Management in Plants; Pandey, G.K., Ed.; Springer: Cham, Switzerland, 2020; pp. 203–232. [Google Scholar] [CrossRef]
- Sardans, J.; Peñuelas, J. Potassium control of plant functions: Ecological and agricultural implications. Plants 2021, 10, 419. [Google Scholar] [CrossRef] [PubMed]
- Vašák, M.; Schnabl, J. Sodium and potassium ions in proteins and enzyme catalysis. Metal. Ions Life Sci. 2016, 16, 259. [Google Scholar] [CrossRef]
- Coskun, D.; Britto, D.T.; Kronzucker, H. The nitrogen-potassium intersection: Membranes, metabolism, and mechanism. Plant Cell Environ. 2017, 40, 2029–2041. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Bhuyan, M.H.M.; Nahar, K.; Hossain, M.S.; Mahmud, J.A.; Hossen, M.S.; Masud, A.A.C.; Moumita Fujita, M. Potassium: A vital regulator of plant responses and tolerance to abiotic stresses. Agronomy 2018, 8, 31. [Google Scholar] [CrossRef]
- Hosseini, S.A.; Maillard, A.; Hajirezaei, M.R.; Ali, N.; Schwarzenberg, A.; Jamois, F.; Yvin, J.C. Induction of Barley Silicon Transporter HvLsi1 and HvLsi2, increased silicon concentration in the shoot and regulated Starch and ABA Homeostasis under Osmotic stress and Concomitant Potassium Deficiency. Front. Plant Sci. 2017, 8, 1359. [Google Scholar] [CrossRef] [PubMed]
- Karimi, R. Potassium-induced freezing tolerance is associated with endogenous abscisic acid, polyamines and soluble sugars changes in grapevine. Sci. Hortic. 2017, 215, 184–194. [Google Scholar] [CrossRef]
- Ho, L.H.; Rode, R.; Siegel, M.; Reinhardt, F.; Neuhaus, H.E.; Yvin, J.C.; Pluchon, S.; Hosseini, S.A.; Pommerrenig, B. Potassium application boosts photosynthesis and sorbitol biosynthesis and accelerates cold acclimation of common plantain (Plantago major L.). Plants 2020, 9, 1259. [Google Scholar] [CrossRef] [PubMed]
- Maheswarappa, H.P.; Thomas, G.V.; Gupta, A.; Bhat, R.; Palaniswami, C. Productivity and nutrient status of coconut (Cocos nucifera) as influenced by integrated nutrient management with vermicomposted coconut leaves. Indian J. Agron. 2014, 59, 455–459. [Google Scholar]
- Kalpana, M.; Gautam, B.; Srinivasulu, B.; Rao, D.V.R.; Arulraj, S.; Jayabose, C. Impact of integrated nutrient management on nut yield and quality of coconut under coastal ecosystem. J. Plant Crops 2008, 36, 249–253. [Google Scholar]
- Lu, L.; Chen, S.; Yang, W.; Wu, Y.; Liu, Y.; Yin, X.; Yang, Y.; Yang, Y. Integrated transcriptomic and metabolomic analyses reveal key metabolic pathways in response to potassium deficiency in coconut (Cocos nucifera L.) seedlings. Front. Plant Sci. 2023, 14, 1112264. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.X.; Liu, L.Y.; Feng, M.L. Development of coconut industry in the world. Acta Agric. Jiangxi 2016, 11, 159–165. (In Chinese) [Google Scholar]
- Lu, L.L.; Liu, R.; Xiao, Y.; Li, J.; Shen, X.J.; Li, H.S.; Fan, H.K. Research Progress on Coconut Germplasm Resources, Cultivation and Utilization. Chin. J. Trop. Crops 2021, 42, 9. (In Chinese) [Google Scholar]
- Peamprasart, T.; Chiewchan, N. Effect of fat content and preheat treatment on the apparent viscosity of coconut milk after homogenization. J. Food Eng. 2006, 77, 653–658. [Google Scholar] [CrossRef]
- Zhao, S.L.; Cao, H.X. Collection, Conservation, Identification, Evaluation and Innovative Utilization of Coconut Germplasm Resources, 1st ed.; China Agriculture Press: Beijing, China, 2012; pp. 112–116. [Google Scholar]
- Cao, H.; Huang, H.; Lei, X.; Zhang, D.; Zhang, R. Difference of the leaf anatomical structure of coconut varieties under low temperature treatments. Chin. J. Trop. Crops 2014, 35, 2420–2425. (In Chinese) [Google Scholar]
- Yang, Y.; Iqbal, A.; Qadri, R. Breeding of coconut (Cocos nucifera L.): The tree of life. In Advances in Plant Breeding Strategies: Fruits; Al-Khayri, J., Jain, S., Johnson, D., Eds.; Springer: Cham, Switzerland, 2018; pp. 673–725. [Google Scholar]
- Cao, H.X.; Lei, X.T.; Liu, Y.J.; Sun, C.X.; Zhang, R.L. Identification and comprehensive evaluation of cold resistance indexes of coconut. Guangdong Agric. Sci. 2016, 2, 49–54. (In Chinese) [Google Scholar] [CrossRef]
- Yang, Y.D.; Mumtaz, A.S.; Walid, B.A.; Zhang, J.; Wu, Y.; Li, J.; Fan, H.K.; Wang, F.Y. iTRAQ-based comparative proteomic analysis of two coconut varieties reveals aromatic coconut cold-sensitive in response to low temperature. J. Proteom. 2020, 220, 103766. [Google Scholar] [CrossRef]
- Li, J.; Wang, F.; Sayed, M.A.; Shen, X.; Zhou, L.; Liu, X.; Sun, X.; Chen, S.; Wu, Y.; Lu, L.; et al. Integrated transcriptomic and metabolomic data reveal the cold stress responses molecular mechanisms of two coconut varieties. Front. Plant Sci. 2024, 15, 1353352. [Google Scholar] [CrossRef]
- Brunin, C.; He, G.L. Diagnosis of coconut leaves-world tropical agriculture information. World Trop. Agric. Inform. 1965, 5, 58–60. [Google Scholar]
- Hoagland, D.R.; Arnon, D.I. The water-culture method for growing plants without soil. Calif. Agric. Exp. Stn. Circ. 1950, 347, 1–32. [Google Scholar]
- Wang, Y.J.; Hu, X.; Jin, G.; Hou, Z.W.; Ning, J.M.; Zhang, Z.Z. Rapid prediction of chlorophylls and carotenoids content in tea leaves under different levels of nitrogen application based on hyperspectral imaging. J. Sci. Food Agric. 2019, 99, 1997–2004. [Google Scholar] [CrossRef]
- Bao, S.D. Agrochemical Analysis of Soil, 3rd ed.; Agricultural Publishing House: Beijing, China, 2000; pp. 23–56. (In Chinese) [Google Scholar]
- Li, H.S. Principles and Techniques of Plant Physiology and Biochemistry Experiment; Higher Education Press: Beijing, China, 2000; pp. 25–63. [Google Scholar]
- Xiao, Y.; Xu, P.X.; Fan, H.K.; Luc, B.; Xia, W.; Stephanie, B.; Xu, J.Y.; Li, Q.; Guo, A.P.; Zhou, L.X.; et al. The genome draft of coconut (Cocos nucifera). GigaSci 2017, 6, 1–17. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Meth. 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A. The COG database: A tool for genome scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef]
- Finn, R.D.; Alex, B.; Jody, C.; Penelope, C.; Eberhardt, R.Y.; Eddy, S.R.; Andreas, H.; Kirstie, H.; Liisa, H.; Jaina, M.; et al. Pfam: The protein families database. Nucleic Acids Res. 2013, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinf. 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Alexa, A.; Rahnenfuhrer, J. topGO: Enrichment Analysis for Gene Ontology; R Package Version 2.8; The Pennsylvania State University: University Park, PA, USA, 2010. [Google Scholar]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real time quantitative PCR and the 2-DDCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.R.; Yan, W.J.; Yang, C.K.; Huang, X.B.; Hu, X.T.; Li, Y.Y.; Yang, J.S.; Xiang, S.P.; Yi, P.F.; Hu, R.S. Integrative analysis of transcriptome and metabolome provides insights into the underlying mechanism of cold stress response and recovery in two tobacco cultivars. Environ. Exp. Bot. 2022, 200, 104920. [Google Scholar] [CrossRef]
- Li, H.; Wang, H.; Wen, W.J.; Yang, G.W. The antioxidant system in Suaeda salsa under salt stress. Plant Signal. Behav. 2020, 15, 1771939. [Google Scholar] [CrossRef]
- Ju, F.; Pang, J.; Sun, L.; Gu, J.; Wang, Z.; Wu, X.; Ali, S.; Wang, Y.; Zhao, W.; Wang, S.; et al. Integrative transcriptomic, metabolomic and physiological analyses revealed the physiological and molecular mechanisms by which potassium regulates the salt tolerance of cotton (Gossypium hirsutum L.) roots. Ind. Crops Prod. 2023, 193, 116177. [Google Scholar] [CrossRef]
- Ahanger, M.A.; Agarwal, R.M. Salinity stress induced alterations in antioxidant metabolism and nitrogen assimilation in wheat (Triticum aestivum L.) as influenced by potassium supplementation. Plant Physiol. Biochem. 2017, 115, 449–460. [Google Scholar] [CrossRef]
- Ahanger, M.A.; Tomar, N.S.; Tittal, M.; Argal, S.; Agarwal, R.M. Plant growth under water/salt stress: ROS production; antioxidants and signifcance of added potassium under such conditions. Physiol. Mol. Biol. Plants 2017, 23, 731–744. [Google Scholar] [CrossRef]
- George, I.S.; Pascovici, D.; Mirzaei, M.; Haynes, P.A. Quantitative proteomic analysis of cabernet sauvignon grape cells exposed to thermal stresses reveals alterations in sugar and phenylpropanoid metabolism. Proteomics 2015, 15, 3048–3060. [Google Scholar] [CrossRef]
- Wang, D.Z.; Jin, Y.N.; Ding, X.H.; Wang, W.J.; Zhai, S.S.; Bai, L.P.; Guo, Z.F. Gene regulation and signal transduction in the ICE–CBF–COR signaling pathway during cold stress in plants. Biochemistry 2017, 82, 1103–1117. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Chen, X.; Dong, S.; Jiang, X.; Wang, L.; Yu, J.; Zhou, Y. Crosstalk of PIF4 and DELLA modulates CBF transcript and hormone homeostasis in cold response in tomato. Plant Biotechnol. J. 2020, 18, 1041–1055. [Google Scholar] [CrossRef]
- Guo, X.Y.; Liu, D.F.; Chong, K. Cold signaling in plants: Insights into mechanisms and regulation. J. Integr. Plant Biol. 2018, 60, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.L.; Yan, J.Y.; Su, Z.Z.; Chang, J.J.; Yang, J.Q.; Wei, C.H.; Zhang, Y.; Ma, J.X.; Zhang, X.; Li, H. Abscisic acid mediates grafting-induced cold tolerance of watermelon via interaction with melatonin and methyl jasmonate. Front. Plant Sci. 2021, 12, 785317. [Google Scholar] [CrossRef]
- Wang, Y.X.; Hu, Y.; Chen, B.H.; Zhu, Y.F.; Dawuda, M.M.; Svetla, S. Physiological mechanisms of resistance to cold stress associated with 10 elite apple rootstocks. J. Integr. Agric. 2018, 17, 857–866. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Lin, Y.; Luo, Y.; Wang, X.; Chen, Q.; Sun, B.; Wang, Y.; Li, M.; Tang, H. A transcriptomic analysis reveals diverse regulatory networks that respond to cold stress in strawberry (Fragaria × ananassa). Int. J. Genom. 2019, 2019, 7106092. [Google Scholar] [CrossRef]
- Sazegari, S.; Niazi, A.; Ahmadi, F.S.A. Study on the regulatory network with promoter analysis for Arabidopsis DREB-genes. Bioinformation 2015, 11, 101. [Google Scholar] [CrossRef]
- Vazquez, M.; Romero, I.; Escribano, M.I.; Merodio, C.; Sanchez-Ballesta, M.T. Deciphering the Role of CBF/DREB Transcription Factorsand Dehydrins in Maintaining the Quality of Table Grapes cv. Autumn Royal Treated with High CO2 Levels and Stored at 0 °C. Front. Plant Sci. 2017, 8, 1591. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Pandit, E.; Nayak, D.K.; Behera, L.; Mohapatra, T. Genes, pathways and transcription factors involved in seedling stage chilling stress tolerance in indica rice through RNA-Seq analysis. BMC Plant Biol. 2019, 19, 352. [Google Scholar] [CrossRef]
- Bao, F.; Ding, A.; Cheng, T.; Wang, J.; Zhang, Q. Genome-wide analysis of members of the WRKY gene family and their cold stress response in Prunus mume. Genes 2019, 10, 911. [Google Scholar] [CrossRef]
- Zhang, C.Y.; Liu, H.C.; Zhang, X.S.; Guo, Q.X.; Bian, S.M.; Wang, J.Y.; Zhai, L.L. VcMYB4a, an R2R3-MYB transcription factor from Vaccinium corymbosum, negatively regulates salt, drought, and temperature stress. Gene 2020, 757, 144935. [Google Scholar] [CrossRef]
- Olate, E.; Jiménez-Gómez, J.M.; Holuigue, L.; Salinas, J. NPR1 mediates a novel regulatory pathway in cold acclimation by interacting with HSFA1 factors. Nat. Plants 2018, 4, 811–823. [Google Scholar] [CrossRef] [PubMed]
- Lang, S.; Liu, X.; Xue, H.; Li, X.; Wang, X. Functional characterization of BnHSFA4a as a heat shock. J. Exp. Bot. 2017, 68, 2361–2375. [Google Scholar] [CrossRef]
- Wei, L.; Sun, X.; Qi, X.; Zhang, Y.; Xu, Y. Dihydromyricetin ameliorates cardiac ischemia/reperfusion injury through Sirt3 activation. BioMed Res. Int. 2019, 2019, 6803943. [Google Scholar] [CrossRef]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.M.; Visser, R.G.F.; Bovy, A. Anthocyanin biosynthesis and degradation mechanisms in solanaceous vegetables: A review. Front. Chem. 2018, 6, 52–58. [Google Scholar] [CrossRef]
- Li, Y.T.; Zhang, J.; Wang, S.J.; Zhang, H.E.; Liu, Y.C.; Yang, M.S. Integrative transcriptomic and metabolomic analyses reveal the flavonoid biosynthesis of Pyrus hopeiensis flowers under cold stress. Hortic. Plant J. 2023, 9, 395–413. [Google Scholar] [CrossRef]
- Zhang, J.; Yin, X.R.; Li, H.; Xu, M.; Zhang, M.X.; Li, S.J.; Liu, X.F.; Shi, Y.N.; Grierson, D.; Chen, K.S. Ethylene response factor EjERF39-EjMYB8 complex activates cold-induced lignification of loquat fruit, via the biosynthetic gene Ej4CL1. J. Exp. Bot. 2020, 71, 3172–3184. [Google Scholar] [CrossRef]
- An, J.P.; Wang, X.F.; Zhang, X.W.; Xu, H.F.; Hao, Y.J. An apple MYB transcription factor regulates cold tolerance and anthocyanin accumulation and undergoes MIEL1-mediated degradation. Plant Biotechnol. J. 2020, 18, 337–353. [Google Scholar] [CrossRef]
- Xie, D.; Li, Z. Transcriptome profile and differentially expressed genes analysis in winter wheat under cold stress conditions. Res. J. Biotechnol. 2015, 10, 73–88. [Google Scholar]


| Cultivation Temperature | K Treatment | SS (mg/g, FW) | SPr (mg/mL, FW) | Pro (μg/g, FW) | MDA (nmol/g, FW) | Total Chlorophyll (mg/g, FW) | SPAD Value | K (g/kg, DW) | Dry Weight/Plant (g) |
|---|---|---|---|---|---|---|---|---|---|
| RT | K0 | 39.496 ± 1.17 aA | 0.100 ± 0.003 aA | 36.126 ± 1.23 aA | 37.371 ± 1.14 aA | 2.69 ± 0.09 aA | 30.5 ± 0.94 aA | 0.66 ± 0.023 aA | 120.38 ± 0.36 aA |
| KL | 34.423 ± 0.89 aA | 0.106 ± 0.005 aA | 41.588 ± 1.14 aA | 21.556 ± 1.02 aA | 3.32 ± 0.12 aA | 36.6 ± 0.77 aA | 0.92 ± 0.025 aA | 139.56 ± 0.56 aA | |
| KM | 33.563 ± 1.02 abAB | 0.119 ± 0.004 abAB | 48.301 ± 1.25 abAB | 17.645 ± 1.04 abAB | 4.56 ± 0.13 abAB | 42.3 ± 0.75 abAB | 1.15 ± 0.032 abAB | 150.46 ± 0.95 abAB | |
| KH | 28.481 ± 0.58 bB | 0.141 ± 0.01 bB | 62.287 ± 1.45 bB | 15.439 ± 0.88 bB | 4.92 ± 0.33 bB | 46.4 ± 0.88 bB | 1.20 ± 0.032 bB | 160.85 ± 1.56 bB | |
| LT | K0 | 57.128 ± 1.20 aA | 0.106 ± 0.005 aA | 38.959 ± 1.12 aA | 40.563 ± 1.24 aA | 1.65 ± 0.05 aA | 16.2 ± 0.68 aA | 0.52 ± 0.011 aA | 90.56 ± 0.45 aA |
| KL | 54.434 ± 1.21 aA | 0.112 ± 0.006 aA | 42.035 ± 1.25 aA | 38.451 ± 1.02 aA | 2.05 ± 0.09 aA | 17.6 ± 0.86 aA | 0.68 ± 0.014 aA | 93.56 ± 0.46 aA | |
| KM | 50.175 ± 1.62 abAB | 0.127 ± 0.009 abAB | 52.003 ± 1.35 abAB | 25.482 ± 1.03 abAB | 2.56 ± 0.05 abAB | 20.3 ± 0.87 abAB | 0.85 ± 0.036 abAB | 111.59 ± 0.63 abAB | |
| KH | 45.258 ± 1.25 bB | 0.151 ± 0.007 bB | 67.939 ± 1.41 bB | 20.564 ± 0.68 bB | 2.88 ± 0.10 bB | 25.4 ± 1.25 bB | 0.95 ± 0.035 bB | 121.85 ± 0.75 bB |
| Ion Mode | Pairs | Up | Down | Total | Fs | AAD | ONC | OA | BD | OOC | PA | LLM | PN | FA | CAD | OHC | CD | As | Others |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pos | RT-K0 vs. RT-KL | 53 | 62 | 115 | 18 | 22 | 1 | 17 | 9 | 8 | 10 | 3 | 3 | 5 | 3 | 1 | 0 | 1 | 14 |
| RT-K0 vs. RT-KM | 67 | 112 | 179 | 20 | 37 | 10 | 15 | 13 | 24 | 5 | 7 | 7 | 4 | 1 | 2 | 2 | 1 | 31 | |
| RT-K0 vs. RT-KH | 8 | 38 | 46 | 4 | 8 | 3 | 5 | 0 | 0 | 1 | 2 | 3 | 3 | 3 | 2 | 1 | 1 | 10 | |
| LT-K0 vs. LT-KL | 32 | 19 | 51 | 14 | 4 | 2 | 5 | 0 | 3 | 5 | 1 | 5 | 0 | 1 | 0 | 2 | 2 | 7 | |
| LT-K0 vs. LT-KM | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | |
| LT-K0 vs. LT-KH | 64 | 22 | 86 | 23 | 9 | 3 | 4 | 0 | 3 | 5 | 5 | 12 | 2 | 3 | 0 | 2 | 5 | 10 | |
| Neg | RT-K0 vs. RT-KL | 18 | 14 | 32 | 2 | 3 | 0 | 5 | 2 | 4 | 3 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 10 |
| RT-K0 vs. RT-KM | 16 | 27 | 43 | 3 | 11 | 0 | 3 | 1 | 6 | 3 | 1 | 1 | 1 | 3 | 1 | 1 | 0 | 8 | |
| RT-K0 vs. RT-KH | 2 | 12 | 14 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 1 | 0 | 2 | 4 | 0 | 0 | 0 | 4 | |
| LT-K0 vs. LT-KL | 2 | 2 | 4 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| LT-K0 vs. LT-KM | 11 | 11 | 22 | 6 | 4 | 0 | 1 | 0 | 2 | 1 | 0 | 2 | 0 | 1 | 0 | 0 | 2 | 4 | |
| LT-K0 vs. LT-KH | 31 | 12 | 43 | 2 | 8 | 0 | 2 | 1 | 5 | 3 | 0 | 4 | 2 | 0 | 0 | 1 | 1 | 14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, L.; Wang, Y.; Sayed, M.A.; Iqbal, A.; Yang, Y. Exploring the Physiological and Molecular Mechanisms by Which Potassium Regulates Low-Temperature Tolerance of Coconut (Cocos nucifera L.) Seedlings. Agronomy 2024, 14, 2983. https://doi.org/10.3390/agronomy14122983
Lu L, Wang Y, Sayed MA, Iqbal A, Yang Y. Exploring the Physiological and Molecular Mechanisms by Which Potassium Regulates Low-Temperature Tolerance of Coconut (Cocos nucifera L.) Seedlings. Agronomy. 2024; 14(12):2983. https://doi.org/10.3390/agronomy14122983
Chicago/Turabian StyleLu, Lilan, Yuping Wang, Md. Abu Sayed, Amjad Iqbal, and Yaodong Yang. 2024. "Exploring the Physiological and Molecular Mechanisms by Which Potassium Regulates Low-Temperature Tolerance of Coconut (Cocos nucifera L.) Seedlings" Agronomy 14, no. 12: 2983. https://doi.org/10.3390/agronomy14122983
APA StyleLu, L., Wang, Y., Sayed, M. A., Iqbal, A., & Yang, Y. (2024). Exploring the Physiological and Molecular Mechanisms by Which Potassium Regulates Low-Temperature Tolerance of Coconut (Cocos nucifera L.) Seedlings. Agronomy, 14(12), 2983. https://doi.org/10.3390/agronomy14122983





