Overexpressing ATP Sulfurylase Improves Fe-Deficiency Tolerance in Apple Calli and Tobacco
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Gene Sequence Analysis
2.3. Quantitative Real-Time PCR
2.4. Gene Cloning and Vector Construction
2.5. Agrobacterium-Mediated Transformation of Tobacco and Apple Calli
2.6. Physiological Index Detection Measurement
2.7. Quantitative Measurement of H2O2 Content and 3,3-Diaminobenzidine (DAB) Staining
2.8. Quantitative Measurement of O2− Content, and Nitro Tetrazolium Blue Chloride (NBT) Staining
2.9. Acidification Capacity Determination
2.10. Statistical Analyses
3. Result
3.1. Analysis of the Physical and Chemical Properties of Apple ATPS Protein
3.2. Subcellular Location Predictions for Four ATPS Proteins
3.3. Cloning of the ATP Sulfurylase Gene from Malus halliana
3.4. Analysis of Cis-Acting Elements of Promoters
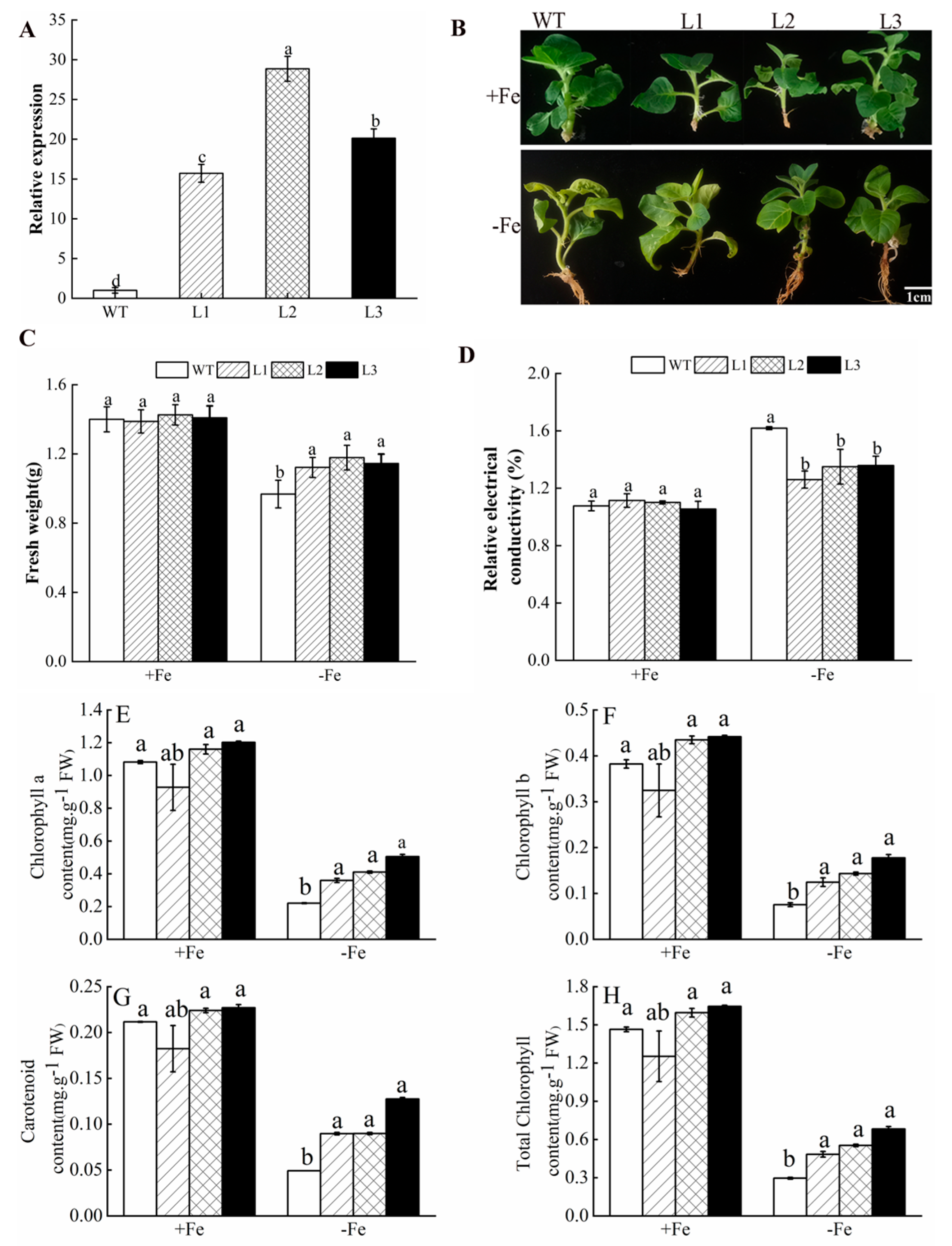
3.5. Overexpression of MhATPS1 in Tobacco Enhances Fe-Deficiency Tolerance
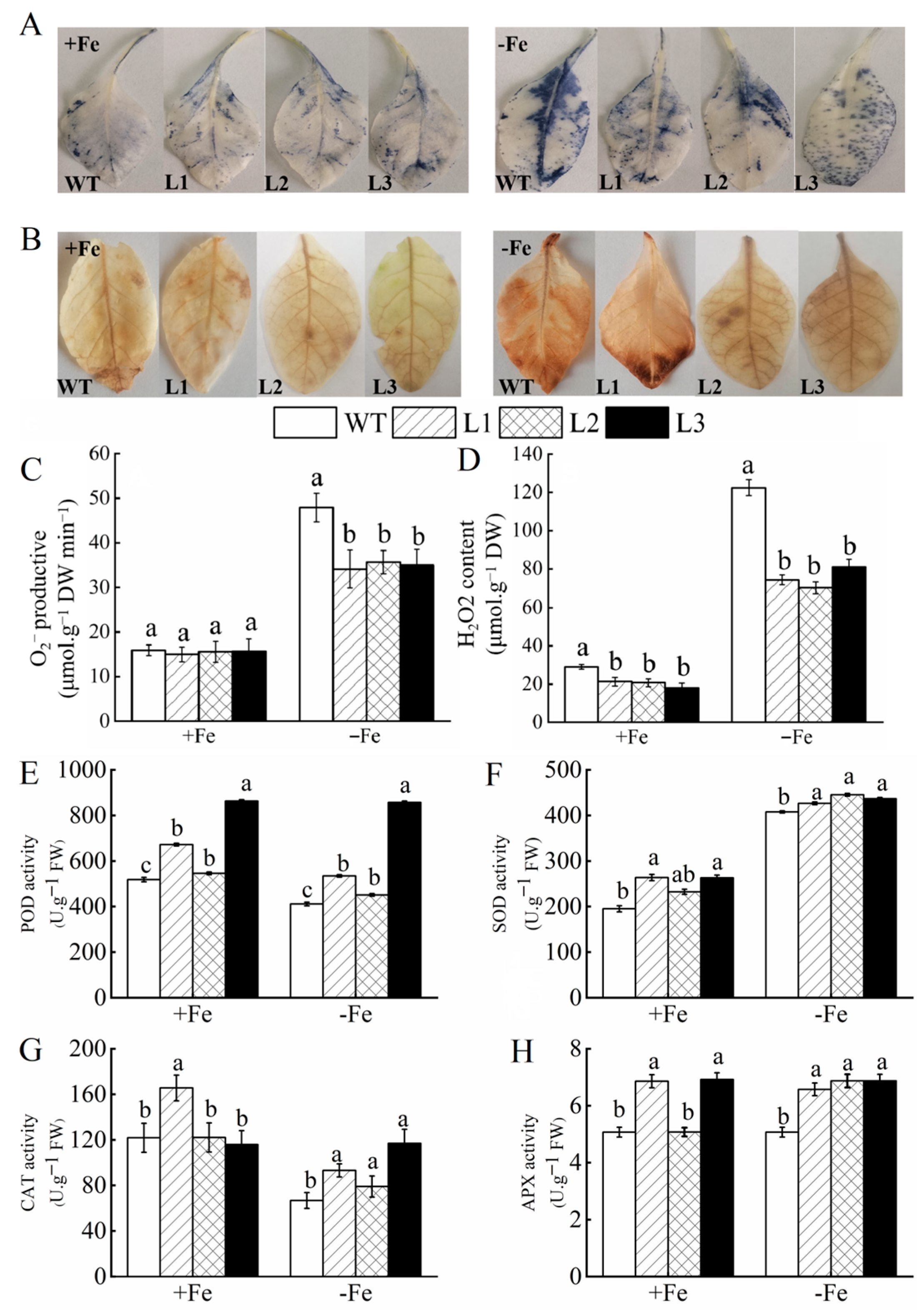
3.6. MhATPS1 Overexpression Enhances Antioxidant Activity in Transgenic Tobacco under Fe Deficiency
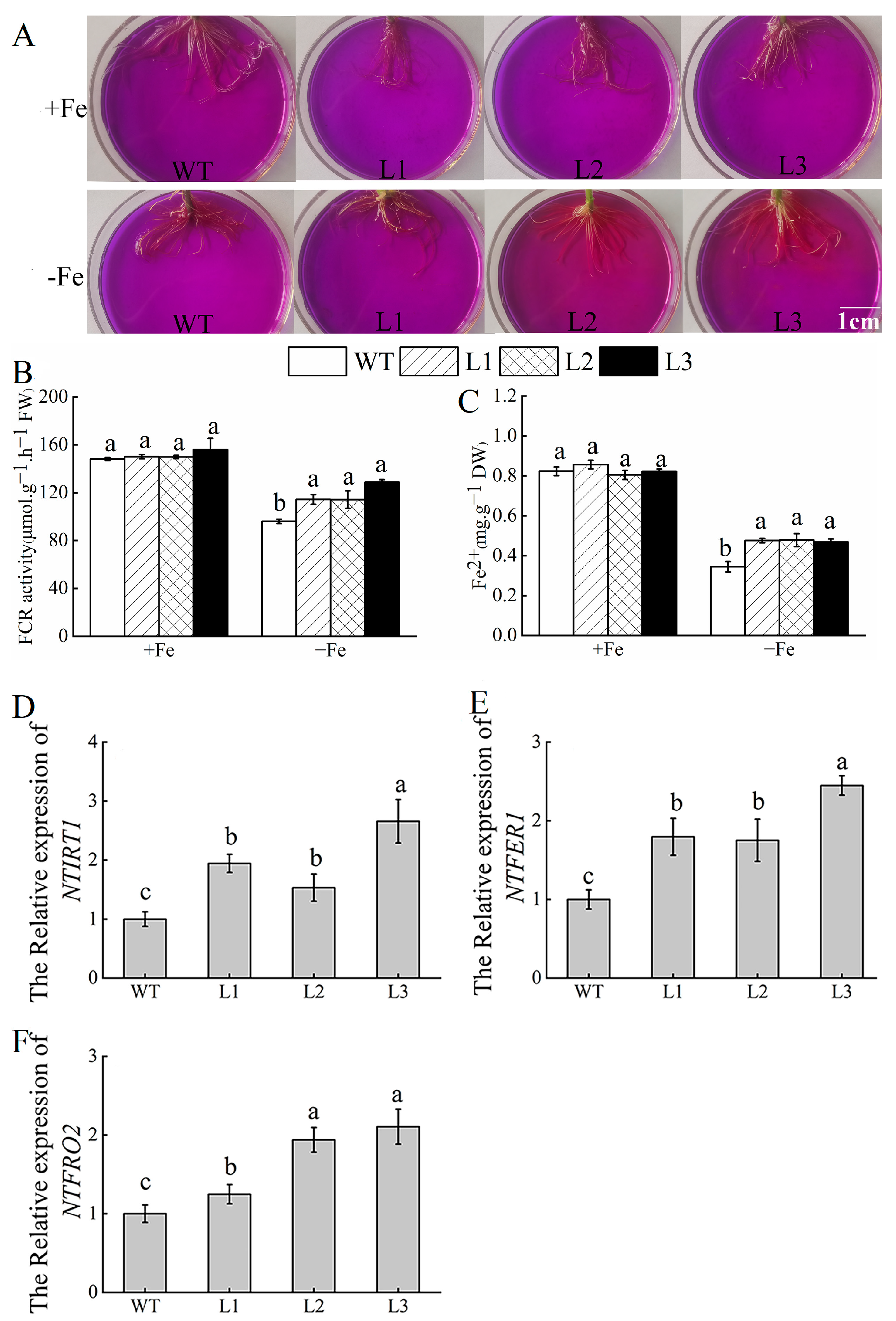
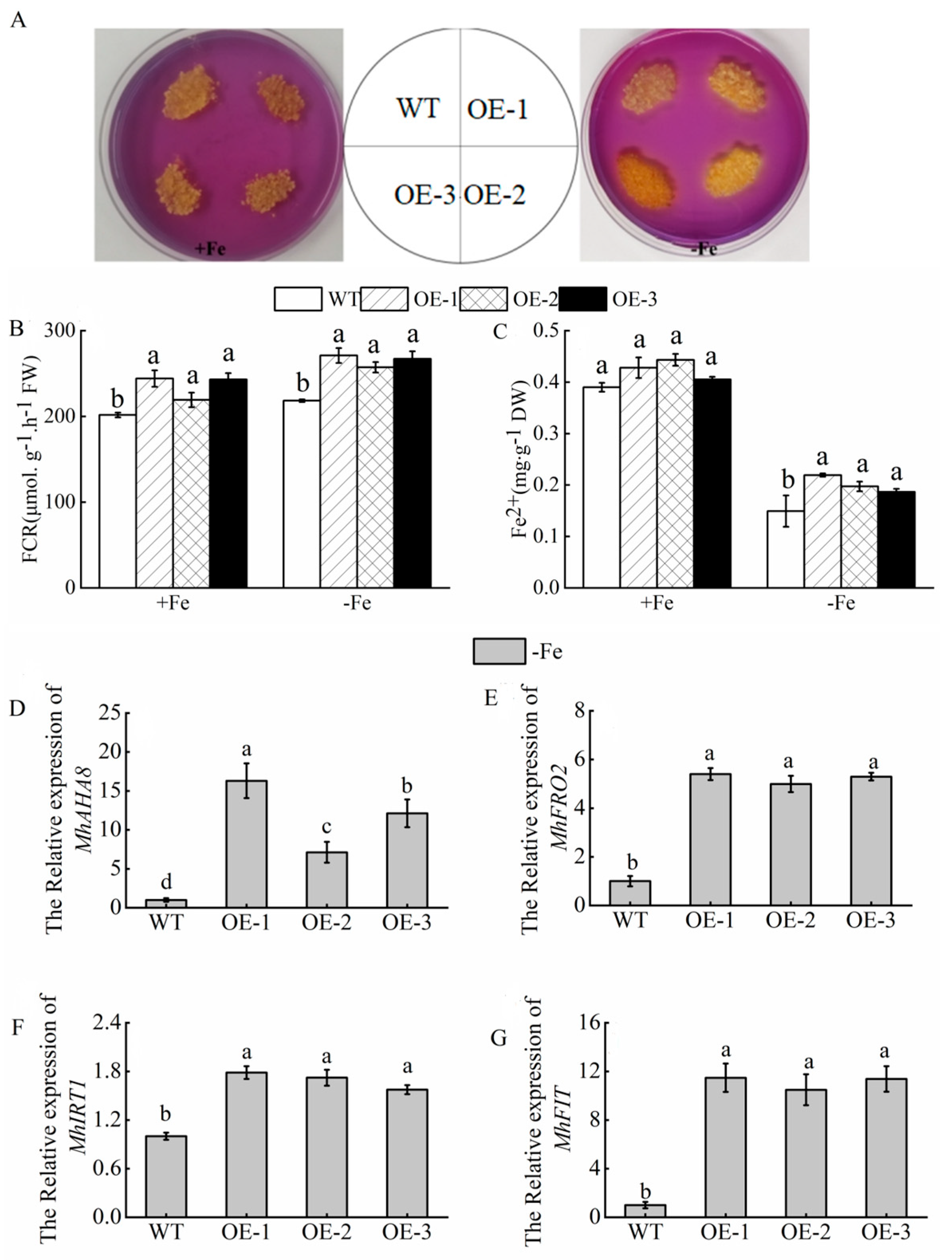
3.7. Overexpression of MhATPS1 Promotes Rhizosphere Acidification and Fe Acquisition in Transgenic Tobacco
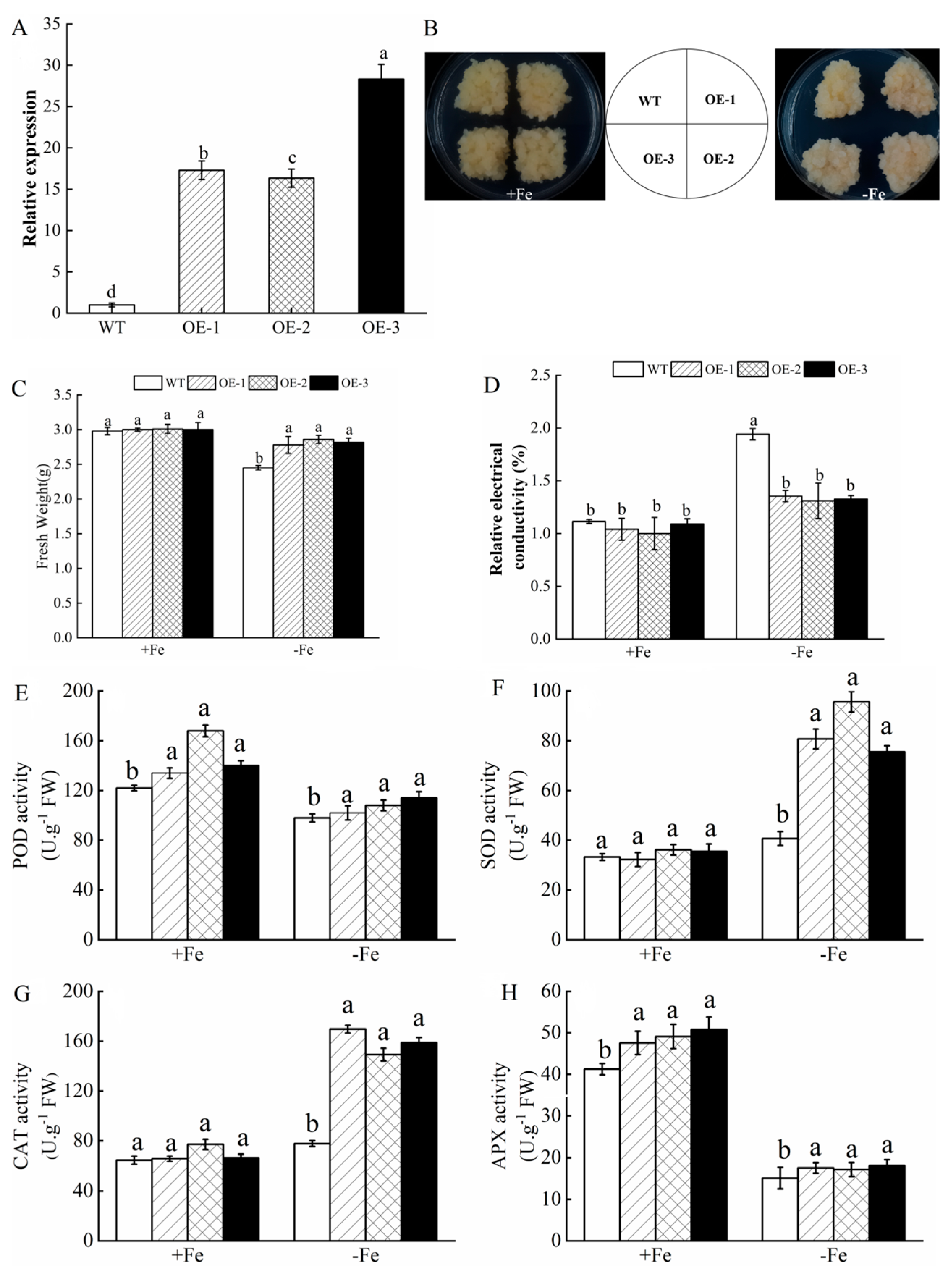
3.8. Tolerance of MhATPS1 Transgenic Apple Calli to Fe Deficiency
3.9. Overexpression of MhATPS1 Promotes Rhizosphere Acidification and Fe Acquisition in Transgenic Calli
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Curie, C.; Panaviene, Z.; Loulergue, C.; Dellaporta, S.L.; Briat, J.F.; Walker, E.-L. Maize yellow stripe1 encodes a membrane protein directly involved in Fe (III) uptake. Nature 2001, 409, 346–349. [Google Scholar] [CrossRef]
- Li, L.; Gao, W.W.; Peng, Q.; Zhou, B.; Kong, Q.H.; Ying, Y.H.; Shou, H.X. Two soybean bHLH factors regulate response to iron deficiency. J. Integr. Plant Biol. 2018, 60, 608–622. [Google Scholar] [CrossRef] [PubMed]
- Li, D.Y.; Sun, Q.; Zhang, G.F.; Zhai, L.M.; Li, K.T.; Feng, Y.; Zhang, X.Z.; Xu, X.F.; Wang, Y.; Han, Z.H. MxMPK6-2-bHLH104 interaction is involved in reactive oxygen species signaling in response to iron deficiency in apple rootstock. J. Exp. Bot. 2021, 72, 1919–1932. [Google Scholar] [CrossRef] [PubMed]
- Santi, S.; Schmidt, W. Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol. 2009, 183, 1072–1084. [Google Scholar] [CrossRef]
- Waters, B.M.; Blevins, D.G.; Eide, D.J. Characterization of FRO1, a pea ferric-chelate reductase involved in root iron acquisition. Plant Physiol. 2002, 129, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Vert, G.; Grot, N.; Dedaldechamp, F.; Gaymard, F.; Guerinot, M.L.; Briat, J.F.; Curie, C. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell. 2003, 14, 1223–1233. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Chiecko, J.C.; Kim, S.A.; Walker, E.L.; Lee, Y.; Guerinot, M.L.; An, G. Disruption of OsYSL15 leads to iron inefficiency in rice plants. Plant Physiol. 2009, 150, 786–800. [Google Scholar] [CrossRef] [PubMed]
- Zuchi, S.; Cesco, S.; Astolfifi, S. High S supply improves Fe accumulation in durum wheat plants grown under Fe limitation. Environ. Exp. Bot. 2012, 77, 25–32. [Google Scholar] [CrossRef]
- Bouranis, D.L.; Chorianopoulou, S.N.; Protonotarios, V.E.; Siyannis, V.F.; Hopkins, L.; Hawkesford, M.J. Leaf response of young iron- inefficient maize plants to sulfur deprivation. J. Plant Nutr. 2003, 26, 1189–1202. [Google Scholar] [CrossRef]
- Zuchi, S.; Cesco, S.; Varanini, Z.; Pinton, R.; Astolfi, S. Sulphur deprivation limits Fe-deficiency responses in tomato plants. Planta 2009, 230, 85–94. [Google Scholar] [CrossRef]
- Khan, N.A.; Anjum, N.A.; Nazar, R.; Iqbal, N. Increased activity of ATP-sulfurylase and increased contents of cysteine and glutathione reduce high cadmium-induced oxidative stress in mustard cultivar with high photosynthetic potential. Russ. J. Plant Physl. 2009, 56, 670–677. [Google Scholar] [CrossRef]
- Logan, H.; Cathala, N.; Grignon, C.; Davidian, J. Cloning of a cDNA encoded by a member of the Arabidopsis thaliana ATP Sulfurylase multigene family expression studies in yeast and in relation to plant sulfur nutrition. J. Biol. Chem. 1996, 271, 12227. [Google Scholar] [CrossRef]
- Bolchi, A.; Petrucco, S.; Tenca, P.L.; Foroni, C.; Ottonello, S. Coordinate modulation of maize sulfate permease and ATP sulfurylase mRNAs in response to variations in sulfur nutritional status: Stereospecific down-regulation by L-cysteine. Plant Mol. Biol. 1999, 39, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.X.; Wirtz, M.; Phua, S.Y.; Estavillo, G.M.; Pogson, B.J. Balancing metabolites in drought: The sulfur assimilation conundrum. Trends Plant Sci. 2013, 18, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Leduc, D.L.; Abdelsamie, M.; Montes-Bayon, M.; Wu, C.P.; Reisinger, S.J.; Terry, N. Overexpressing both ATP sulfurylase and selenocysteine methyltransferase enhances selenium phytoremediation traits in Indian mustard. Environ. Pollut. 2006, 144, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.H.; Wu, T.; Zhai, L.M.; Li, D.Y.; Zhang, X.Z.; Xu, X.F.; Ma, H.Q.; Wang, Y.; Han, Z.H. Reactive oxygen species function to mediate the Fe deficiency response in an Fe-efficient apple genotype: An early response mechanism for enhancing reactive oxygen production. Front. Plant Sci. 2016, 7, 1726. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Ren, Y.R.; Wang, Q.J.; Yao, Y.X.; You, C.X.; Hao, Y.J. Overexpression of MdbHLH104 gene enhances the tolerance to iron deficiency in apple. Plant Biotechnol. J. 2016, 14, 1633–1645. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Z.; Cheng, J.; Wang, S.C.; Gao, Y.L.; Xian, X.L.; Li, C.L.; Wang, Y.X. Molecular cloning and functional characterization of MhHEC2-like genes in Malus halliana reveals it enhances Fe (iron) deficiency tolerance. Funct. Integr. Genom. 2022, 22, 1283–1295. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.F.; Yang, C.Q.; You, Y.; Liang, W.; Wang, N.; Ma, F.W.; Li, C.Y. Validation of reference genes for qRT-PCR analysis in peel and flesh of six apple cultivars (Malus domestica) at diverse stages of fruit development. Sci. Hortic. 2019, 244, 165–171. [Google Scholar] [CrossRef]
- Zhao, Q.; Fan, Z.H.; Qiu, L.N.; Che, Q.Q.; Wang, Y.Z. MdbHLH130, an Apple bHLH transcription factor, confers water stress resistance by regulating stomatal closure and ROS homeostasis in transgenic tobacco. Front. Plant Sci. 2020, 11, 543696. [Google Scholar] [CrossRef]
- Li, T.Y.; Wang, Y.; Zhang, X.Z.; Han, Z.H. Isolation and characterization of ARRO-1 genes from apple rootstocks in response to auxin treatment. Plant Mol. Biol. Rep. 2012, 30, 1408–1414. [Google Scholar] [CrossRef]
- Xie, B.X.; Li, S.; Zhang, R.F.; Zhao, J.; Chen, Y.C.; Zhao, Q.; Hao, Y.J. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 2012, 35, 884–1897. [Google Scholar] [CrossRef]
- Gong, J.M.; Lee, D.A.; Schroeder, J.I. Long-distance root-to-shoot transport of phytochelatins and cadmium in Arabidopsis. Proc. Natl. Acad. Sci. USA 2003, 100, 10118–10123. [Google Scholar] [CrossRef]
- Chazaux, M.; Schiphorst, C.; Lazzari, G.; Caffarri, S. Precise estimation of chlorophyll a, b and carotenoid content by deconvolution of the absorption spectrum and new simultaneous equations for Chl determination. Plant J. 2022, 109, 1630–1648. [Google Scholar] [CrossRef]
- Fitter, D.W.; Martin, D.J.; Copley, M.J.; Scotland, R.W.; Langdale, J.A. GLK gene pairs regulate chloroplast development in diverse plant species. Plant J. 2002, 31, 713–727. [Google Scholar] [CrossRef] [PubMed]
- Ying, Y.Q.; Song, L.L.; Jacobs, D.F.; Mei, L.; Liu, P.; Jin, S.H.; Wu, J.S. Physiological response to drought stress in Camptotheca acuminata seedlings from two provenances. Front. Plant Sci. 2015, 6, 361. [Google Scholar] [CrossRef] [PubMed]
- Schikora, A.; Schmidt, W. Iron stress-induced changes in root epidermal cell fate are regulated independently from physiological responses to low iron availability. Plant Physiol. 2001, 125, 1679–1687. [Google Scholar] [CrossRef] [PubMed]
- Sairam, R.K.; Srivastava, G.C. Changes in antioxidant activity in sub-cellular fractions of tolerant and susceptible wheat genotypes in response to long term salt stress. Plant Sci. 2002, 162, 897–904. [Google Scholar] [CrossRef]
- Liang, Y.; Ma, F.; Li, B.; Guo, C.; Hu, T.; Zhang, M.; Zhan, X. A bHLH transcription factor, SlbHLH96, promotes drought tolerance in tomato. Hortic. Res. 2022, 9, uhac198. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Ren, Y.R.; Wang, Q.J.; Wang, X.F.; You, C.X.; Hao, Y.J. Ubiquitination-related MdBT scaffold proteins target a bHLH transcription factor for iron homeostasis. Plant Physiol. 2016, 172, 1973–1988. [Google Scholar] [CrossRef] [PubMed]
- Rampey, R.A.; Woodward, A.W.; Hobbs, B.N.; Tierney, M.P.; Lahner, B.; Salt, D.E.; Bartel, B. An Arabidopsis basic helix-loop-helix leucine zipper protein modulates metal homeostasis and auxin conjugate responsiveness. Genetics 2006, 174, 1841–1857. [Google Scholar] [CrossRef]
- Phartiyal, P.; Kim, W.S.; Cahoon, R.E.; Jez, J.M.; Krishnan, H.B. Soybean ATP sulfurylase, a homodimeric enzyme involved in sulfur assimilation, is abundantly expressed in roots and induced by cold treatment. Arch. Biochem. Biophys. 2006, 450, 20–29. [Google Scholar] [CrossRef]
- Pilon-Smits, E.; Hwang, S.; Mel-Lytle, C.; Zhu, Y.L.; Tai, J.C.; Chen, Y.C.; Leustek, T.; Terry, N.; Bravo, R.C. Overexpression of ATP sulfurylase in Indian mustard leads to increased selenate uptake, reduction, and tolerance. Plant Physiol. 1999, 119, 123–132. [Google Scholar] [CrossRef]
- Takahashi, H.; Watanabe-Takahashi, A.; Smith, F.W.; Blake-Kalff, M.; Hawkesford, M.J.; Saito, K. The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. Plant J. 2000, 23, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.A.; Nazar, R.; Anjum, N.A. Growth, photosynthesis and antioxidant metabolism in mustard (Brassica juncea L.) cultivars differing in ATP-sulfurylase activity under salinity stress. Sci. Hotic. 2009, 122, 455–460. [Google Scholar] [CrossRef]
- Halliwell, B.; Gutteridge, J. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem. J. 1984, 219, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Wang, H.; Wu, Y.; Yang, N.; Yang, J.; Zhang, P. H+-pyrophosphatase IbVP1 promotes efficient iron use in sweet potato [Ipomoea batatas (L.) Lam.]. Plant Biotechnol. J. 2017, 15, 698–712. [Google Scholar] [CrossRef] [PubMed]
- Yamauch, I.M.; Peng, X.X. Iron toxicity and stress-induced ethylene production in rice leaves. Plant Soil. 1995, 173, 21–28. [Google Scholar] [CrossRef]
- Huang, X.S.; Wang, W.; Zhang, Q.; Liu, J.H. A basic helix-loop-helix transcription factor, PtrbHLH, of Poncirus trifoliata confers cold tolerance and modulates peroxidase-mediated scavenging of hydrogen peroxide. Plant Physiol. 2013, 162, 1178–1194. [Google Scholar] [CrossRef] [PubMed]
- Jelali, N.; Donnini, S.; Dell‘Orto, M.; Abdelly, C.; Gharsalli, M.; Zocchi, G. Root antioxidant responses of two Pisum sativum cultivars to direct and induced Fe deficiency. Plant Biol. 2014, 16, 607–614. [Google Scholar] [CrossRef]
- Wu, T.; Zhang, H.T.; Wang, Y.; Jia, W.S.; Xu, X.F.; Zhang, X.Z.; Han, Z.H. Induction of root Fe (lll) reductase activity and proton extrusion by iron deficiency is mediated by auxin-based systemic signalling in Malus xiaojinensis. J. Exp. Bot. 2012, 63, 859–870. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Luo, J.; Feng, P.; Yang, F.; Liu, Y.; Lisng, J.; Wang, H.; Zou, Y.; Ma, F.; Zhao, T. MbHY5-MbYSL7 mediates chlorophyll synthesis and iron transport under iron deficiency in Malus baccata. Front. Plant Sci. 2022, 13, 1035233. [Google Scholar] [CrossRef] [PubMed]
- Palmgren, M.G. Plant plasma membrane H+-ATPases: Powerhouses for nutrient uptake. Annu. Rev. Plant Biol. 2001, 52, 817–845. [Google Scholar] [CrossRef] [PubMed]
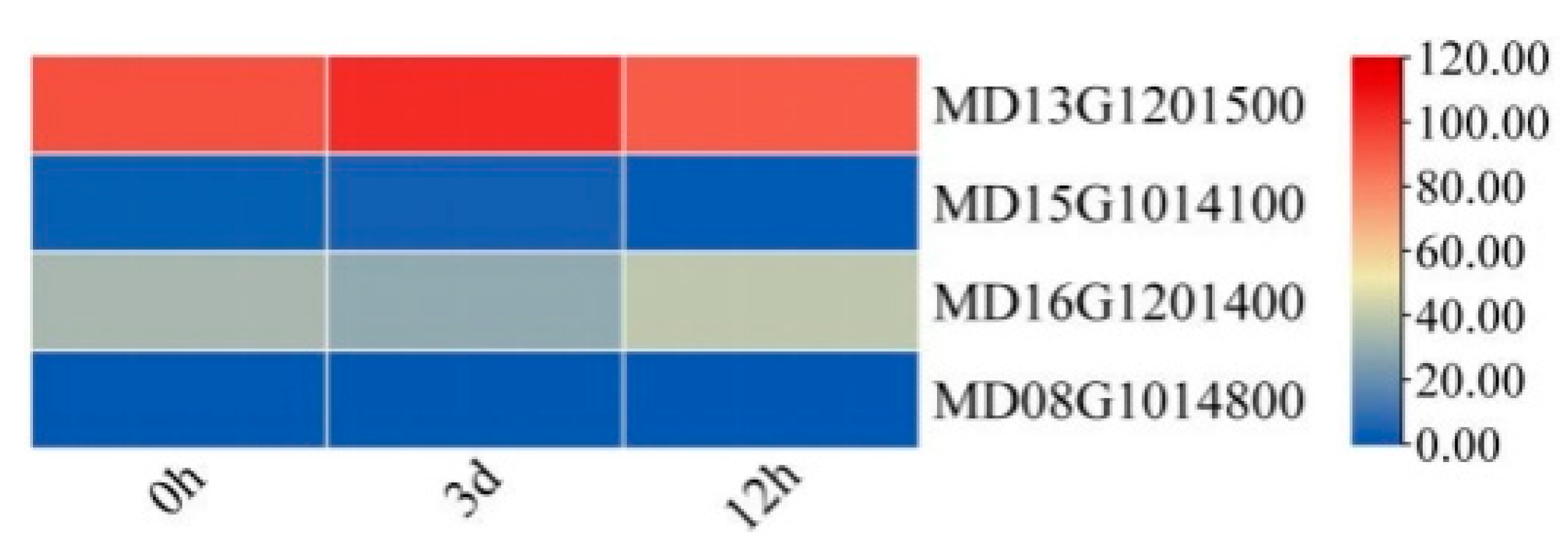
| Accession No | Gene Name | Conserved Domain | Amino Acid | Molecular Mass (kDa) | pI | Positive Residues | Negative Residues | Aliphatic Index | Protein Hydrophobicity | Alpha Helix (%) | Random Coil (%) | Instability | Grand Average Hydropathicity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MD13G1201500 | LOC103410737 | ATPS | 465 | 52.28 | 8.82 | 61 | 57 | 84.75 | −0.397 | 32.90% | 49.89% | 48.71 | −0.397 |
| MD15G1014100 | LOC103450392 | ATPS | 486 | 54.20 | 7.01 | 57 | 58 | 89.47 | −0.340 | 30.86% | 47.94% | 48.09 | −0.340 |
| MD16G1201400 | LOC103403953 | ATPS | 465 | 52.21 | 7.81 | 59 | 58 | 83.91 | −0.375 | 30.54% | 51.61% | 48.96 | −0.375 |
| MD08G1014800 | LOC103440334 | ATPS | 486 | 54.39 | 7.34 | 59 | 59 | 88.87 | −0.370 | 30.86% | 48.56% | 48.25 | −0.370 |
| Gene | Cytoplasmic | Chloroplast | Nuclear | Mitochondrial | Periplast | Nuclear and Cytoplasmic | Golgi Apparatus | Nuclear and Plasma Membrane |
|---|---|---|---|---|---|---|---|---|
| MD13G1201500 | 11 | |||||||
| MD15G1014100 | 1 | 11 | ||||||
| MD16G1201400 | 10 | |||||||
| MD08G1014800 | 5 | 8 |
| Regulator Sequence | Position | Matrix Score | Sequence | Function of Site |
|---|---|---|---|---|
| GATA-motif | 244 | 9 | AAGGATAAGG | part of a light-responsive element |
| TGA-box | 57 | 8 | TGACGTAA | Part of an auxin-responsive element |
| LTR | 968;1382;1335;1429 | 6 | CCGAAA | Cis-acting element involved in low-temperature responsiveness |
| ABR1 | 1562 | 7 | TACGGTC | Cis-acting element involved in the abscisic acid responsiveness |
| MBSI | 1907 | 11 | TTTTTACGGTTA | MYB binding site involved in flavonoid biosynthetic gene regulation |
| TGACG-motif | 60;73;1487;60 | 5 | TGACG | Cis-acting regulatory element involved in the MeJA-responsiveness |
| TATC-box | 1359 | 7 | TATCCCA | Cis-acting element involved in gibberellin-responsiveness |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, J.; Zhang, Z.; Gao, Y.; Dong, Y.; Xian, X.; Li, C.; Ding, L.; Wang, Y. Overexpressing ATP Sulfurylase Improves Fe-Deficiency Tolerance in Apple Calli and Tobacco. Agronomy 2024, 14, 404. https://doi.org/10.3390/agronomy14030404
Cheng J, Zhang Z, Gao Y, Dong Y, Xian X, Li C, Ding L, Wang Y. Overexpressing ATP Sulfurylase Improves Fe-Deficiency Tolerance in Apple Calli and Tobacco. Agronomy. 2024; 14(3):404. https://doi.org/10.3390/agronomy14030404
Chicago/Turabian StyleCheng, Jiao, Zhongxing Zhang, Yanlong Gao, Yongjuan Dong, Xulin Xian, Cailong Li, Liang Ding, and Yanxiu Wang. 2024. "Overexpressing ATP Sulfurylase Improves Fe-Deficiency Tolerance in Apple Calli and Tobacco" Agronomy 14, no. 3: 404. https://doi.org/10.3390/agronomy14030404
APA StyleCheng, J., Zhang, Z., Gao, Y., Dong, Y., Xian, X., Li, C., Ding, L., & Wang, Y. (2024). Overexpressing ATP Sulfurylase Improves Fe-Deficiency Tolerance in Apple Calli and Tobacco. Agronomy, 14(3), 404. https://doi.org/10.3390/agronomy14030404





