Genome-Wide Association Studies of Three-Dimensional (3D) Cassava Root Crowns and Agronomic Traits Using Partially Inbred Populations
Abstract
1. Introduction
2. Materials and Methods
2.1. Germplasm, Experimental Design and Cultivation Conditions
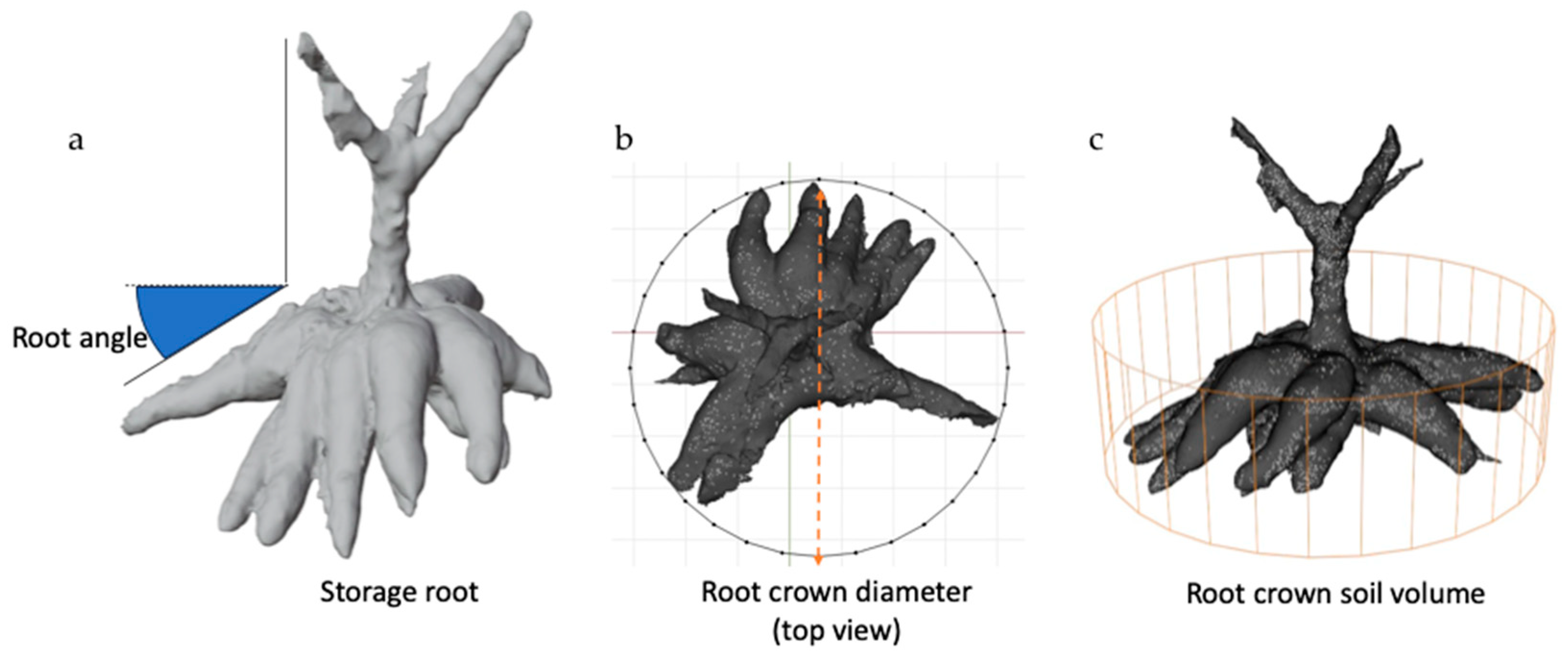
2.2. Data Collection and 3D Analysis
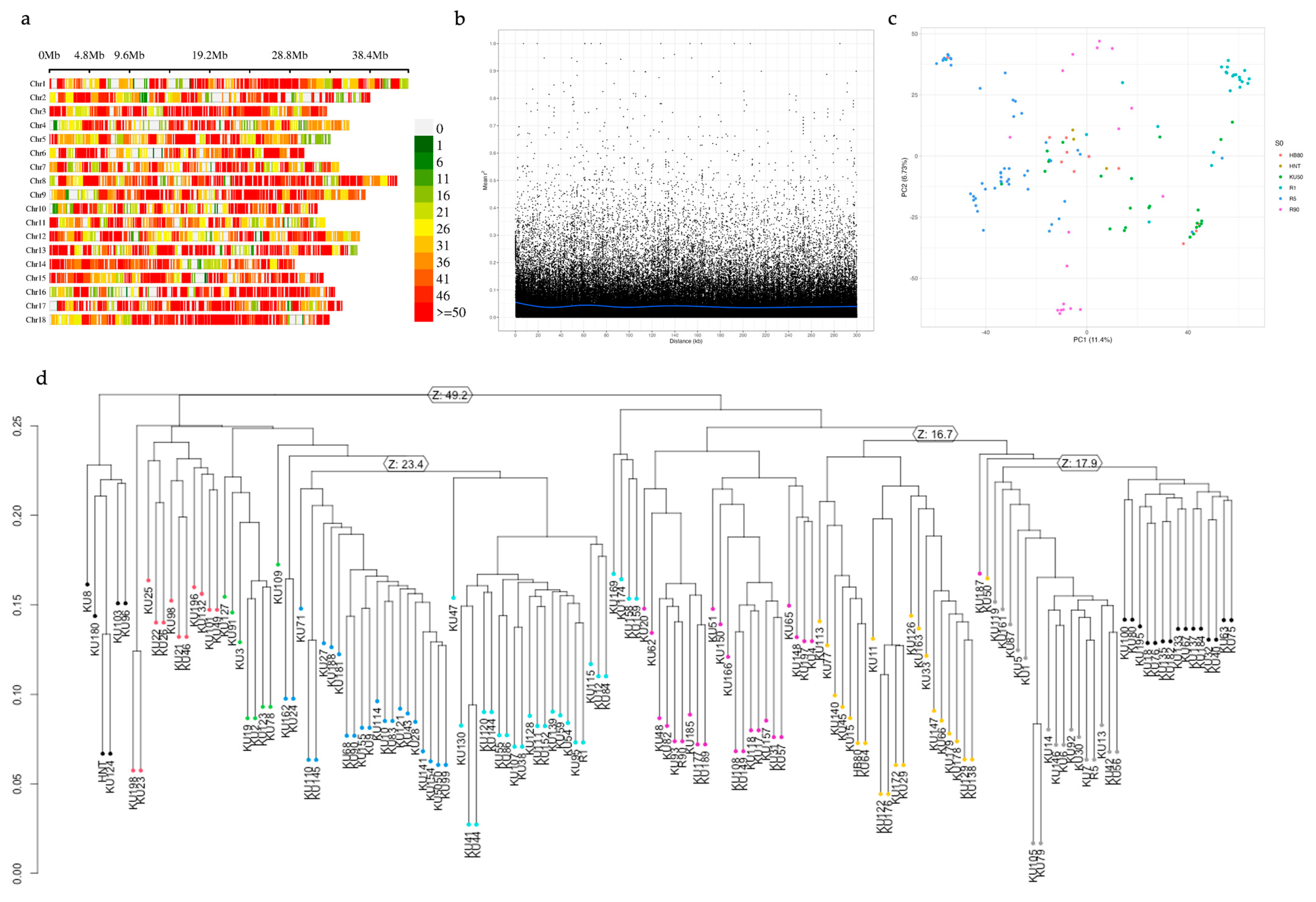
2.3. DNA Isolation and Sequence Analysis
2.4. Data Analysis
3. Results
3.1. Phenotypic Variability of Cassava Root Traits in Partially Inbred Populations
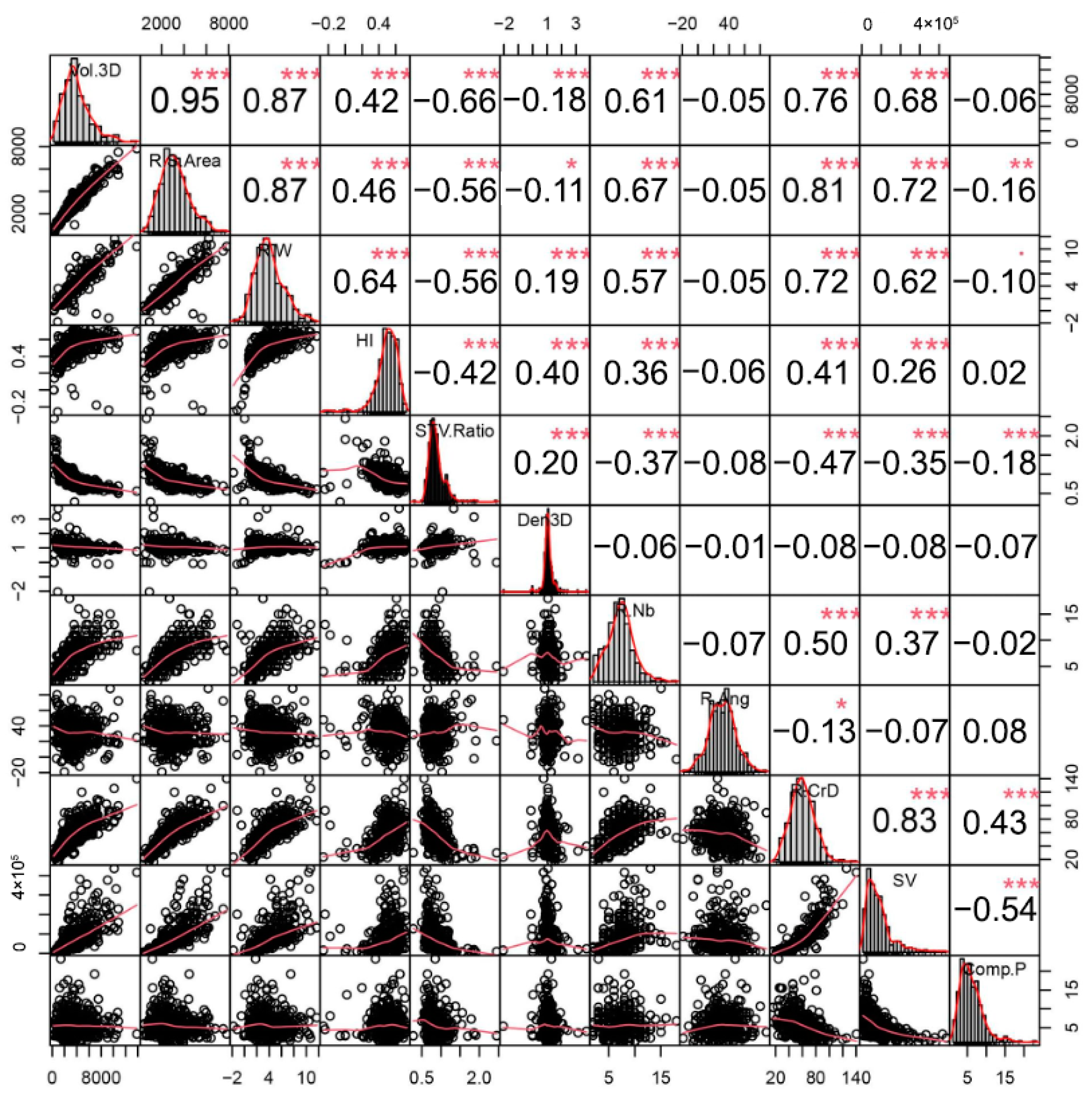
3.2. Correlation Analysis of the 11 Root Traits in Partially Inbred Populations
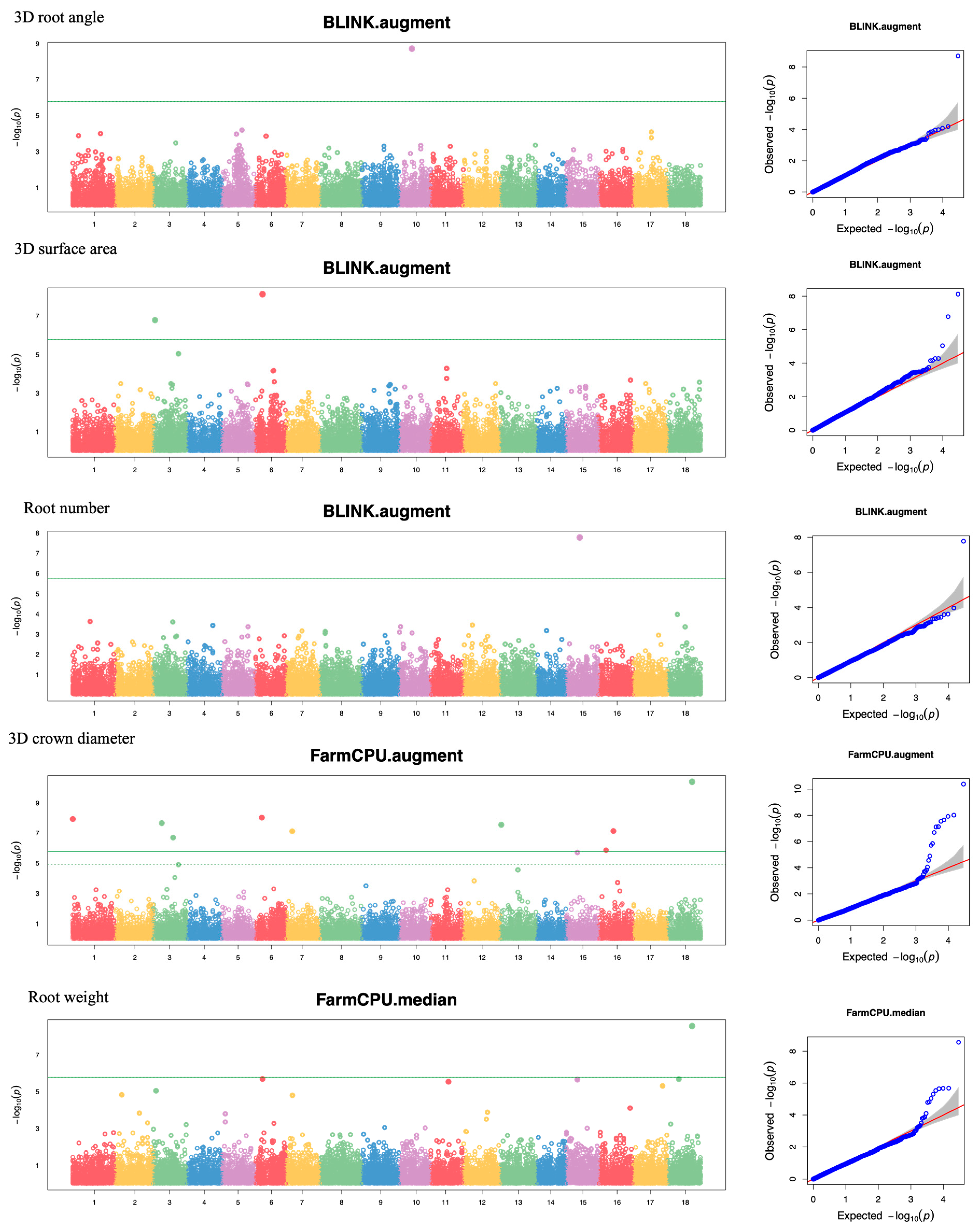
3.3. GWAS of Yield and 3D Root Crown Phenotypes
3.4. Candidate Gene Identification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, S.; Cui, Y.; Zhou, Y.; Luo, Z.; Liu, J.; Zhao, M. The industrial applications of cassava: Current status, opportunities and prospects. J. Sci. Food Agric. 2017, 97, 2282–2290. [Google Scholar] [CrossRef]
- Parmar, A.; Sturm, B.; Hensel, O. Crops that feed the world: Production and improvement of cassava for food, feed, and industrial uses. Food Secur. 2017, 9, 907–927. [Google Scholar] [CrossRef]
- Malik, A.I.; Kongsil, P.; Nguyễn, V.A.; Ou, W.; Srean, P.; López-Lavalle, L.A.; Utsumi, Y.; Lu, C.; Kittipadakul, P.; Nguyễn, H.H.; et al. Cassava breeding and agronomy in Asia: 50 years of history and future directions. Breed. Sci. 2020, 70, 145–166. [Google Scholar] [CrossRef]
- Ceballos, H.; Kawuki, R.S.; Gracen, V.E.; Yencho, G.C.; Hershey, C.H. Conventional breeding, marker-assisted selection, genomic selection and inbreeding in clonally propagated crops: A case study for cassava. Theor. Appl. Genet. 2015, 128, 1647–1667. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, M.D.; Kulakow, P.; Rabbi, I.Y.; Jannink, J.-L. Marker-Based Estimates Reveal Significant Nonadditive Effects in Clonally Propagated Cassava (Manihot esculenta): Implications for the Prediction of Total Genetic Value and the Selection of Varieties. G3 Genes Genomes Genet. 2016, 6, 3497–3506. [Google Scholar] [CrossRef] [PubMed]
- Sunvittayakul, P.; Kittipadakul, P.; Wonnapinij, P.; Chanchay, P.; Wannitikul, P.; Sathitnaitham, S.; Phanthanong, P.; Changwitchukarn, K.; Suttangkakul, A.; Ceballos, H.; et al. Cassava root crown phenotyping using three-dimension (3D) multi-view stereo reconstruction. Sci. Rep. 2022, 12, 10030. [Google Scholar] [CrossRef] [PubMed]
- Alseekh, S.; Kostova, D.; Bulut, M.; Fernie, A.R. Genome-wide association studies: Assessing trait characteristics in model and crop plants. Cell. Mol. Life Sci. 2021, 78, 5743–5754. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chen, X.; Lu, C.; Ye, J.; Zou, M.; Lu, K.; Feng, S.; Pei, J.; Liu, C.; Zhou, X.; et al. Genome-wide association studies of 11 agronomic traits in cassava (Manihot esculenta Crantz). Front. Plant Sci. 2018, 9, 503. [Google Scholar] [CrossRef] [PubMed]
- Rabbi, I.Y.; Kayondo, S.I.; Bauchet, G.; Yusuf, M.; Aghogho, C.I.; Ogunpaimo, K.; Uwugiaren, R.; Smith, I.A.; Peteti, P.; Agbona, A.; et al. Genome-wide association analysis reveals new insights into the genetic architecture of defensive, agro-morphological and quality-related traits in cassava. Plant Mol. Biol. 2022, 109, 195–213. [Google Scholar] [CrossRef] [PubMed]
- Yonis, B.O.; del Carpio, D.P.; Wolfe, M.; Jannink, J.-L.; Kulakow, P.; Rabbi, I. Improving root characterisation for genomic prediction in cassava. Sci. Rep. 2020, 10, 8003. [Google Scholar] [CrossRef]
- dos Santos Silva, P.P.; Sousa, M.B.E.; de Oliveira, E.J.; Morgante, C.V.; de Oliveira, C.R.S.; Vieira, S.L.; Borel, J.C. Genome-wide association study of drought tolerance in cassava. Euphytica 2021, 217, 1–26. [Google Scholar] [CrossRef]
- Hu, W.; Ji, C.; Liang, Z.; Ye, J.; Ou, W.; Ding, Z.; Zhou, G.; Tie, W.; Yan, Y.; Yang, J.; et al. Resequencing of 388 cassava accessions identifies valuable loci and selection for variation in heterozygosity. Genome Biol. 2021, 22, 1–23. [Google Scholar] [CrossRef]
- Community, B.O. Blender—A 3D Modelling and Rendering Package; Blender Foundation, Stichting Blender Foundation: Amsterdam, The Netherlands, 2018. [Google Scholar]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef] [PubMed]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- R Core Team R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. 2022. Available online: https://www.R-project.org/ (accessed on 11 August 2023).
- Aravind, J.; Mukesh Sankar, S.; Wankhede, D.P.; Kaur, V. augmentedRCBD: Analysis of Augmented Randomized Complete Block Designs. R package version 0.1.7.9000. 2020. Available online: https://aravind-j.github.io/augmentedRCBD/ (accessed on 11 August 2023).
- Peterson, B.G.; Carl, P.; Boudt, K.; Bennett, R.; Ulrich, J.; Zivot, E.; Cornilly, D.; Hung, E.; Lestel, M.; Balkissoon, K.; et al. Package ‘Performanceanalytics’. 2018. pp. 13–14. Available online: https://github.com/braverock/PerformanceAnalytics (accessed on 11 August 2023).
- Gilmour, A.R.; Thompson, R.; Cullis, B.R. Average Information REML: An Efficient Algorithm for Variance Parameter Estimation in Linear Mixed Models. Biometrics 1995, 51, 1440. [Google Scholar] [CrossRef]
- Kruijer, W.; Boer, M.P.; Malosetti, M.; Flood, P.J.; Engel, B.; Kooke, R.; Keurentjes, J.J.B.; van Eeuwijk, F.A. Marker-Based Estimation of Heritability in Immortal Populations. Genetics 2015, 199, 379–398. [Google Scholar] [CrossRef] [PubMed]
- Speed, D.; Hemani, G.; Johnson, M.R.; Balding, D.J. Improved Heritability Estimation from Genome-wide SNPs. Am. J. Hum. Genet. 2012, 91, 1011–1021. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Horikoshi, M.; Li, W. ggfortify: Unified Interface to Visualize Statistical Results of Popular R Packages. R J. 2016, 8, 474–485. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. Available online: https://ggplot2.tidyverse.org (accessed on 11 August 2023).
- Endelman, J.B. Ridge Regression and Other Kernels for Genomic Selection with R Package rrBLUP. Plant Genome 2011, 4, 255–258. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Z. GAPIT Version 3: Boosting Power and Accuracy for Genomic Association and Prediction. Genom. Proteom. Bioinform. 2021, 19, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Dong, S.-S.; Xu, J.-Y.; He, W.-M.; Yang, T.-L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, P.A.; Suarez-Vega, A.; Marras, G.; Cánovas, Á. GALLO: An R package for genomic annotation and integration of multiple data sources in livestock for positional candidate loci. GigaScience 2020, 9, giaa149. [Google Scholar] [CrossRef] [PubMed]
- Hohenfeld, C.S.; Passos, A.R.; de Carvalho, H.W.L.; de Oliveira, S.A.S.; de Oliveira, E.J. Genome-wide association study and selection for field resistance to cassava root rot disease and productive traits. PLoS ONE 2022, 17, e0270020. [Google Scholar] [CrossRef]
- Okeke, U.G.; Akdemir, D.; Rabbi, I.; Kulakow, P.; Jannink, J. Regional Heritability Mapping Provides Insights into Dry Matter Content in African White and Yellow Cassava Populations. Plant Genome 2018, 11, 170050. [Google Scholar] [CrossRef] [PubMed]
- Rabbi, I.Y.; Udoh, L.I.; Wolfe, M.; Parkes, E.Y.; Gedil, M.A.; Dixon, A.; Ramu, P.; Jannink, J.L.; Kulakow, P. Genome-wide association mapping of correlated traits in cassava: Dry matter and total carotenoid content. Plant Genome 2017, 10, 1–14. [Google Scholar] [CrossRef]
- Wilson, M.C.; Mutka, A.M.; Hummel, A.W.; Berry, J.; Chauhan, R.D.; Vijayaraghavan, A.; Taylor, N.J.; Voytas, D.F.; Chitwood, D.H.; Bart, R.S. Gene expression atlas for the food security crop cassava. New Phytol. 2017, 213, 1632–1641. [Google Scholar] [CrossRef]
- Zang, Y.; Pei, Y.; Cong, X.; Ran, F.; Liu, L.; Wang, C.; Wang, D.; Min, Y. Single-cell RNA-sequencing profiles reveal the developmental landscape of the Manihot esculenta Crantz leaves. Plant Physiol. 2024, 194, 456–474. [Google Scholar] [CrossRef]
- Esuma, W.; Herselman, L.; Labuschagne, M.T.; Ramu, P.; Lu, F.; Baguma, Y.; Buckler, E.S.; Kawuki, R.S. Genome-wide association mapping of provitamin A carotenoid content in cassava. Euphytica 2016, 212, 97–110. [Google Scholar] [CrossRef]
- Ozimati, A.; Kawuki, R.; Esuma, W.; Kayondo, S.I.; Pariyo, A.; Wolfe, M.; Jannink, J.-L. Genetic Variation and Trait Correlations in an East African Cassava Breeding Population for Genomic Selection. Crop Sci. 2019, 59, 460–473. [Google Scholar] [CrossRef]
- Phumichai, C.; Aiemnaka, P.; Nathaisong, P.; Hunsawattanakul, S.; Fungfoo, P.; Rojanaridpiched, C.; Vichukit, V.; Kongsil, P.; Kittipadakul, P.; Wannarat, W.; et al. Genome-wide association mapping and genomic prediction of yield-related traits and starch pasting properties in cassava. Theor. Appl. Genet. 2022, 135, 145–171. [Google Scholar] [CrossRef] [PubMed]
- Brito, A.C.; Oliveira, S.A.S.; Oliveira, E.J. Genome-wide association study for resistance to cassava root rot. J. Agric. Sci. 2017, 155, 1424–1441. [Google Scholar] [CrossRef]
| Progenitor | Remarks | S1 Family | S2 Family |
|---|---|---|---|
| R1 | Landrace | 24 | 1 |
| R5 | Released by RFCRC 1 | 34 | 16 |
| R90 | Released by RFCRC | 11 | 7 |
| KU50 | Released by KU 2, DOA 3, and CIAT 4 | 18 | 7 |
| HB80 | Released by TTDI 5 and KU | 10 | - |
| HNT | Landrace | 2 | - |
| Total | 99 | 31 |
| Trait | Unit | S0 Data Range | S1 Data Range | S2 Data Range | h2 | |
|---|---|---|---|---|---|---|
| 1 | Root weight (RW) | Kg | 0.87–12.10 | 0.067–12.45 | 0.10–7.50 | 0.325 |
| 2 | 3D volume (Vol3D) | cm3 | 779–12,573 | 268–9678 | 340–8050 | 0.434 |
| 3 | 3D density (Den3D) | kg/L | 0.57–1.66 | 0.049–1.94 | 0.22–1.42 | 0.046 |
| 4 | 3D surface area (RS Area) | cm2 | 983–7333 | 556–7122 | 451–5888 | 0.438 |
| 5 | 3D surface-to-volume ratio (SV Ratio) | non-unit index | 0.53–1.62 | 0.07–2.07 | 0.69–1.50 | 0.334 |
| 6 | 3D crown diameter (RCrD) | cm | 27.79–105.11 | 23.61–113.89 | 19.30–88.74 | 0.303 |
| 7 | Root number (Nb) | Root | 4.60–12.20 | 2.00–13.40 | 3–13.40 | 0.320 |
| 8 | 3D root angle (RAng) | Degree | 0–49.00 | 0.20–45.80 | 1.6–48 | 0.307 |
| 9 | Harvest index (HI) | non-unit index | 0.4–0.72 | 0.03–0.70 | 0.02–0.75 | 0.149 |
| 10 | 3D cylinder soil volume (SV) | cm3 | 42,125–277,043 | 12,796–288,618 | 34,133–94,025 | 0.392 |
| 11 | 3D CRC compactness (CompP) | % | 2.33–12.35 | 1.69–13.24 | 4.45–7.01 | 0.455 |
| Trait | SNP ID | Chr | Position | Major/Minor | MAF | Method | p Value | Phenotype Variance Explained |
|---|---|---|---|---|---|---|---|---|
| Root angle_aug | 15531 | 10 | 13,042,872 | A/T | 0.177 | BLINK | 1.96 × 10−9 | 38.99 |
| RS Area_aug | 3560 | 3 | 1,556,906 | A/G | 0.288 | BLINK | 1.70 × 10−7 | 11.05 |
| RS Area_aug | 8401 | 6 | 7,187,383 | A/C | 0.333 | BLINK | 7.66 × 10−9 | 24.77 |
| RS Area_mean | 411 | 1 | 12,282,357 | G/A | 0.153 | BLINK | 2.18 × 10−7 | 37.91 |
| Nb_aug | 23002 | 15 | 13,707,725 | G/A | 0.304 | BLINK | 1.68 × 10−8 | 30.32 |
| Nb_mean | 22998 | 15 | 13,707,707 | A/G | 0.383 | BLINK | 4.47 × 10−8 | 36.48 |
| RCrD_aug | 22 | 1 | 78,953 | C/T | 0.038 | FarmCPU | 1.21 × 10−8 | 10.20 |
| RCrD_aug | 4647 | 3 | 19,714,017 | G/A | 0.441 | FarmCPU | 2.05 × 10−7 | 0.90 |
| RCrD_aug | 3863 | 3 | 8,392,427 | G/A | 0.064 | FarmCPU | 2.27 × 10−8 | 9.98 |
| RCrD_aug | 8343 | 6 | 6,490,768 | T/G | 0.076 | FarmCPU | 9.67 × 10−9 | 10.74 |
| RCrD_aug | 9751 | 7 | 6,638,776 | C/T | 0.025 | FarmCPU | 7.73 × 10−8 | 7.07 |
| RCrD_aug | 19334 | 13 | 918,716 | A/T | 0.462 | FarmCPU | 2.93 × 10−8 | 1.99 |
| RCrD_aug | 24488 | 16 | 14,895,032 | G/A | 0.267 | BLINK | 1.47 × 10−6 | 10.37 |
| RCrD_aug | 24488 | 16 | 14,895,032 | G/A | 0.267 | FarmCPU | 7.46 × 10−8 | 2.63 |
| RCrD_aug | 24235 | 16 | 7,411,599 | G/A | 0.220 | FarmCPU | 1.40 × 10−6 | 0.99 |
| RCrD_aug | 28919 | 18 | 24,956,060 | A/G | 0.208 | FarmCPU | 4.20 × 10−11 | 7.42 |
| RCrD_aug | 28919 | 18 | 24,956,060 | A/G | 0.208 | BLINK | 9.47 × 10−9 | 16.37 |
| RW_mean | 3559 | 3 | 1,556,895 | C/T | 0.286 | BLINK | 1.59 × 10−6 | 9.93 |
| RW_mean | 22840 | 15 | 11,311,049 | G/A | 0.339 | BLINK | 1.63 × 10−11 | 18.46 |
| RW_median | 1674 | 1 | 38,467,214 | T/A | 0.395 | BLINK | 1.06 × 10−6 | 9.87 |
| RW_median | 22840 | 15 | 11,311,049 | G/A | 0.339 | BLINK | 1.34 × 10−11 | 18.46 |
| RW_median | 28919 | 18 | 24,956,060 | A/G | 0.198 | FarmCPU | 2.76 × 10−9 | 7.42 |
| Vol3D_aug | 3425 | 2 | 38,192,800 | G/C | 0.233 | BLINK | 1.38 × 10−11 | 8.98 |
| Vol3D_aug | 3559 | 3 | 1,556,895 | C/T | 0.275 | BLINK | 1.31 × 10−7 | 9.93 |
| Vol3D_aug | 15263 | 10 | 5,847,582 | G/T | 0.237 | BLINK | 1.68 × 10−9 | 6.91 |
| Vol3D_aug | 25301 | 16 | 27,856,247 | C/T | 0.064 | BLINK | 4.71 × 10−8 | 15.24 |
| Vol3D_mean | 4986 | 3 | 25,180,853 | T/C | 0.314 | BLINK | 8.85 × 10−7 | 13.09 |
| Vol3D_mean | 411 | 1 | 12,282,357 | G/A | 0.153 | BLINK | 7.60 × 10−9 | 18.51 |
| Vol3D_median | 411 | 1 | 12,282,357 | G/A | 0.153 | BLINK | 8.94 × 10−8 | 8.61 |
| Vol3D_median | 25301 | 16 | 27,856,247 | C/T | 0.064 | BLINK | 4.86 × 10−9 | 24.55 |
| Vol3D_median | 26510 | 17 | 16,630,621 | G/A | 0.051 | BLINK | 8.48 × 10−7 | 16.78 |
| Traits | Loci | Description | Wilson et al. [32] | Zang et al. [33] |
|---|---|---|---|---|
| Root number | Manes_15G164200 | ras-related protein Rab7 | V, C, M | |
| 3D crown diameter | Manes_06G028100 | probable 1-deoxy-D-xylulose-5-phosphate synthase 2, chloroplastic | F | V |
| Manes_13G003500 | ER membrane protein complex subunit 10 | M | ||
| Manes_13G003400 | calreticulin-3-like | V | ||
| Manes_13G003300 | acyl carrier protein 2, mitochondrial | V, C, M | ||
| Manes_16G044600 | probable Galacturonosyltransferase 10 isoform X1 | V | ||
| Manes_16G048700 | 18.1 kDa class I heat shock protein-like | S | V | |
| 3D area | Manes_03G018100 | probable histone H2A variant 3 | V | |
| Manes_03G018400 | V-type proton ATPase subunit c″1 | V, C, M | ||
| Manes_03G018600 | pleiotropic drug resistance 12 | F | - | |
| Manes_03G018800 | probable protein phosphatase 2C 22 | V | ||
| Manes_03G018900 | Acetylesterase | V, C | ||
| Root weight | Manes_01G214700 | monothiol glutaredoxin-S9-like | F | C |
| Manes_01G215300 | Phototropic-responsive NPH3 family protein | F, L | - | |
| Manes_01G215100 | monothiol glutaredoxin-S1 | F | - | |
| Manes_01G215500 | peptide chain release factor APG3, chloroplastic | F | - | |
| Manes_01G215600 | PREDICTED: CW-type | C | ||
| Manes_01G214800 | Thioredoxin superfamily protein | L | - | |
| Manes_01G215800 | potassium transporter 5-like | V | ||
| Manes_01G214400 | zinc finger protein CONSTANS-LIKE 13 | F | - | |
| Manes_01G215700 | nuclear factor Y, subunit B5 | L | - | |
| Manes_03G018100 | probable histone H2A variant 3 | V | ||
| Manes_03G018400 | V-type proton ATPase subunit c″1 | V, C, M | ||
| Manes_03G018600 | pleiotropic drug resistance 12 | F | - | |
| Manes_03G018800 | probable protein phosphatase 2C 22 | V | ||
| Manes_03G018900 | Acetylesterase | V, C | ||
| Manes_15G139400 | non-functional NADPH-dependent codeinone reductase 2-like | S | V, M | |
| Manes_15G139600 | transmembrane 9 superfamily member 2 | L | - | |
| 3D volume | Manes_03G018100 | probable histone H2A variant 3 | V | |
| Manes_03G018400 | V-type proton ATPase subunit c″1 | V, C, M | ||
| Manes_03G018600 | pleiotropic drug resistance 12 | F | - | |
| Manes_03G018800 | probable protein phosphatase 2C 22 | V | ||
| Manes_03G018900 | Acetylesterase | V, C | ||
| Manes_03G128000 | early nodulin-like protein 9 | F | - | |
| Manes_03G127800 | glucose-6-phosphate/phosphate translocator 1, chloroplastic-like | L | V | |
| Manes_03G127100 | protein MOS2-like | V | ||
| Manes_10G051600 | Protein of unknown function (DUF1635) | F | - | |
| Manes_16G075000 | peptidyl-tRNA hydrolase 2, mitochondrial | V | ||
| Manes_16G075700 | carotenoid cleavage dioxygenase 8 | S | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sunvittayakul, P.; Wonnapinij, P.; Chanchay, P.; Wannitikul, P.; Sathitnaitham, S.; Phanthanong, P.; Changwitchukarn, K.; Suttangkakul, A.; Ceballos, H.; Gomez, L.D.; et al. Genome-Wide Association Studies of Three-Dimensional (3D) Cassava Root Crowns and Agronomic Traits Using Partially Inbred Populations. Agronomy 2024, 14, 591. https://doi.org/10.3390/agronomy14030591
Sunvittayakul P, Wonnapinij P, Chanchay P, Wannitikul P, Sathitnaitham S, Phanthanong P, Changwitchukarn K, Suttangkakul A, Ceballos H, Gomez LD, et al. Genome-Wide Association Studies of Three-Dimensional (3D) Cassava Root Crowns and Agronomic Traits Using Partially Inbred Populations. Agronomy. 2024; 14(3):591. https://doi.org/10.3390/agronomy14030591
Chicago/Turabian StyleSunvittayakul, Pongsakorn, Passorn Wonnapinij, Pornchanan Chanchay, Pitchaporn Wannitikul, Sukhita Sathitnaitham, Phongnapha Phanthanong, Kanokpoo Changwitchukarn, Anongpat Suttangkakul, Hernan Ceballos, Leonardo D. Gomez, and et al. 2024. "Genome-Wide Association Studies of Three-Dimensional (3D) Cassava Root Crowns and Agronomic Traits Using Partially Inbred Populations" Agronomy 14, no. 3: 591. https://doi.org/10.3390/agronomy14030591
APA StyleSunvittayakul, P., Wonnapinij, P., Chanchay, P., Wannitikul, P., Sathitnaitham, S., Phanthanong, P., Changwitchukarn, K., Suttangkakul, A., Ceballos, H., Gomez, L. D., Kittipadakul, P., & Vuttipongchaikij, S. (2024). Genome-Wide Association Studies of Three-Dimensional (3D) Cassava Root Crowns and Agronomic Traits Using Partially Inbred Populations. Agronomy, 14(3), 591. https://doi.org/10.3390/agronomy14030591







