Cross-Regional Crop Classification Based on Sentinel-2
Abstract
1. Introduction
2. Materials and Methods
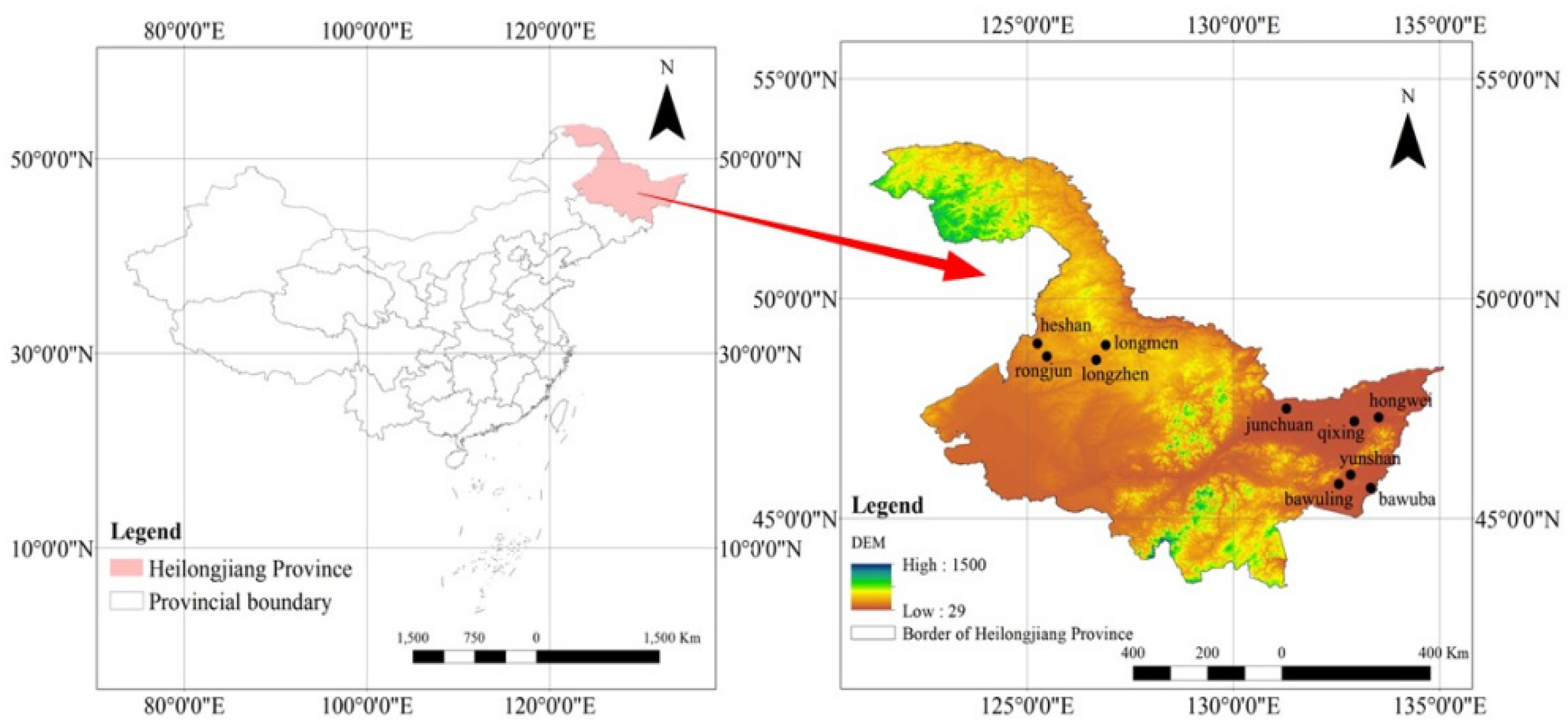
2.1. Study Area
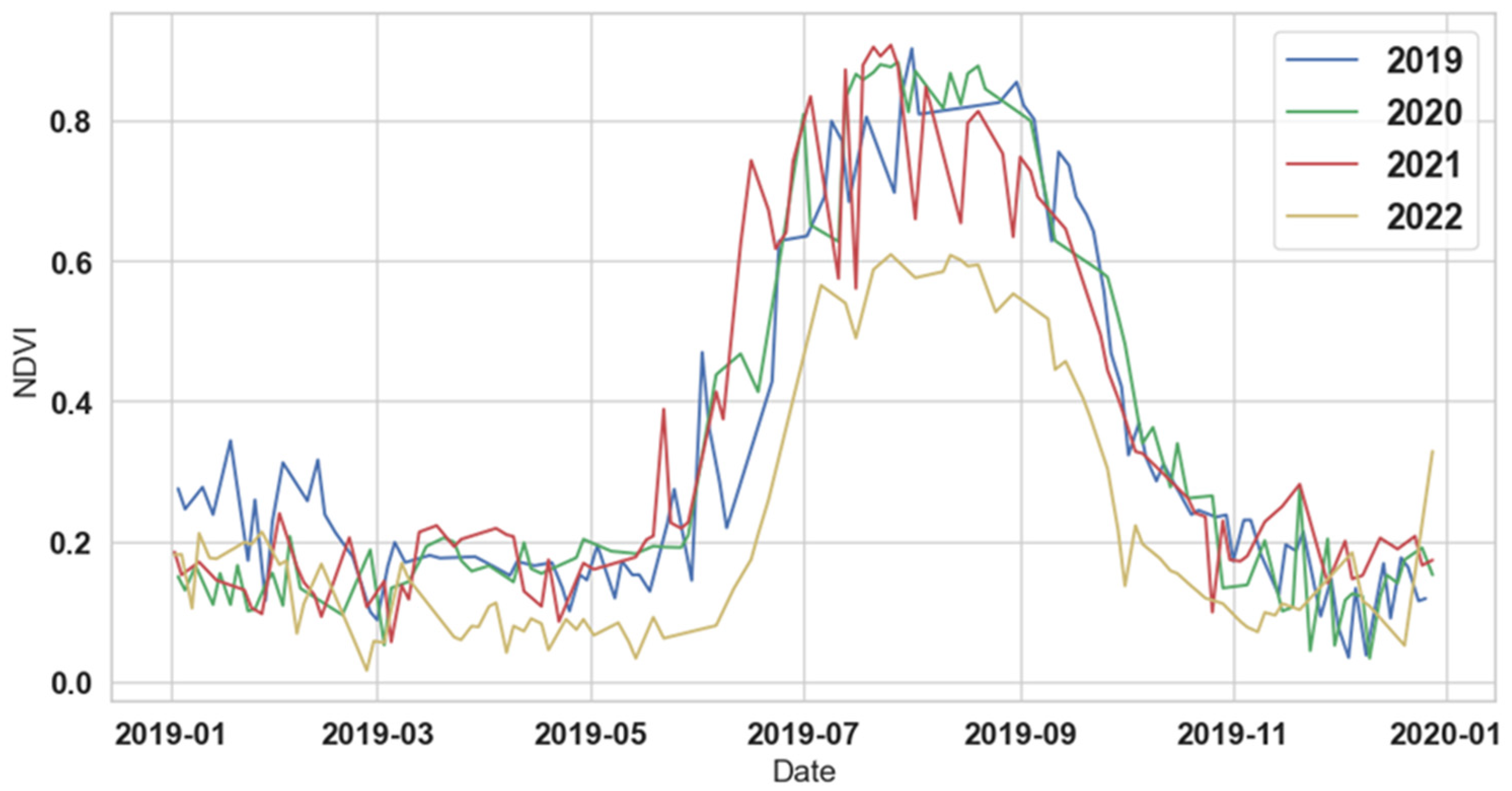
2.2. Image Sources and Input Feature
3. Model Development and Evaluation
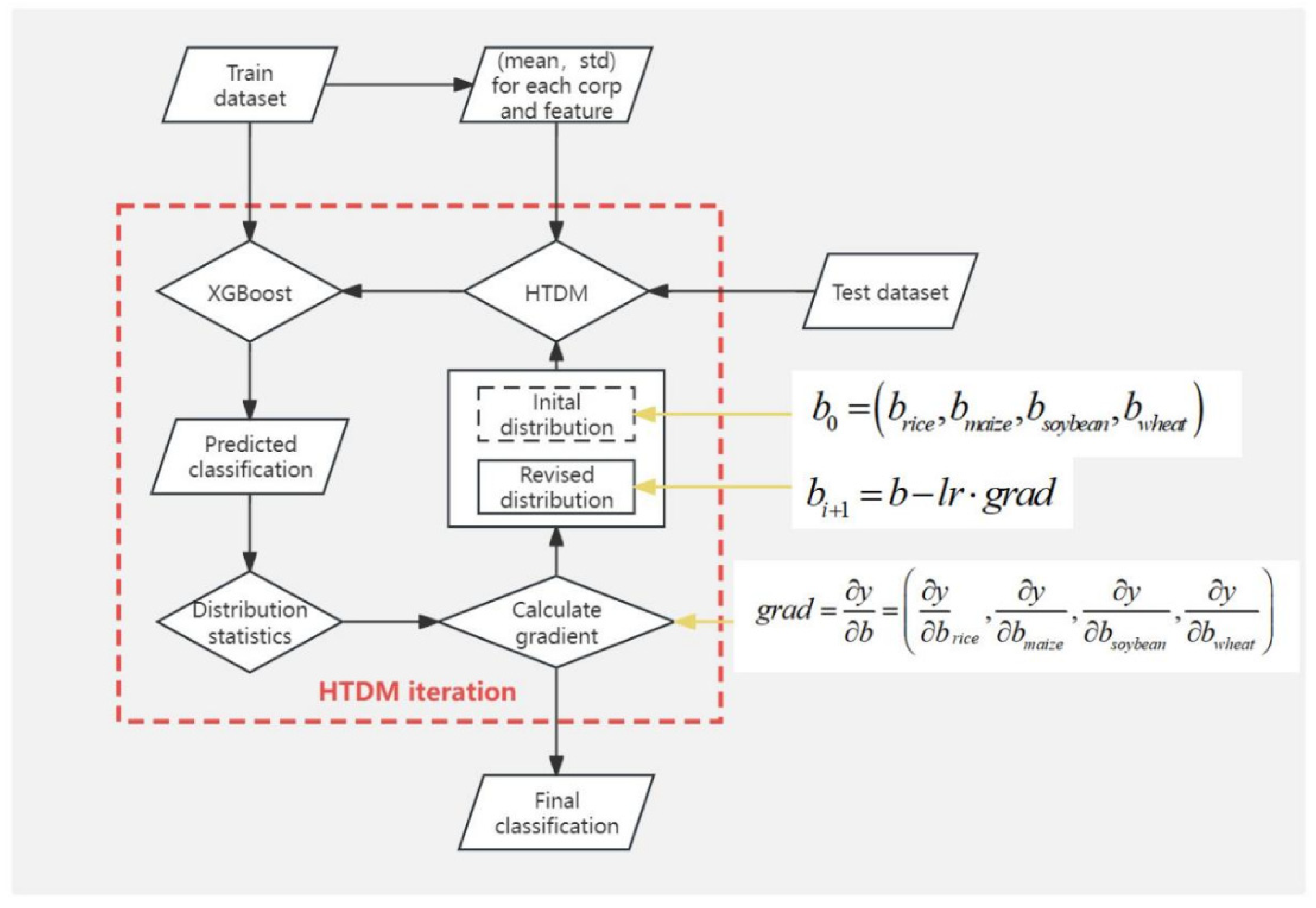
3.1. Hypothesis Testing Distribution Method
3.2. Gradient Descent
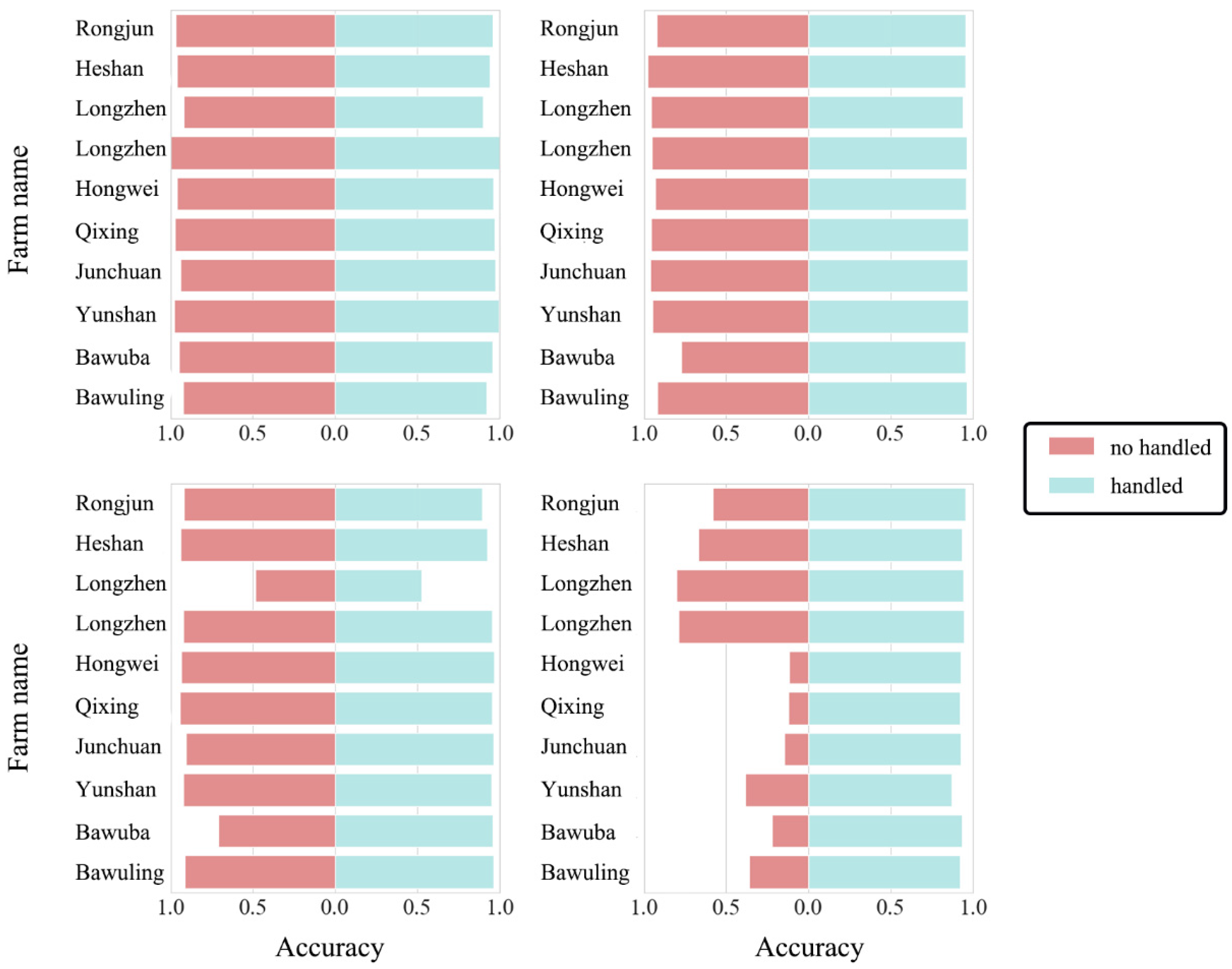
3.3. Method for Evaluating Model Prediction Accuracy
3.4. Input Feature Evaluation
4. Results and Discussion
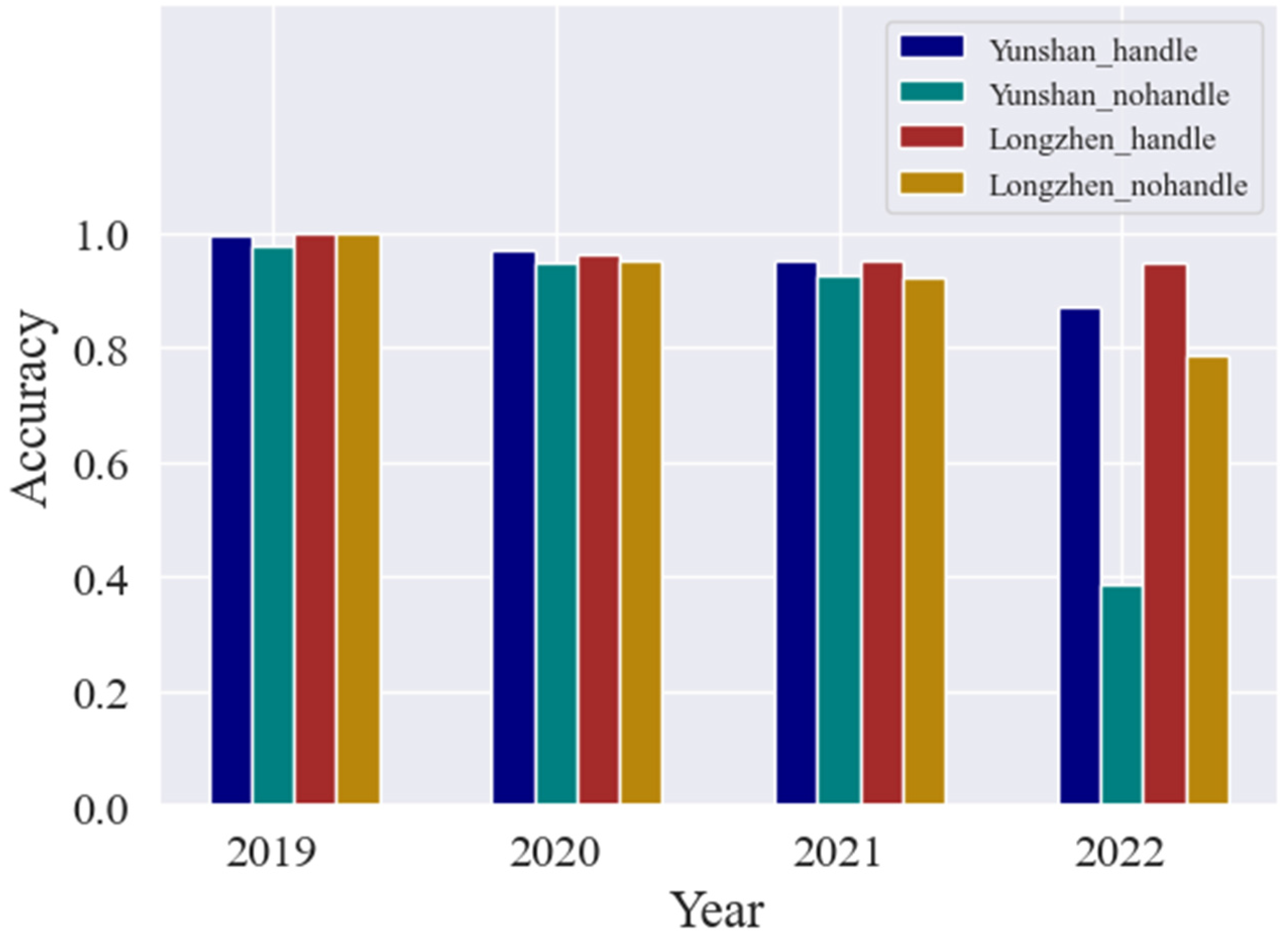
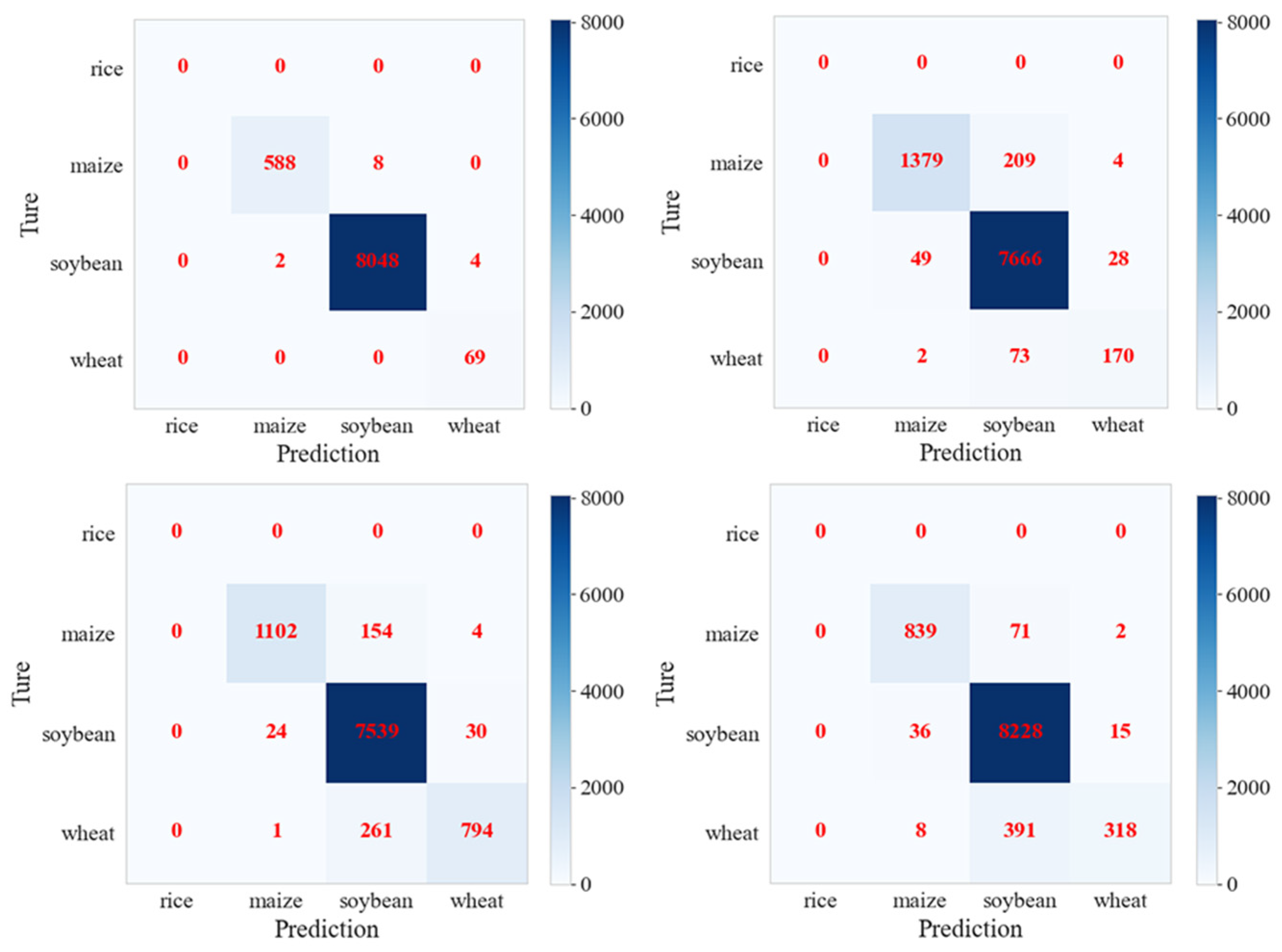
4.1. Comparison of the Improvement in Forecast Accuracy across Time Domains with the HTDM
4.2. Robustness of Hypothetical Distribution Test Method for Different Crop Distributions
4.3. SHAP Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jiang, Y.; Lu, Z.; Li, S.; Lei, Y.; Chu, Q.; Yin, X.; Chen, F. Large-Scale and High-Resolution Crop Mapping in China Using Sentinel-2 Satellite Imagery. Agriculture 2020, 10, 433. [Google Scholar] [CrossRef]
- Luo, N.; Meng, Q.; Feng, P.; Qu, Z.; Yu, Y.; Liu, D.L.; Müller, C.; Wang, P. China Can Be Self-Sufficient in Maize Production by 2030 with Optimal Crop Management. Nat. Commun. 2023, 14, 2637. [Google Scholar] [CrossRef]
- Davis, K.F.; Rulli, M.C.; Seveso, A.; D’Odorico, P. Increased Food Production and Reduced Water Use through Optimized Crop Distribution. Nat. Geosci. 2017, 10, 919–924. [Google Scholar] [CrossRef]
- Tittonell, P.; Shepherd, K.D.; Vanlauwe, B.; Giller, K.E. Unravelling the Effects of Soil and Crop Management on Maize Productivity in Smallholder Agricultural Systems of Western Kenya—An Application of Classification and Regression Tree Analysis. Agric. Ecosyst. Environ. 2008, 123, 137–150. [Google Scholar] [CrossRef]
- Guo, W.; Fu, Y.; Ruan, B.; Ge, H.; Zhao, N. Agricultural Non-Point Source Pollution in the Yongding River Basin. Ecol. Indic. 2014, 36, 254–261. [Google Scholar] [CrossRef]
- Sun, B.; Zhang, L.; Yang, L.; Zhang, F.; Norse, D.; Zhu, Z. Agricultural Non-Point Source Pollution in China: Causes and Mitigation Measures. Ambio 2012, 41, 370–379. [Google Scholar] [CrossRef]
- Xie, W.; Zhu, A.; Ali, T.; Zhang, Z.; Chen, X.; Wu, F.; Huang, J.; Davis, K.F. Crop Switching Can Enhance Environmental Sustainability and Farmer Incomes in China. Nature 2023, 616, 300–305. [Google Scholar] [CrossRef]
- You, N.; Dong, J.; Huang, J.; Du, G.; Zhang, G.; He, Y.; Yang, T.; Di, Y.; Xiao, X. The 10-m Crop Type Maps in Northeast China during 2017. Sci. Data 2021, 8, 41. [Google Scholar] [CrossRef]
- Pereira, L.S.; Paredes, P.; Hunsaker, D.J.; López-Urrea, R.; Shad, Z.M. Standard Single and Basal Crop Coefficients for Field Crops. Updates and Advances to the FAO56 Crop Water Requirements Method. Agric. Water Manag. 2021, 243, 106466. [Google Scholar] [CrossRef]
- Novotny, V. Water Quality: Prevention, Identification and Management of Diffuse Pollution; Van Nostrand-Reinhold Publishers: New York, NY, USA, 1994; ISBN 0-442-00559-8. [Google Scholar]
- Sun, X.; Ritzema, H.; Huang, X.; Bai, X.; Hellegers, P. Assessment of Farmers’ Water and Fertilizer Practices and Perceptions in the North China Plain. Irrig. Drain. 2022, 71, 980–996. [Google Scholar] [CrossRef]
- Karthikeyan, L.; Chawla, I.; Mishra, A.K. A Review of Remote Sensing Applications in Agriculture for Food Security: Crop Growth and Yield, Irrigation, and Crop Losses. J. Hydrol. 2020, 586, 124905. [Google Scholar] [CrossRef]
- Vyas, S.; Dalhaus, T.; Kropff, M.; Aggarwal, P.; Meuwissen, M.P. Mapping Global Research on Agricultural Insurance. Environ. Res. Lett. 2021, 16, 103003. [Google Scholar] [CrossRef]
- Cai, Y.; Guan, K.; Peng, J.; Wang, S.; Seifert, C.; Wardlow, B.; Li, Z. A High-Performance and in-Season Classification System of Field-Level Crop Types Using Time-Series Landsat Data and a Machine Learning Approach. Remote Sens. Environ. 2018, 210, 35–47. [Google Scholar] [CrossRef]
- Mountrakis, G.; Im, J.; Ogole, C. Support Vector Machines in Remote Sensing: A Review. ISPRS J. Photogramm. Remote Sens. 2011, 66, 247–259. [Google Scholar] [CrossRef]
- Conrad, C.; Colditz, R.R.; Dech, S.; Klein, D.; Vlek, P.L. Temporal Segmentation of MODIS Time Series for Improving Crop Classification in Central Asian Irrigation Systems. Int. J. Remote Sens. 2011, 32, 8763–8778. [Google Scholar] [CrossRef]
- Tatsumi, K.; Yamashiki, Y.; Torres, M.A.C.; Taipe, C.L.R. Crop Classification of Upland Fields Using Random Forest of Time-Series Landsat 7 ETM+ Data. Comput. Electron. Agric. 2015, 115, 171–179. [Google Scholar] [CrossRef]
- Fan, C.; Lu, R. UAV Image Crop Classification Based on Deep Learning with Spatial and Spectral Features. In Proceedings of the IOP Conference Series: Earth and Environmental Science, Zhangjiajie, China, 23–25 April 2021; IOP Publishing: Bristol, UK, 2021; Volume 783, p. 012080. [Google Scholar]
- Yang, S.; Gu, L.; Li, X.; Jiang, T.; Ren, R. Crop Classification Method Based on Optimal Feature Selection and Hybrid CNN-RF Networks for Multi-Temporal Remote Sensing Imagery. Remote Sens. 2020, 12, 3119. [Google Scholar] [CrossRef]
- Agilandeeswari, L.; Prabukumar, M.; Radhesyam, V.; Phaneendra, K.L.N.B.; Farhan, A. Crop Classification for Agricultural Applications in Hyperspectral Remote Sensing Images. Appl. Sci. 2022, 12, 1670. [Google Scholar] [CrossRef]
- Chakhar, A.; Ortega-Terol, D.; Hernández-López, D.; Ballesteros, R.; Ortega, J.F.; Moreno, M.A. Assessing the Accuracy of Multiple Classification Algorithms for Crop Classification Using Landsat-8 and Sentinel-2 Data. Remote Sens. 2020, 12, 1735. [Google Scholar] [CrossRef]
- Neetu; Ray, S.S. Exploring Machine Learning Classification Algorithms for Crop Classification Using Sentinel 2 Data. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2019, 42, 573–578. [Google Scholar]
- Shi, M.; Xie, F.; Zi, Y.; Yin, J. Cloud Detection of Remote Sensing Images by Deep Learning. In Proceedings of the 2016 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Beijing, China, 10–15 July 2016; pp. 701–704. [Google Scholar]
- Kussul, N.; Lavreniuk, M.; Skakun, S.; Shelestov, A. Deep Learning Classification of Land Cover and Crop Types Using Remote Sensing Data. IEEE Geosci. Remote Sens. Lett. 2017, 14, 778–782. [Google Scholar] [CrossRef]
- Shen, H.; Li, H.; Qian, Y.; Zhang, L.; Yuan, Q. An Effective Thin Cloud Removal Procedure for Visible Remote Sensing Images. ISPRS J. Photogramm. Remote Sens. 2014, 96, 224–235. [Google Scholar] [CrossRef]
- Lu, X.; Gong, T.; Zheng, X. Multisource Compensation Network for Remote Sensing Cross-Domain Scene Classification. IEEE Trans. Geosci. Remote Sens. 2020, 58, 2504–2515. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, H.; Wang, C.; Zhang, B.; Liu, M. Crop Classification Based on Temporal Information Using Sentinel-1 SAR Time-Series Data. Remote Sens. 2018, 11, 53. [Google Scholar] [CrossRef]
- Chakhar, A.; Hernández-López, D.; Ballesteros, R.; Moreno, M.A. Improving the Accuracy of Multiple Algorithms for Crop Classification by Integrating Sentinel-1 Observations with Sentinel-2 Data. Remote Sens. 2021, 13, 243. [Google Scholar] [CrossRef]
- Li, H.; Zhang, C.; Zhang, S.; Atkinson, P.M. Crop Classification from Full-Year Fully-Polarimetric L-Band UAVSAR Time-Series Using the Random Forest Algorithm. Int. J. Appl. Earth Obs. Geoinf. 2020, 87, 102032. [Google Scholar] [CrossRef]
- Argenti, F.; Lapini, A.; Bianchi, T.; Alparone, L. A Tutorial on Speckle Reduction in Synthetic Aperture Radar Images. IEEE Geosci. Remote Sens. Mag. 2013, 1, 6–35. [Google Scholar] [CrossRef]
- Joshi, N.; Baumann, M.; Ehammer, A.; Fensholt, R.; Grogan, K.; Hostert, P.; Jepsen, M.; Kuemmerle, T.; Meyfroidt, P.; Mitchard, E.; et al. A Review of the Application of Optical and Radar Remote Sensing Data Fusion to Land Use Mapping and Monitoring. Remote Sens. 2016, 8, 70. [Google Scholar] [CrossRef]
- Ulaby, F.T.; Long, D.G.; Blackwell, W.; Elachi, C.; Zebker, H. Microwave Radar and Radiometric Remote Sensing; University of Michigan Press: Ann Arbor, MI, USA, 2015. [Google Scholar]
- Kwak, G.-H.; Park, N.-W. Impact of Texture Information on Crop Classification with Machine Learning and UAV Images. Appl. Sci. 2019, 9, 643. [Google Scholar] [CrossRef]
- Reedha, R.; Dericquebourg, E.; Canals, R.; Hafiane, A. Transformer Neural Network for Weed and Crop Classification of High Resolution UAV Images. Remote Sens. 2022, 14, 592. [Google Scholar] [CrossRef]
- Bégué, A.; Arvor, D.; Bellon, B.; Betbeder, J.; De Abelleyra, D.; PD Ferraz, R.; Lebourgeois, V.; Lelong, C.; Simões, M.; Verón, S.R. Remote Sensing and Cropping Practices: A Review. Remote Sens. 2018, 10, 99. [Google Scholar] [CrossRef]
- Li, J.; Shen, Y.; Yang, C. An Adversarial Generative Network for Crop Classification from Remote Sensing Timeseries Images. Remote Sens. 2021, 13, 65. [Google Scholar] [CrossRef]
- Zhong, L.; Hu, L.; Zhou, H. Deep Learning Based Multi-Temporal Crop Classification. Remote Sens. Environ. 2019, 221, 430–443. [Google Scholar] [CrossRef]
- Ji, S.; Zhang, C.; Xu, A.; Shi, Y.; Duan, Y. 3D Convolutional Neural Networks for Crop Classification with Multi-Temporal Remote Sensing Images. Remote Sens. 2018, 10, 75. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, H.; He, W.; Zhang, L. Phenology Alignment Network: A Novel Framework for Cross-Regional Time Series Crop Classification. In Proceedings of the 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Nashville, TN, USA, 19–25 June 2021; pp. 2934–2943. [Google Scholar]
- Sonobe, R.; Tani, H.; Wang, X.; Kobayashi, N.; Shimamura, H. Parameter Tuning in the Support Vector Machine and Random Forest and Their Performances in Cross- and Same-Year Crop Classification Using TerraSAR-X. Int. J. Remote Sens. 2014, 35, 7898–7909. [Google Scholar] [CrossRef]
- Muhammad, S.; Zhan, Y.; Wang, L.; Hao, P.; Niu, Z. Major Crops Classification Using Time Series MODIS EVI with Adjacent Years of Ground Reference Data in the US State of Kansas. Optik 2016, 127, 1071–1077. [Google Scholar] [CrossRef]
- Xue, J.; Su, B. Significant Remote Sensing Vegetation Indices: A Review of Developments and Applications. J. Sens. 2017, 2017, 1353691. [Google Scholar] [CrossRef]
- Chandrasekar, K.; Sesha Sai, M.V.R.; Roy, P.S.; Dwevedi, R.S. Land Surface Water Index (LSWI) Response to Rainfall and NDVI Using the MODIS Vegetation Index Product. Int. J. Remote Sens. 2010, 31, 3987–4005. [Google Scholar] [CrossRef]
- Li, H.; Shi, Q.; Wan, Y.; Shi, H.; Imin, B. Using Sentinel-2 Images to Map the Populus Euphratica Distribution Based on the Spectral Difference Acquired at the Key Phenological Stage. Forests 2021, 12, 147. [Google Scholar] [CrossRef]
- Wang, Z.; Zheng, Y.-C.; Li, J.-F.; Wang, Y.-Z.; Rong, L.-S.; Wang, J.-X.; Jiang, D.-C.; Qi, W.-C. Study on GLI Values of Polygonatum Odoratum Base on Multi-Temporal of Unmanned Aerial Vehicle Remote Sensing. Zhongguo Zhong Yao Za Zhi Zhongguo Zhongyao Zazhi China J. Chin. Mater. Medica 2020, 45, 5663–5668. [Google Scholar]
- Chen, J.; Jönsson, P.; Tamura, M.; Gu, Z.; Matsushita, B.; Eklundh, L. A Simple Method for Reconstructing a High-Quality NDVI Time-Series Data Set Based on the Savitzky–Golay Filter. Remote Sens. Environ. 2004, 91, 332–344. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Xgboost: A Scalable Tree Boosting System. arXiv 2016, arXiv:1603.02754. [Google Scholar]
- Mousa, S.R.; Bakhit, P.R.; Ishak, S. An Extreme Gradient Boosting Method for Identifying the Factors Contributing to Crash/near-Crash Events: A Naturalistic Driving Study. Can. J. Civ. Eng. 2019, 46, 712–721. [Google Scholar] [CrossRef]
- Zhang, J. Gradient Descent Based Optimization Algorithms for Deep Learning Models Training. arXiv 2019, arXiv:1903.03614. [Google Scholar]
- Chmura Kraemer, H.; Periyakoil, V.S.; Noda, A. Kappa Coefficients in Medical Research. Stat. Med. 2002, 21, 2109–2129. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J. A Coefficient of Agreement for Nominal Scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.-I. A Unified Approach to Interpreting Model Predictions. arXiv 2017, arXiv:1705.07874. [Google Scholar]
- Iqbal, N.; Mumtaz, R.; Shafi, U.; Zaidi, S.M.H. Gray Level Co-Occurrence Matrix (GLCM) Texture Based Crop Classification Using Low Altitude Remote Sensing Platforms. PeerJ Comput. Sci. 2021, 7, e536. [Google Scholar] [CrossRef]
- Chong, K.L.; Lai, S.H.; Ahmed, A.N.; Zaafar, W.Z.W.; Rao, R.V.; Sherif, M.; Sefelnasr, A.; El-Shafie, A. Review on Dam and Reservoir Optimal Operation for Irrigation and Hydropower Energy Generation Utilizing Meta-Heuristic Algorithms. IEEE Access 2021, 9, 19488–19505. [Google Scholar] [CrossRef]
- Vuolo, F.; Neuwirth, M.; Immitzer, M.; Atzberger, C.; Ng, W.-T. How Much Does Multi-Temporal Sentinel-2 Data Improve Crop Type Classification? Int. J. Appl. Earth Obs. Geoinf. 2018, 72, 122–130. [Google Scholar] [CrossRef]
- Xiang, K.; Yuan, W.; Wang, L.; Deng, Y. An LSWI-Based Method for Mapping Irrigated Areas in China Using Moderate-Resolution Satellite Data. Remote Sens. 2020, 12, 4181. [Google Scholar] [CrossRef]
- Owusu-Mensah, E.; Oduro, I.; Sarfo, K.J. Steeping: A way of improving the malting of rice grain: Improving the malting of rice grain. J. Food Biochem. 2011, 35, 80–91. [Google Scholar] [CrossRef]
- Wang, L.; Liu, J.; Yang, L.; Yang, F.; Fu, C. Application of Random Forest Method in Maize-Soybean Accurate Identification. Acta Agron. Sin. 2018, 44, 569–580. [Google Scholar] [CrossRef]



| Farm Name | Year | Proportion of Rice | Proportion of Maize | Proportion of Soybeans | Proportion of Wheat | AREA (m2) |
|---|---|---|---|---|---|---|
| Bawuling | 2019 | 0.751 | 0.201 | 0.048 | 0 | 788,374,229.43 |
| 2020 | 0.749 | 0.178 | 0.073 | 0 | 821,538,859.59 | |
| 2021 | 0.752 | 0.222 | 0.026 | 0 | 820,802,304.13 | |
| 2022 | 0.72 | 0.165 | 0.115 | 0 | 820,989,318.27 | |
| Bawuba | 2019 | 0.936 | 0.025 | 0.039 | 0 | 801,388,296.55 |
| 2020 | 0.939 | 0.015 | 0.046 | 0 | 801,942,534.54 | |
| 2021 | 0.927 | 0.031 | 0.041 | 0 | 801,667,709.59 | |
| 2022 | 0.895 | 0.027 | 0.078 | 0 | 806,883,923.12 | |
| Yunshan | 2019 | 0.525 | 0.171 | 0.305 | 0 | 560,784,029.20 |
| 2020 | 0.528 | 0.215 | 0.257 | 0 | 560,773,507.85 | |
| 2021 | 0.528 | 0.315 | 0.158 | 0 | 561,091,394.98 | |
| 2022 | 0.516 | 0.165 | 0.319 | 0 | 563,253,919.40 | |
| Junchuan | 2019 | 0.827 | 0.158 | 0.016 | 0 | 842,704,046.51 |
| 2020 | 0.827 | 0.129 | 0.044 | 0 | 842,557,865.42 | |
| 2021 | 0.829 | 0.159 | 0.012 | 0 | 842,484,184.65 | |
| 2022 | 0.822 | 0.093 | 0.085 | 0 | 845,874,235.93 | |
| Qixing | 2019 | 0.858 | 0.055 | 0.088 | 0 | 1,489,302,362.59 |
| 2020 | 0.847 | 0.062 | 0.092 | 0 | 1,494,554,545.85 | |
| 2021 | 0.825 | 0.081 | 0.094 | 0 | 1,493,962,739.02 | |
| 2022 | 0.832 | 0.076 | 0.092 | 0 | 1,494,063,878.37 | |
| Hongwei | 2019 | 0.944 | 0.008 | 0.048 | 0 | 621,707,476.94 |
| 2020 | 0.939 | 0.009 | 0.052 | 0 | 625,380,146.97 | |
| 2021 | 0.928 | 0.035 | 0.038 | 0 | 625,145,749.98 | |
| 2022 | 0.849 | 0.061 | 0.089 | 0 | 633,873,572.45 | |
| Longmen | 2019 | 0 | 0.068 | 0.924 | 0.008 | 401,765,003.04 |
| 2020 | 0 | 0.166 | 0.808 | 0.026 | 403,031,764.76 | |
| 2021 | 0 | 0.127 | 0.766 | 0.107 | 404,177,043.18 | |
| 2022 | 0 | 0.092 | 0.836 | 0.072 | 404,716,699.72 | |
| Longzhen | 2019 | 0.018 | 0.362 | 0.619 | 0 | 684,490,977.34 |
| 2020 | 0.021 | 0.303 | 0.676 | 0 | 680,682,141.26 | |
| 2021 | 0 | 0.2 | 0.729 | 0.071 | 680,930,624.84 | |
| 2022 | 0.017 | 0.321 | 0.661 | 0 | 681,397,904.22 | |
| Heshan | 2019 | 0 | 0.374 | 0.626 | 0 | 1,071,868,559.92 |
| 2020 | 0 | 0.377 | 0.622 | 0.001 | 1,067,989,960.08 | |
| 2021 | 0 | 0.379 | 0.621 | 0.001 | 1,067,959,640.31 | |
| 2022 | 0 | 0.318 | 0.681 | 0.001 | 1,064,687,937.30 | |
| Rongjun | 2019 | 0.001 | 0.297 | 0.694 | 0.008 | 6,695,068,01.95 |
| 2020 | 0.001 | 0.3 | 0.687 | 0.012 | 6,717,733,51.38 | |
| 2021 | 0 | 0.422 | 0.568 | 0.01 | 6,860,031,09.95 | |
| 2022 | 0.001 | 0.236 | 0.761 | 0.002 | 6,861,598,02.83 |
| Year | Farm Name | Kappa | Kappa of Rice | Kappa of Maize | Kappa of Soybeans | Kappa of Wheat |
|---|---|---|---|---|---|---|
| 2019 | Yunshan | 0.958 | 0.998 | 0.987 | 0.989 | - |
| 2020 | Yunshan | 0.949 | 0.974 | 0.937 | 0.929 | - |
| 2021 | Yunshan | 0.917 | 0.963 | 0.905 | 0.864 | - |
| 2022 | Yunshan | 0.793 | 0.793 | 0.967 | 0.695 | - |
| 2019 | Longzhen | 0.989 | - | 0.991 | 0.989 | 0.972 |
| 2020 | Longzhen | 0.875 | - | 0.896 | 0.874 | 0.755 |
| 2021 | Longzhen | 0.868 | - | 0.913 | 0.860 | 0.827 |
| 2022 | Longzhen | 0.794 | - | 0.928 | 0.790 | 0.585 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, J.; Zeng, W.; Ao, C.; Xing, W.; Gaiser, T.; Srivastava, A.K. Cross-Regional Crop Classification Based on Sentinel-2. Agronomy 2024, 14, 1084. https://doi.org/10.3390/agronomy14051084
He J, Zeng W, Ao C, Xing W, Gaiser T, Srivastava AK. Cross-Regional Crop Classification Based on Sentinel-2. Agronomy. 2024; 14(5):1084. https://doi.org/10.3390/agronomy14051084
Chicago/Turabian StyleHe, Jie, Wenzhi Zeng, Chang Ao, Weimin Xing, Thomas Gaiser, and Amit Kumar Srivastava. 2024. "Cross-Regional Crop Classification Based on Sentinel-2" Agronomy 14, no. 5: 1084. https://doi.org/10.3390/agronomy14051084
APA StyleHe, J., Zeng, W., Ao, C., Xing, W., Gaiser, T., & Srivastava, A. K. (2024). Cross-Regional Crop Classification Based on Sentinel-2. Agronomy, 14(5), 1084. https://doi.org/10.3390/agronomy14051084









