Comprehensive Evaluation of Apple Germplasm Genetic Diversity on the Basis of 26 Phenotypic Traits
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Data Collection
2.3. Data Analysis
2.3.1. Phenotypic Trait Diversity Analysis
2.3.2. Comprehensive Evaluation of Phenotypic Traits
3. Results
3.1. Diversity Analysis of Morphological Traits in Apple Germplasm Resources
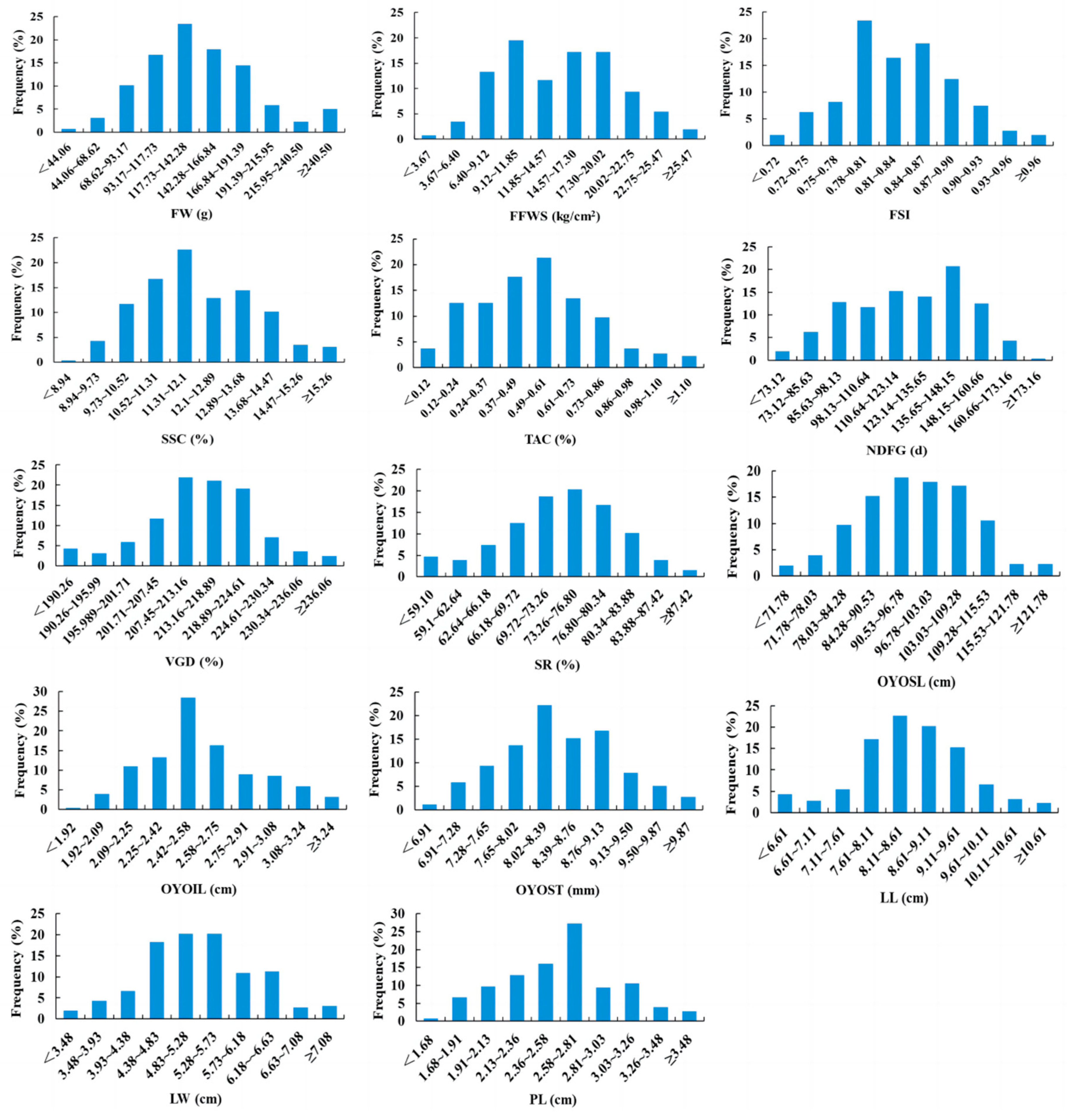
3.2. Diversity Analysis of Numerical Traits in Apple Germplasm Resources
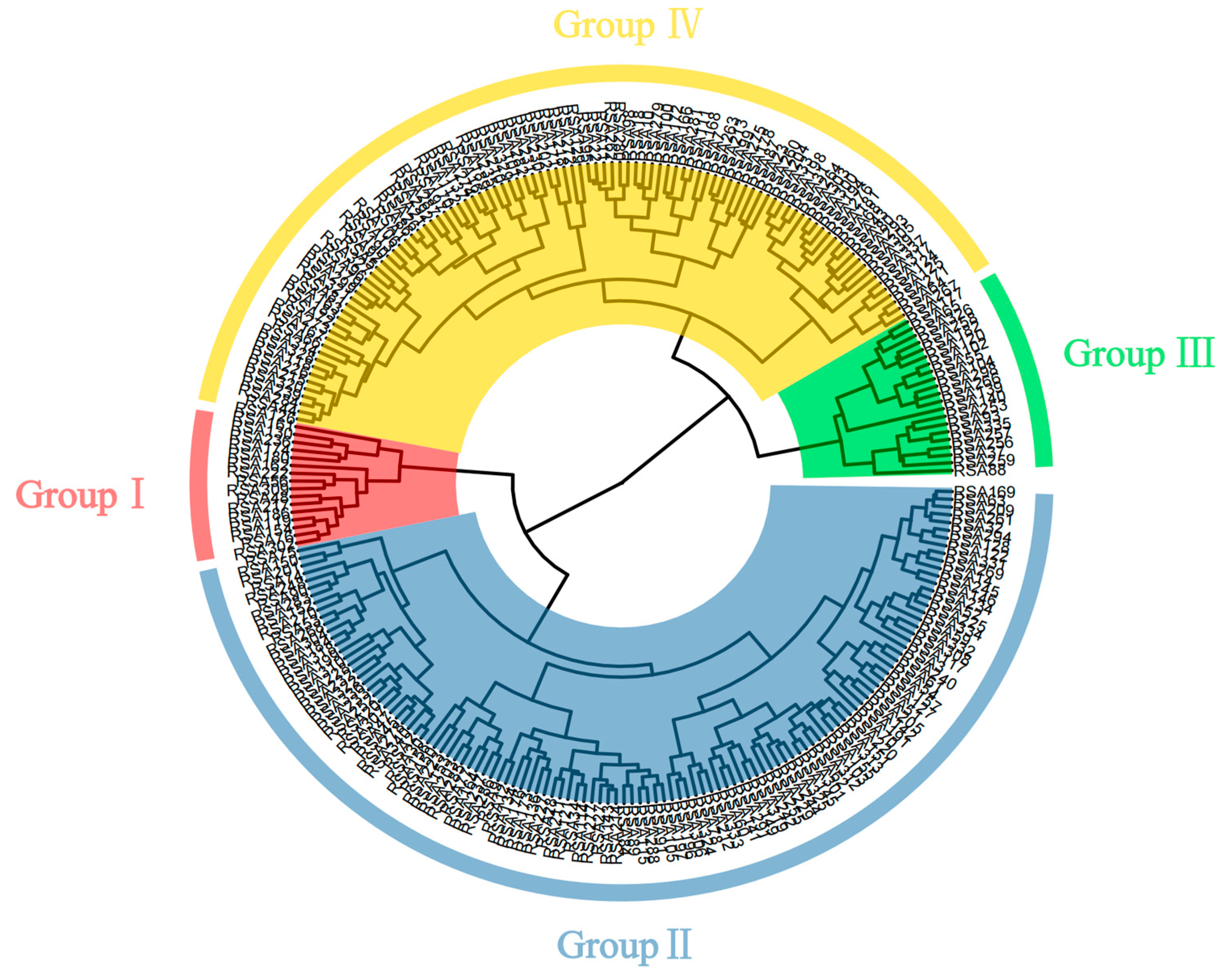
3.3. Cluster Analysis of Phenotypic Traits in Apple Germplasm Resources
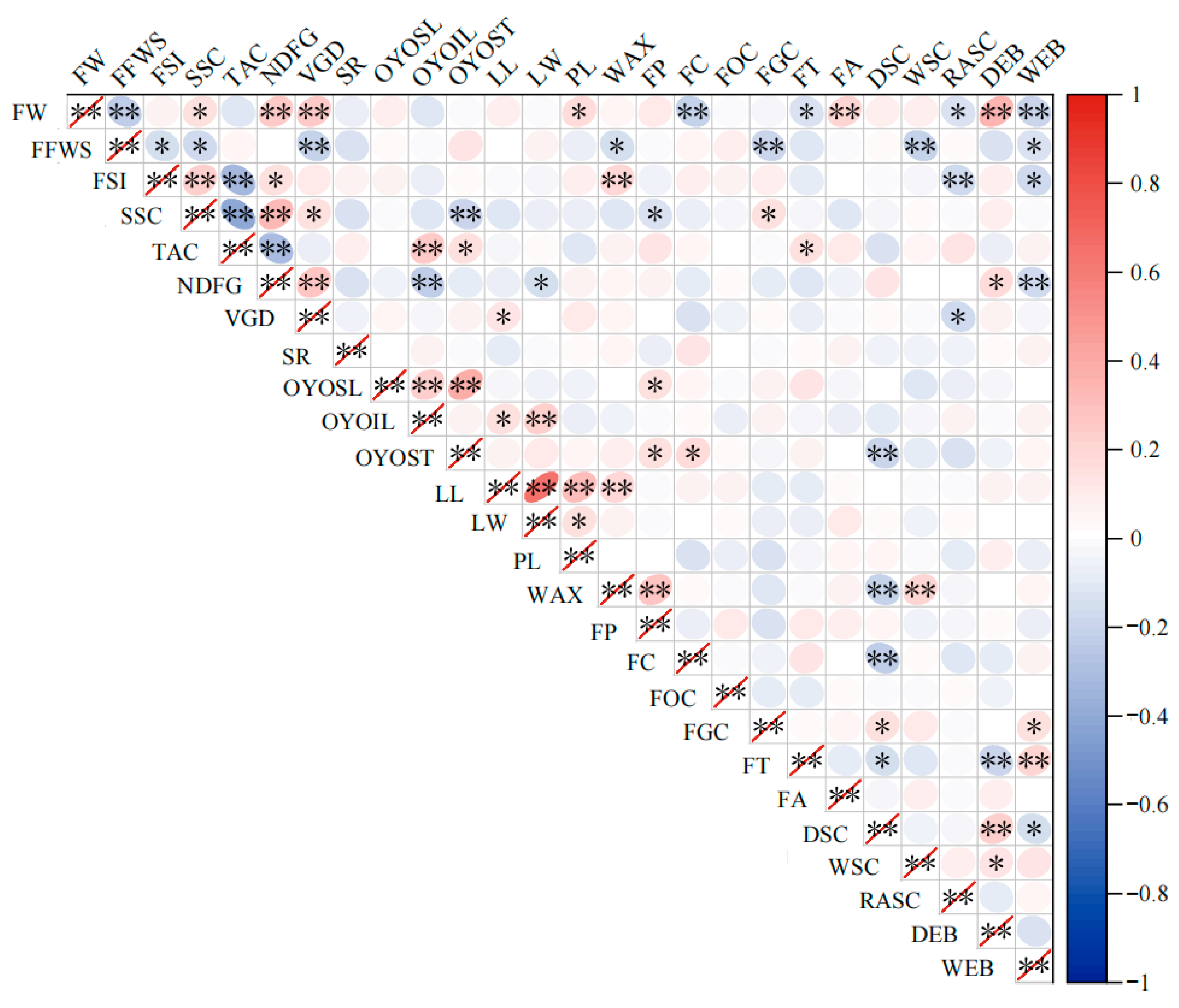
3.4. Correlation Analysis of Phenotypic Traits in Apple Germplasm Resources
3.5. Principal Component Analysis of Phenotypic Traits in Apple Germplasm Resources
3.6. Comprehensive Evaluation of the Phenotypic Traits of Apple Germplasm Resources and Identification of Excellent Germplasms
Comprehensive Evaluation of the Phenotypic Traits of Apple Germplasm Resources
0.10U13 + 0.13U14 + 0U15 − 0.02U16 − 0.19U17 − 0.01U18 + 0.04U19 − 0.19U20 + 0.01U21 + 0.19U22 + 0.06U23 − 0.11U24 +
0.27U25 − 0.19U26
3.7. Regression Model Establishment and Identification of Key Indices
0.006X9 + 0.002X6 + 0.156X21 + 0.041X4 + 0.128X15 + 0.030X22
4. Discussion
4.1. Genetic Diversity Associated with the Phenotypic Traits of Apple Germplasm Resources
4.2. Comprehensive Evaluation of Phenotypic Traits and Screening for Key Traits in Apple Germplasm Resources
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Song, J.; Amyotte, B.; Yu, C.H.; Campbell-Palmer, L.; Vinqvist-Tymchuk, M.; Rupasingh, H.P. Untargeted metabolomics analysis reveals the biochemical variations of polyphenols in a diverse apple population. Fruit Res. 2023, 3, 29. [Google Scholar] [CrossRef]
- Yuan, J.; Wang, Z.; Wang, X.; Zhang, C.; Ma, F.; Li, M. Research advances in genetic quality of sugar content in apples. Fruit Res. 2023, 3, 13. [Google Scholar] [CrossRef]
- Chen, X.; Wang, N.; Peng, F.; Mao, Z.; Yin, C.; Jiang, Y.; Ge, S.; Hu, D.; Li, Y.; Du, Y.; et al. Advances in quality and maturity breeding of important deciduous fruit trees in China. Acta Hortic. Sin. 2024, 51, 8–26. [Google Scholar]
- Mirheidari, F.; Khadivi, A.; Moradi, Y.; Paryan, S. Phenotypic characterization of Prunus haussknechtii Bornm., P. elaeagnifolia Spach, and P. orientalis Mill. Sci. Hortic. 2020, 265, 109273. [Google Scholar] [CrossRef]
- Zhang, Y.; Cao, Y.F.; Huo, H.L.; Xu, J.Y.; Tian, L.M.; Dong, X.G.; Qi, D.; Liu, C. An assessment of the genetic diversity of pear (Pyrus L.) germplasm resources based on the fruit phenotypic traits. J. Integr. Agric. 2022, 21, 2275–2290. [Google Scholar] [CrossRef]
- Sun, X.; Jiao, C.; Schwaninger, H.; Chao, C.T.; Ma, Y.; Duan, N.; Khan, A.; Ban, S.; Xu, K.; Cheng, L. Phased diploid genome assemblies and pan-genomes provide insights into the genetic history of apple domestication. Nat. Genet. 2020, 52, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Boquete, M.T.; Muyle, A.; Alonso, C. Plant epigenetics: Phenotypic and functional diversity beyond the DNA sequence. Am. J. Bot. 2021, 108, 553–558. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.C.; Ren, H.L.; Tang, S.M.; Zhu, L.; Zhang, S.J.; Ren, B. Genetic diversity analysis of phentypic traits in 160 germplasm resources of Malus sieversii (Ledeb.) M.Roem. from Tianshan in Ili. J. Plant Genet. Resour. 2021, 22, 1521–1530. [Google Scholar]
- Ganopoulos, I.; Tourvas, N.; Xanthopoulou, A.; Aravanopoulos, F.A.; Avramidou, E.; Zambounis, A.; Tsaftaris, A.; Madesis, P.; Sotiropoulos, T.; Koutinas, N. Phenotypic and molecular characterization of apple (Malus × domestica Borkh) genetic resources in Greece. Sci. Agric. 2018, 75, 509–518. [Google Scholar] [CrossRef]
- Tang, J.; Lu, L.; Ma, Y.F.; Tao, J.; Shi, Y. Factor analysis and evaluation index selection of fruit quality of apple varieties. China Fruits 2024, 01, 24–29. [Google Scholar]
- Noiton, D.A.; Alspach, P.A. Founding clones, inbreeding, coancestry, and status number of modern apple cultivars. J. Am. Soc. Hortic. Sci. 1996, 5, 773–782. [Google Scholar] [CrossRef]
- Ordidge, M.; Kirdwichai, P.; Baksh, M.F.; Venison, E.P.; Gibbings, J.G.; Dunwell, J.M. Genetic analysis of a major international collection of cultivated apple varieties reveals previously unknown historic heteroploid and inbred relationships. PLoS ONE 2018, 13, e020245. [Google Scholar] [CrossRef] [PubMed]
- Migicovsky, Z.; Gardner, K.M.; Richards, C.; Chao, C.T.; Schwaninger, H.R.; Fazio, G.; Zhong, G.Y.; Myles, S. Genomic consequences of apple improvement. Hortic. Res. 2021, 8, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Minamikawa, M.F.; Kunihisa, M.; Noshita, K.; Moriya, S.; Abe, K.; Hayashi, T.; Katayose, Y.; Matsumoto, T.; Nishitani, C.; Terakami, S.; et al. Tracing founder haplotypes of Japanese apple varieties: Application in genomic prediction and genome-wide association study. Hortic. Res. 2021, 8, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Verda, A.; Miguez, J.M.; Gallardo, M. Melatonin stimulates postharvest ripening of apples by up-regulating gene expression of ethylene synthesis enzymes. Postharvest Biol. Technol. 2023, 195, 112113. [Google Scholar] [CrossRef]
- Farneti, B.; Di Guardo, M.; Khomenko, I.; Cappellin, L.; Biasioli, F.; Velasco, R.; Costa, F. Genome-wide association study unravels the genetic control of the apple volatilome and its interplay with fruit texture. J. Exp. Bot. 2017, 7, 1467–1478. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Molloy, C.; Hunt, M.; Deng, C.H.; Wiedow, C.; Andre, C.; Dare, A.; McGhie, T. GWAS provides new insights into the genetic mechanisms of phytochemicals production and red skin colour in apple. Hortic. Res. 2022, 9, uhac218. [Google Scholar] [CrossRef]
- Goeltz, J.C.; Cuevas, L.A. Guided inquiry activity for teaching titration through total titratable Acidity in a general chemistry laboratory course. J. Chem. Educ. 2021, 98, 882–887. [Google Scholar] [CrossRef]
- Wang, K.; Liu, F.Z.; Cao, Y.F. Descriptors and Date Standard for Apple (Malus spp. Mill.); China Agriculture Press: Beijing, China, 2005; pp. 13–56. [Google Scholar]
- Niu, X.J.; Wang, X.D.; Wang, J.P.; Sun, J.; Qie, Y.M.; Wang, L.N.; Geng, L.G. Genetic diversity and comprehensive evaluation of Sorhum germplasm based on phenotypic traits. J. Plant Genet. Resour. 2024, 1, 1–17. [Google Scholar]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.Y. ggtree: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Moradi, Y.; Khadivi, A.; Mirheidari, F. Multivariate analysis of oriental apple (Malus orientalis Uglitzk.) based on phenotypic and pomological characterizations. Food Sci. Nutr. 2022, 10, 2532–2541. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.J.; Qi, Y.J.; Xing, X.H.; Tong, F.; Wang, X. Analysis and Evaluation of agronomic and quality traits in Soybean germplasms from Huang-Huai-Hai region. J. Plant Genet. Resour. 2022, 23, 468–480. [Google Scholar]
- Rao, Q.L.; Jiang, M.; Liu, X.Y.; Lu, J.W.; Hu, T.H.; Cheng, L.Q.; Wang, J.H.; Wang, J. Genetic diversity and comprehensive evaluation of 296 Peanuts germplasm resources in Guizhou. J. Plant Genet. Resour. 2024, 25, 1–18. [Google Scholar]
- Luo, P.S.; Huang, L.J.; Lu, M.Y.; Li, W.Y.; Jiang, J.J.; Wei, Y.; Yan, Z.L.; Zhao, J.; Zhou, J. Fruit traits diversity analysis and comprehensive evaluation of Sterculia monosperma Vent. germplasms. J. Plant Genet. Resour. 2024, 1, 1–12. [Google Scholar]
- Liu, D.; Wang, X.; Li, W.; Li, J.; Tan, W.; Xing, W. Genetic Diversity Analysis of the Phenotypic Traits of 215 Sugar Beet Germplasm Resources. Sugar Tech 2022, 24, 1790–1800. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y. Genetic diversity analysis of phenotypic traits among 37 Xanthoceras sorbifolium elite germplasms. J. For. Res. 2022, 27, 140–147. [Google Scholar] [CrossRef]
- Alizadeh, K.; Fatholahi, S.; Teixeira Da Silva, J.A. Variation in the fruit characteristics of local pear (Pyrus spp.) in the Northwest of Iran. Genet. Resour. Crop Evol. 2015, 62, 635–641. [Google Scholar] [CrossRef]
- Shuaib, M.; Bahadur, S.; Hussain, F. Enumeration of genetic diversity of wild rice through phenotypic trait analysis. Gene Rep. 2020, 21, 100797. [Google Scholar] [CrossRef]
- Zargar, S.A.; Wani, A.A.; Saggoo, M. Analysis of phenotypic diversity of apricot (Prunus armeniaca L.) accessions from Jammu and Kashmir, India. Plant Genet. Resour. 2021, 19, 203–215. [Google Scholar] [CrossRef]
- Li, Y.; Liu, H.C.; Shi, X.X.; Shi, L.; Han, X.; Liu, J.; Wei, Y.F. Phenotypic diversity analysis of 398 Naked Barley germplasm resources. J. Plant Genet. Resour. 2023, 24, 1311–1320. [Google Scholar]
- Mbovora, S.M.; Musvosvi, C.; Gasura, E. Morphological diversity among accessions of apple tree (Malus × Domestica Borkh). Adv. Agric. 2021, 2021, 705856. [Google Scholar] [CrossRef]
- Zhou, T.; Ning, K.; Zhang, W.; Chen, H.; Lu, X.; Zhang, D.; El-Kassaby, Y.A.; Bian, J. Phenotypic variation of floral organs in flowering crabapples and its taxonomic significance. BMC Plant Biol. 2021, 21, 503. [Google Scholar]
- Sun, S.M.; Gao, Y.; Wang, K.; Wang, D.J.; Li, L.W. Fruit quality analysis of 23 apple germplasm resources. J. Agric. Big Data 2022, 4, 13–19. [Google Scholar]
- Watts, S.; Migicovsky, Z.; Mcclure, K.A.; Yu, C.H.; Amyotte, B.; Baker, T.; Bowlby, D.; Burgher-Maclellan, K.; Butler, L.; Donald, R.; et al. Quantifying apple diversity: A phenomic characterization of Canada’s Apple Biodiversity Collection. Plants People Planet 2021, 3, 747–760. [Google Scholar] [CrossRef]
- Reig, G.; Blanco, Á.; Castillo, A.M.; Gogorcena, Y.; Moreno, M.Á. Phenotypic diversity of Spanish apple (Malus × domestica Borkh) accessions grown at the vulnerable climatic conditions of the Ebro Valley, Spain. Sci. Hortic. 2015, 185, 200–210. [Google Scholar] [CrossRef]
- Debbarma, P.; Hazarika, T. Genetic diversity of Bael [Aegle marmelos (L.) Corr.] accessions from north-east India based on principal component and cluster analysis. Genet. Resour. Crop Evol. 2024, 71, 253–277. [Google Scholar] [CrossRef]
- Kaiser, H.F. The varimax criterion for analytic rotation in factor analysis. Psychometrika 1958, 23, 187–200. [Google Scholar] [CrossRef]
- Mir, J.; Jan, A.; Rashid, M.; Singh, D.; Raja, W.; Sharma, O.; Sharma, A.; Lal, S.; Kumawat, K.; Nabi, S.U. Genetic variability studies for various morphological and quality traits in apple. Indian J. Hortic. 2020, 77, 227–236. [Google Scholar] [CrossRef]
- Chen, T.; Liu, L.; Zhou, Y.; Zheng, Q.; Luo, S.; Xiang, T.; Zhou, L.; Feng, S.; Yang, H.; Ding, C. Characterization and comprehensive evaluation of phenotypic characters in wild Camellia oleifera germplasm for conservation and breeding. Front. Plant Sci. 2023, 14, 1052890. [Google Scholar] [CrossRef]
- Zhang, W.; Lu, T.; Zeng, Q.; Wang, Z.; Yang, R.; Zhao, F. Genetic diversity analysis and screening of excellent germplasm of upland cotton resources. J. Plant Genet. Resour. 2024, 1, 1–14. [Google Scholar]
- Zhou, C.; Wu, H.; Sheng, Q.; Cao, F.; Zhu, Z. Study on the phenotypic diversity of 33 Ornamental Xanthoceras sorbifolium Cultivars. Plants 2023, 12, 2448. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Yao, Z.; Yang, Y.; Wang, Z.; Zou, H.; Fang, B.; Huang, L. Genetic fingerprint construction and genetic diversity analysis of sweet potato (Ipomoea batatas) germplasm resources. BMC Plant Biol. 2023, 23, 355. [Google Scholar] [CrossRef] [PubMed]
- Hasan, N.; Choudhary, S.; Naaz, N.; Sharma, N.; Laskar, R.A. Recent advancements in molecular marker-assisted selection and applications in plant breeding programmes. J. Genet. Eng. Biotechnol. 2021, 19, 128. [Google Scholar] [CrossRef] [PubMed]
- De Mori, G.; Cipriani, G. Marker-assisted selection in breeding for fruit trait improvement: A review. Int. J. Mol. Sci. 2023, 24, 8984. [Google Scholar] [CrossRef]
| Traits | Classification Standard |
|---|---|
| Wax | Absent = 0; Present = 1 |
| Fruit powder | Absent = 0; Present = 1 |
| Fruit core | Small = 3; Medium = 5; Large = 7 |
| Fruit over color | Red = 1; Light red = 2; Strong red = 3; Absent = 4 |
| Fruit ground color | Green = 1; Yellow–green = 2; Yellow = 3; Orange–yellow = 4 |
| Flesh texture | Crisp = 1; Spongy = 2; Tough = 3 |
| Fruit arris | Absent = 0; Present = 1 |
| Depth of stalk cavity | Shallow = 3; Medium = 5; Deep = 7 |
| Width of stalk cavity | Narrow = 3; Medium = 5; Broad = 7 |
| Russet amount on stalk cavity | Absent = 1; Little = 3; Medium = 5; Much = 7 |
| Depth of eye basin | Shallow = 3; Medium = 5; Deep = 7 |
| Width of eye basin | Narrow = 3; Medium = 5; Broad = 7 |
| Trait | Distribution Percentage (%) | CV/% | H′ | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 7 | |||
| WAX | 46.48 | 53.52 | - | - | - | - | - | 92.59 | 0.69 |
| FP | 77.34 | 22.66 | - | - | - | - | - | 182.17 | 0.54 |
| FC | - | - | - | 40.63 | - | 46.88 | 12.5 | 30.34 | 0.98 |
| FOC | - | 17.97 | 22.66 | 40.23 | 19.14 | - | - | 38.01 | 1.33 |
| FGC | - | 41.02 | 50.00 | 8.20 | 0.78 | - | - | 38.70 | 0.96 |
| FT | - | 84.38 | 10.16 | 5.47 | - | - | - | 43.47 | 0.53 |
| FA | 79.69 | 20.31 | - | - | - | - | - | 201.5 | 0.51 |
| DSC | - | - | - | 23.44 | - | 27.34 | 49.22 | 29.49 | 1.04 |
| WSC | - | - | - | 19.92 | - | 53.52 | 26.56 | 26.51 | 1.01 |
| RASC | - | 40.23 | - | 45.31 | - | 13.28 | 1.17 | 57.65 | 1.05 |
| DEB | - | - | - | 57.42 | - | 26.17 | 16.41 | 36.20 | 0.97 |
| WEB | - | - | - | 7.03 | - | 57.81 | 35.16 | 21.10 | 0.87 |
| Trait | Mean | Maximum | Minimum | SD | CV/% | H′ |
|---|---|---|---|---|---|---|
| FW (g) | 142.28 | 290.30 | 28.30 | 49.11 | 34.52 | 2.01 |
| FFWS (kg/cm2) | 14.57 | 28.3 | 0.90 | 5.45 | 37.41 | 2.06 |
| FSI | 0.84 | 1.02 | 0.69 | 0.06 | 7.14 | 2.04 |
| SSC (%) | 12.10 | 16.80 | 8.40 | 1.58 | 13.06 | 2.05 |
| TAC (%) | 0.52 | 1.24 | 0.06 | 0.26 | 50.00 | 2.03 |
| NDFG (d) | 123.14 | 174.00 | 69.00 | 25.01 | 20.3 | 2.08 |
| VGD (d) | 213.16 | 245.00 | 177.00 | 11.45 | 5.37 | 2.03 |
| SR (%) | 73.26 | 90.50 | 55.40 | 7.08 | 9.66 | 2.08 |
| OYOSL (cm) | 96.78 | 134.70 | 58.90 | 12.50 | 12.92 | 2.06 |
| OYOIL (cm) | 2.58 | 3.70 | 1.90 | 0.33 | 12.79 | 2.01 |
| OYOST (mm) | 8.39 | 10.60 | 6.50 | 0.74 | 8.82 | 2.08 |
| LL (cm) | 8.61 | 11.50 | 5.40 | 1.00 | 11.61 | 2.02 |
| LW (cm) | 5.28 | 8.10 | 2.80 | 0.90 | 17.05 | 2.05 |
| PL (cm) | 2.58 | 3.92 | 1.50 | 0.45 | 17.44 | 2.04 |
| Group | I | II | III | IV |
|---|---|---|---|---|
| FW (g) | 256.25 a | 166.3 b | 65.01 d | 110.95 c |
| FFWS (kg/cm2) | 11.13 c | 13.72 b | 14.58 b | 16.16 a |
| FSI | 0.82 a | 0.84 a | 0.82 a | 0.84 a |
| SSC (%) | 12.87 a | 12.31 ab | 12.31 ab | 11.66 c |
| TAC (%) | 0.50 b | 0.45 c | 0.53 ab | 0.58 a |
| NDFG (d) | 123.69 b | 130.43 a | 112.29 c | 116.06 c |
| VGD (d) | 213.25 ab | 216.38 a | 208.24 c | 210.28 bc |
| SR (%) | 71.03 a | 73.33 a | 71.60 a | 73.88 a |
| OYOSL (cm) | 98.83 a | 97.28 a | 91.04 b | 97.20 a |
| OYOIL (cm) | 2.59 a | 2.54 a | 2.63 a | 2.63 a |
| OYOST (mm) | 8.39 ab | 8.33 b | 8.35 ab | 8.47 a |
| LL (cm) | 8.39 a | 8.69 a | 8.73 a | 8.52 a |
| LW (cm) | 5.04 b | 5.24 ab | 5.51 a | 5.31 ab |
| PL (cm) | 2.54 ab | 2.65 a | 2.39 b | 2.55 ab |
| Traits | Principal Components | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| FW (g) | 0.553 | 0.333 | 0.121 | −0.001 | 0.338 | 0.166 | 0.021 | −0.019 | 0.045 | −0.191 |
| FFWS (kg/cm2) | −0.269 | −0.045 | −0.482 | −0.507 | −0.083 | −0.063 | −0.097 | 0.172 | 0.092 | −0.237 |
| FSI | 0.397 | 0.056 | 0.359 | −0.103 | −0.339 | −0.133 | −0.376 | 0.149 | −0.015 | 0.277 |
| SSC (%) | 0.542 | −0.290 | 0.147 | 0.098 | −0.345 | 0.011 | 0.219 | 0.229 | −0.018 | −0.018 |
| TAC (%) | −0.589 | 0.127 | 0.014 | 0.104 | 0.467 | 0.118 | 0.010 | −0.045 | −0.133 | −0.083 |
| NDFG (d) | 0.623 | −0.052 | 0.010 | −0.244 | 0.025 | −0.169 | 0.225 | 0.105 | −0.092 | −0.039 |
| VGD (d) | 0.396 | 0.241 | 0.250 | 0.003 | −0.064 | 0.151 | 0.298 | −0.119 | −0.065 | −0.103 |
| SR (%) | −0.170 | 0.010 | 0.214 | 0.140 | 0.003 | −0.004 | −0.522 | −0.305 | −0.211 | 0.353 |
| OYOSL (cm) | −0.131 | 0.133 | 0.416 | −0.379 | −0.030 | 0.499 | 0.061 | 0.258 | −0.013 | 0.049 |
| OYOIL (cm) | −0.376 | 0.216 | 0.004 | 0.169 | −0.134 | 0.444 | 0.008 | 0.299 | −0.427 | 0.102 |
| OYOST (mm) | −0.295 | 0.342 | 0.373 | −0.403 | −0.058 | 0.171 | 0.019 | 0.147 | 0.139 | −0.232 |
| LL (cm) | −0.041 | 0.747 | −0.218 | 0.189 | −0.365 | −0.065 | 0.158 | 0.020 | 0.081 | 0.045 |
| LW (cm) | −0.158 | 0.658 | −0.302 | 0.189 | −0.354 | 0.029 | 0.060 | 0.102 | 0.068 | 0.073 |
| PL (cm) | 0.214 | 0.460 | −0.099 | 0.017 | −0.124 | −0.040 | 0.044 | −0.484 | 0.028 | 0.008 |
| WAX | −0.002 | 0.385 | 0.380 | 0.068 | 0.199 | −0.476 | 0.049 | 0.140 | −0.184 | 0.187 |
| FP | −0.035 | 0.270 | 0.182 | −0.345 | 0.499 | −0.060 | 0.230 | 0.053 | 0.052 | 0.315 |
| FC | −0.317 | −0.013 | 0.303 | −0.085 | −0.320 | −0.245 | −0.314 | 0.070 | −0.017 | −0.282 |
| FOC | −0.023 | 0.071 | −0.106 | −0.206 | 0.065 | −0.252 | −0.088 | 0.355 | 0.401 | 0.416 |
| FGC | 0.060 | −0.231 | 0.207 | 0.401 | −0.089 | 0.455 | −0.077 | 0.185 | 0.332 | 0.083 |
| FT | −0.310 | −0.174 | 0.357 | −0.052 | −0.030 | 0.030 | 0.361 | −0.403 | 0.153 | 0.127 |
| FA | 0.014 | 0.256 | 0.035 | 0.165 | 0.337 | −0.030 | −0.339 | 0.027 | 0.457 | −0.210 |
| DSC | 0.320 | −0.027 | −0.399 | −0.023 | 0.152 | 0.434 | −0.116 | −0.079 | 0.161 | 0.273 |
| WSC | 0.106 | 0.038 | 0.160 | 0.530 | 0.266 | −0.238 | −0.013 | 0.300 | −0.025 | −0.263 |
| RASC | −0.189 | −0.160 | −0.300 | 0.238 | 0.174 | −0.126 | 0.282 | 0.318 | −0.165 | 0.187 |
| DEB | 0.448 | 0.232 | −0.037 | 0.141 | 0.246 | 0.215 | −0.268 | 0.064 | −0.162 | −0.073 |
| WEB | −0.315 | −0.073 | 0.253 | 0.432 | −0.117 | −0.025 | 0.27 | −0.027 | 0.352 | 0.052 |
| Eigenvalue | 2.719 | 2.126 | 1.758 | 1.682 | 1.59 | 1.467 | 1.295 | 1.201 | 1.044 | 1.028 |
| Contribution rate (%) | 10.456 | 8.175 | 6.761 | 6.468 | 6.114 | 5.644 | 4.98 | 4.618 | 4.016 | 3.953 |
| Cumulative contribution rate (%) | 10.456 | 18.632 | 25.393 | 31.861 | 37.975 | 43.619 | 48.599 | 53.217 | 57.233 | 61.186 |
| Germplasm Number | Principal Component Score | Scores | Ranking | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCA1 | PCA2 | PCA3 | PCA4 | PCA5 | PCA6 | PCA7 | PCA8 | PCA9 | PCA10 | |||
| Fuhong | 3.71 | 0.25 | 3.10 | 1.89 | 0.83 | 0.71 | −1.14 | −0.34 | 0.84 | 0.04 | 1.30 | 1 |
| Red Delicious | 1.37 | 2.72 | 3.74 | −1.46 | 1.90 | 1.23 | −1.19 | 1.35 | 1.37 | −0.37 | 1.23 | 2 |
| Huamei | 0.82 | 1.18 | 3.13 | 2.29 | 1.26 | 1.06 | −0.59 | −0.11 | 0.96 | −0.21 | 1.10 | 3 |
| Weixi | 3.10 | −0.85 | 4.87 | −0.63 | −1.89 | 1.88 | 1.66 | 0.79 | 1.28 | −1.79 | 1.03 | 4 |
| Lvguang | 1.35 | 3.00 | 3.56 | −1.40 | −0.69 | −0.65 | 1.43 | 0.07 | 1.47 | 0.88 | 1.02 | 5 |
| Beni Shogun | 3.04 | −0.48 | 2.60 | 0.42 | −0.98 | 1.55 | 0.83 | 0.36 | 0.12 | 0.48 | 0.97 | 6 |
| Aixian | 0.90 | 2.84 | 0.73 | −0.29 | 3.42 | −0.30 | −0.69 | 0.17 | 0.55 | 0.03 | 0.89 | 7 |
| Dailv | 0.63 | 3.94 | 1.50 | 1.91 | −0.95 | −1.00 | 0.61 | −0.13 | 0.71 | −0.48 | 0.87 | 8 |
| Anweii Spurdei | 2.35 | 0.34 | −0.07 | −0.08 | 1.24 | 0.00 | 1.48 | 0.25 | 0.97 | 1.15 | 0.83 | 9 |
| Qingxiang | 3.47 | 1.17 | 0.76 | −0.17 | −1.17 | −1.36 | 1.06 | 0.96 | 0.61 | 0.32 | 0.79 | 10 |
| Trait | Correlation Coefficient | Trait | Correlation Coefficient |
|---|---|---|---|
| FW | 0.71 ** | WAX | 0.36 ** |
| FFWS | −0.51 ** | FP | 0.27 ** |
| FSI | 0.26 ** | FC | −0.33 ** |
| SSC | 0.38 ** | FOC | 0.132 * |
| TAC | −0.25 ** | FGC | 0.28 ** |
| NDFG | 0.41 ** | FT | −0.14 * |
| VGD | 0.69 ** | FA | 0.21 ** |
| SR | −0.13 * | DSC | 0.30 ** |
| OYOSL | 0.27 ** | WSC | 0.25 ** |
| OYOIL | −0.036 | RASC | −0.081 |
| OYOST | 0.037 | DEB | 0.53 ** |
| LL | 0.30 ** | WEB | 0.034 |
| LW | 0.182 * | ||
| PL | 0.27 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, W.; Li, Z.; Wang, L.; Sun, S.; Wang, D.; Wang, K.; Wang, G.; Liu, Z.; Lu, X.; Feng, J.; et al. Comprehensive Evaluation of Apple Germplasm Genetic Diversity on the Basis of 26 Phenotypic Traits. Agronomy 2024, 14, 1264. https://doi.org/10.3390/agronomy14061264
Tian W, Li Z, Wang L, Sun S, Wang D, Wang K, Wang G, Liu Z, Lu X, Feng J, et al. Comprehensive Evaluation of Apple Germplasm Genetic Diversity on the Basis of 26 Phenotypic Traits. Agronomy. 2024; 14(6):1264. https://doi.org/10.3390/agronomy14061264
Chicago/Turabian StyleTian, Wen, Zichen Li, Lin Wang, Simiao Sun, Dajiang Wang, Kun Wang, Guangyi Wang, Zhao Liu, Xiang Lu, Jianrong Feng, and et al. 2024. "Comprehensive Evaluation of Apple Germplasm Genetic Diversity on the Basis of 26 Phenotypic Traits" Agronomy 14, no. 6: 1264. https://doi.org/10.3390/agronomy14061264





