Rapid Identification of Rhizobia Nodulating Soybean by a High-Resolution Melting Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reference Strains and DNA Isolation
2.2. Crude Extract Preparation from Soybean Nodules
2.3. HRM-PCR Conditions
2.4. Verification of HRM Results
2.5. Statistical Analysis
3. Results
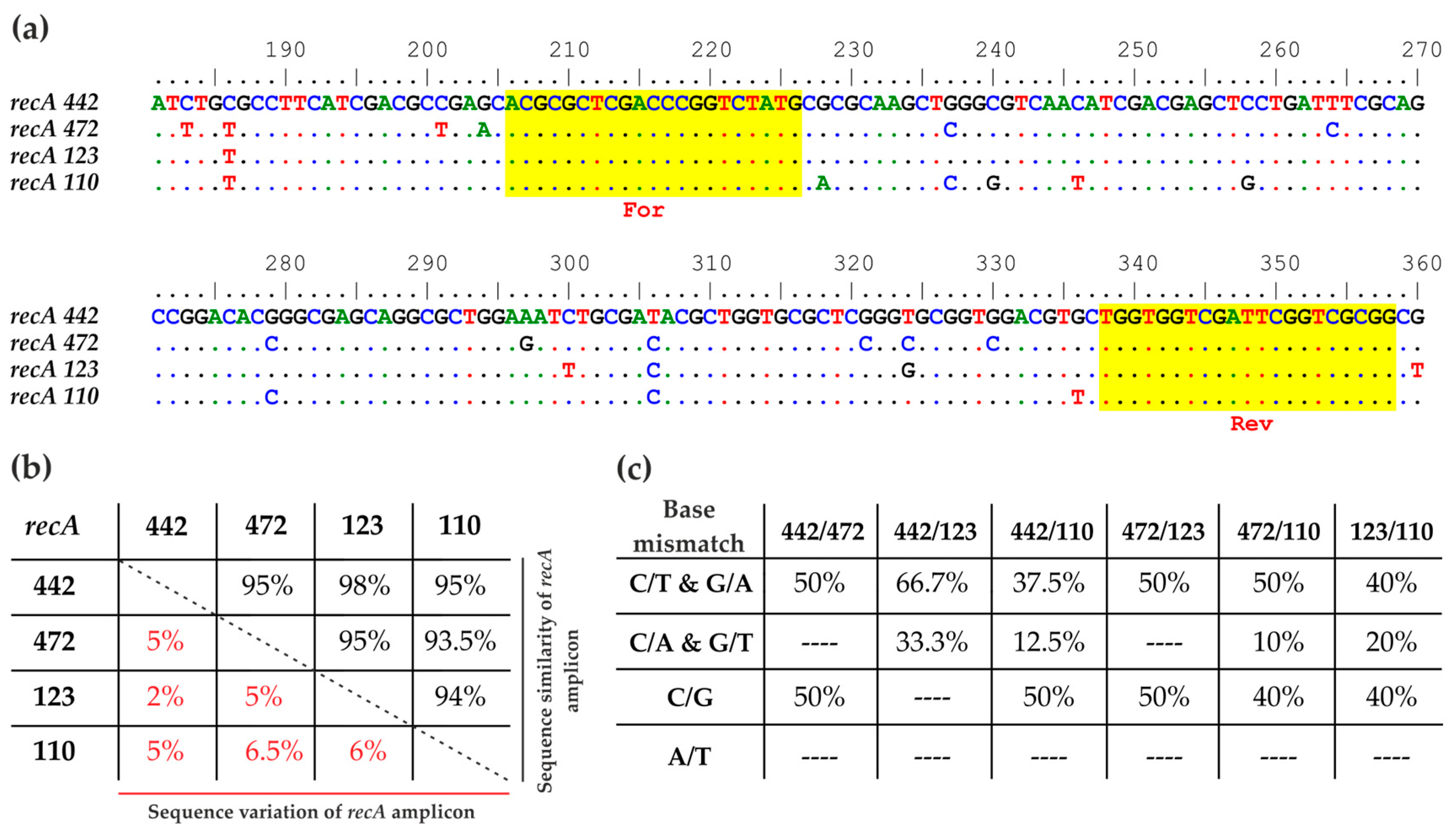
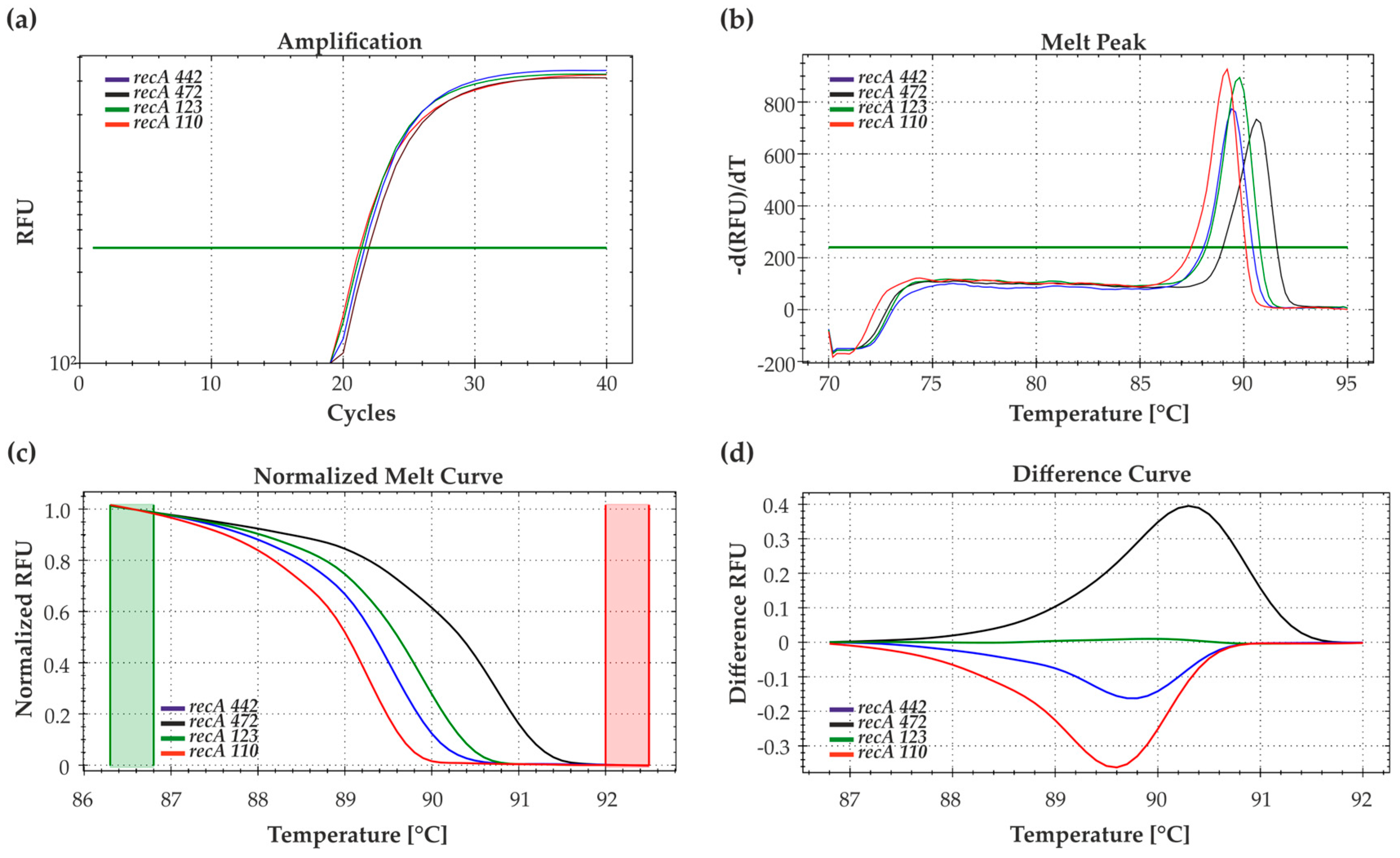
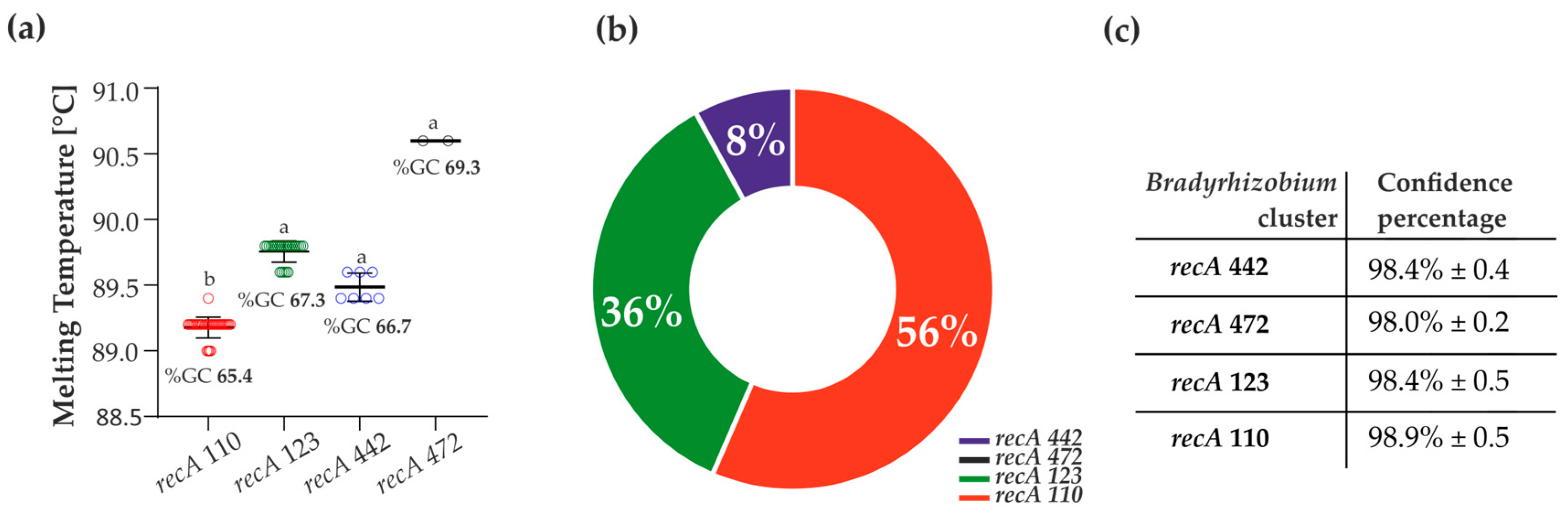
3.1. HRM-PCR Analysis of the Bradyrhizobium Reference Strains
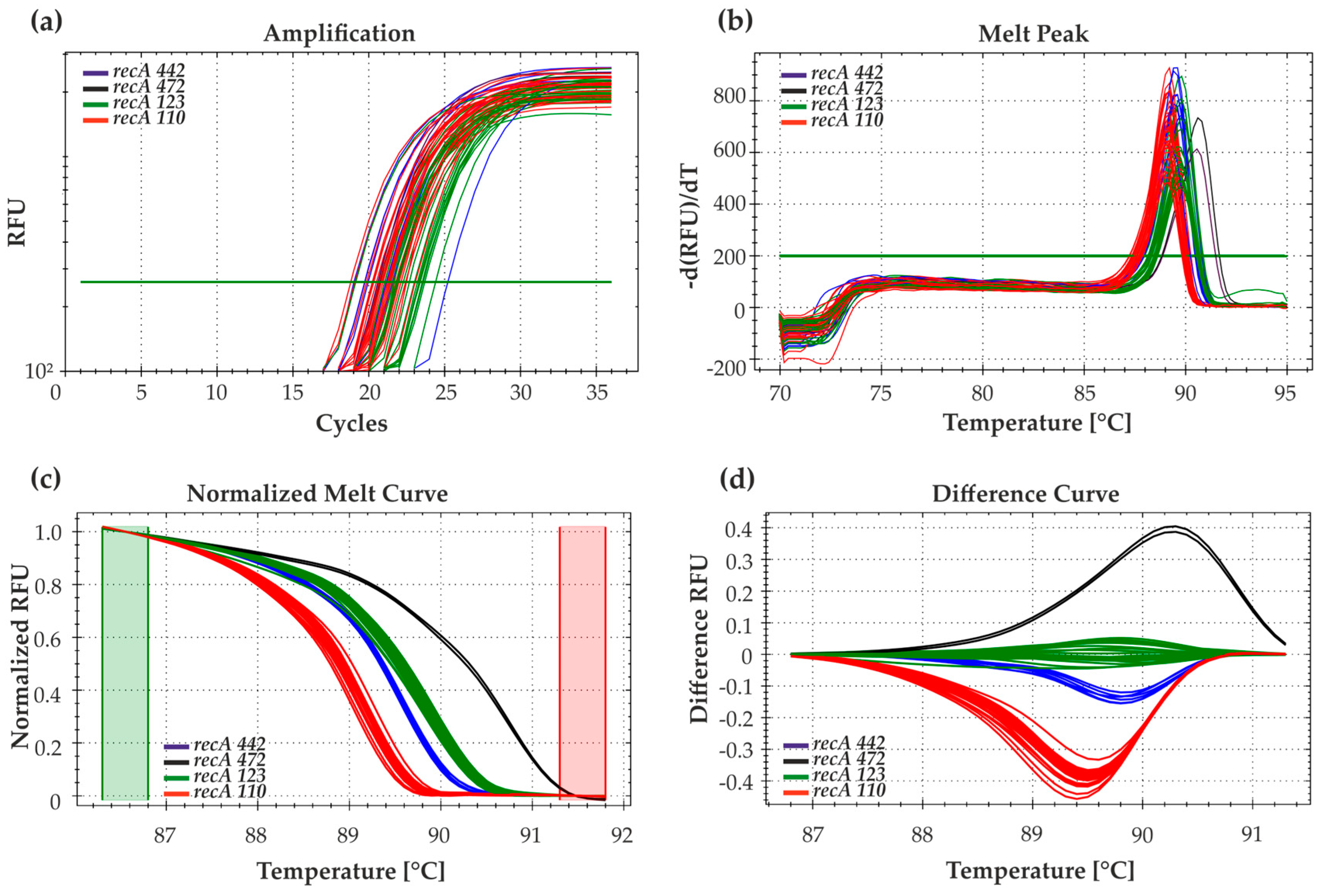
3.2. Detection of Soybean-Nodulating Bacteria Using HRM-PCR
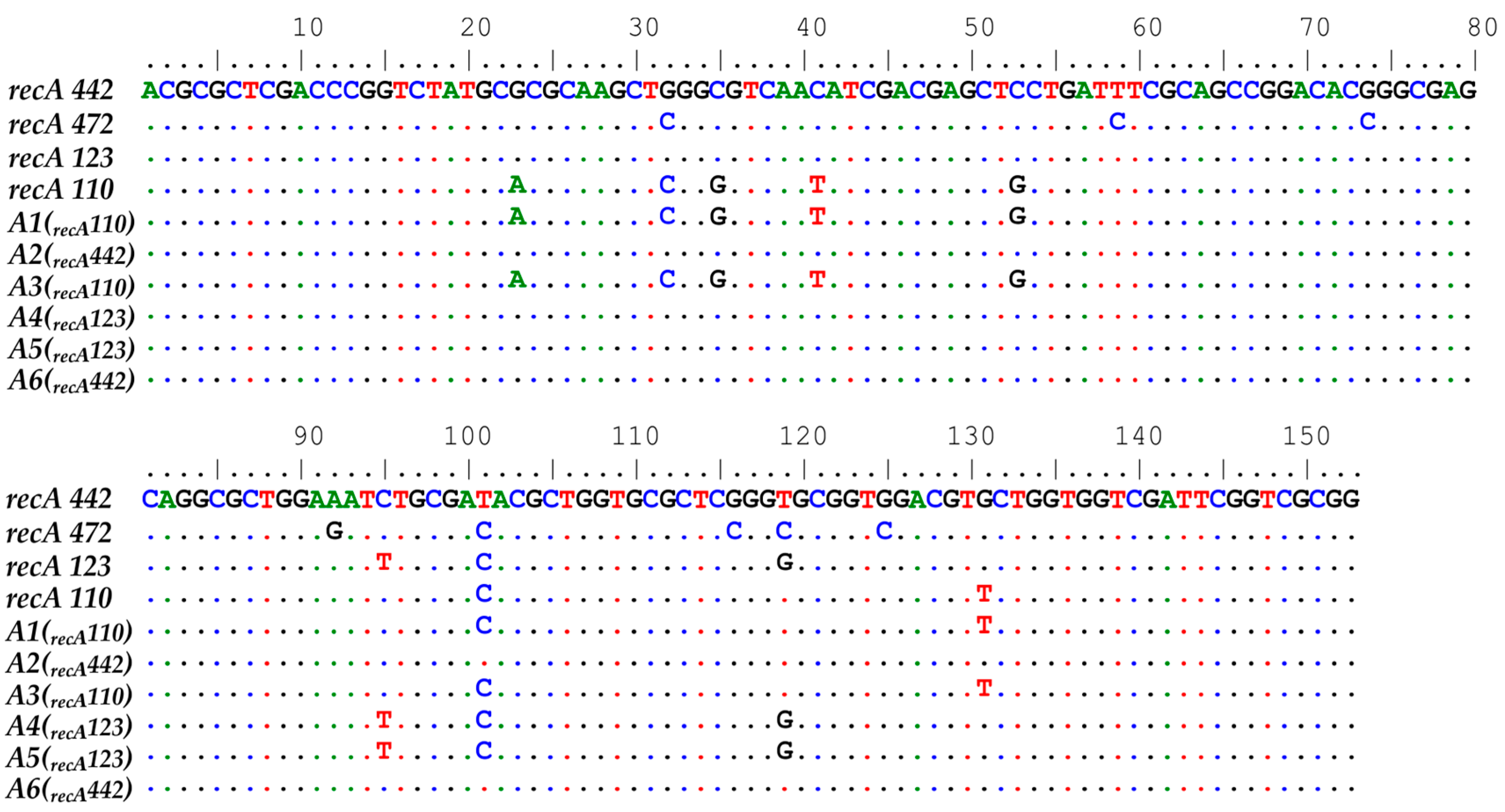
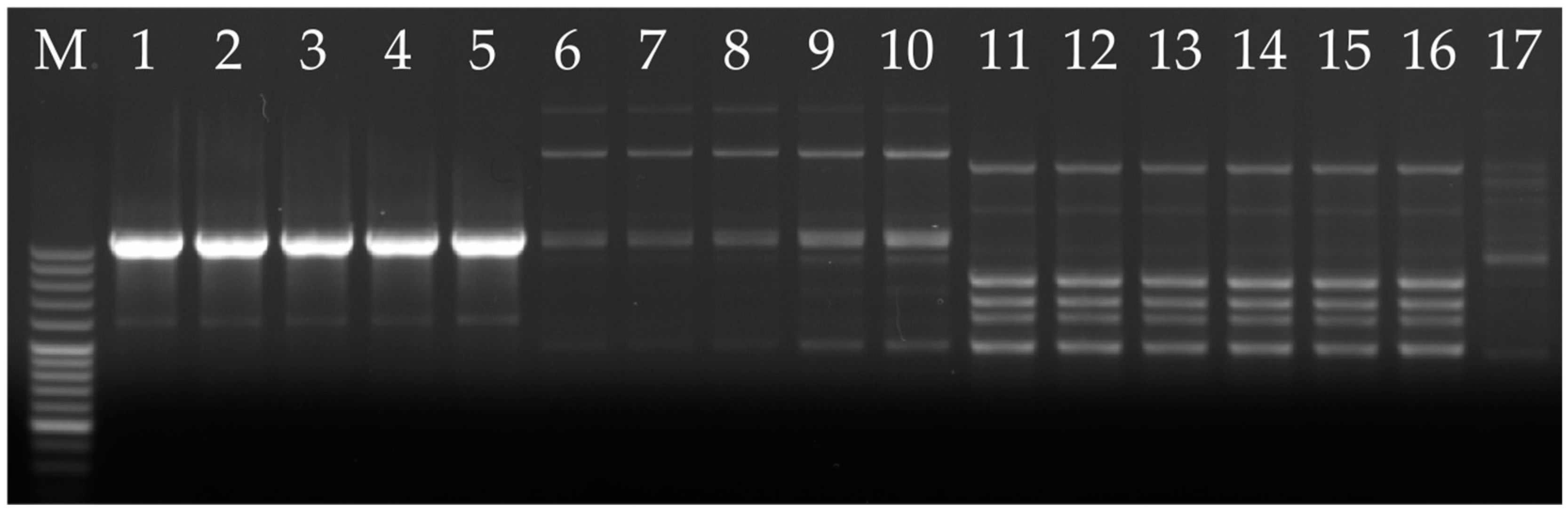
3.3. Verification of the HRM Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nendel, C.; Reckling, M.; Debaeke, P.; Schulz, S.; Berg-Mohnicke, M.; Constantin, J.; Fronzek, S.; Hoffmann, M.; Jakšić, S.; Kersebaum, K.-C.; et al. Future Area Expansion Outweighs Increasing Drought Risk for Soybean in Europe. Glob. Chang. Biol. 2023, 29, 1340–1358. [Google Scholar] [CrossRef]
- Karges, K.; Bellingrath-Kimura, S.D.; Watson, C.A.; Stoddard, F.L.; Halwani, M.; Reckling, M. Agro-Economic Prospects for Expanding Soybean Production beyond Its Current Northerly Limit in Europe. Eur. J. Agron. 2022, 133, 126415. [Google Scholar] [CrossRef]
- Roy, S.; Liu, W.; Nandety, R.S.; Crook, A.; Mysore, K.S.; Pislariu, C.I.; Frugoli, J.; Dickstein, R.; Udvardi, M.K. Celebrating 20 Years of Genetic Discoveries in Legume Nodulation and Symbiotic Nitrogen Fixation. Plant Cell 2020, 32, 15–41. [Google Scholar] [CrossRef] [PubMed]
- Debaeke, P.; Forslund, A.; Guyomard, H.; Schmitt, B.; Tibi, A. Could Domestic Soybean Production Avoid Europe’s Protein Imports in 2050? OCL 2022, 29, 38. [Google Scholar] [CrossRef]
- Wójcik-Gront, E.; Gozdowski, D.; Derejko, A.; Pudełko, R. Analysis of the Impact of Environmental and Agronomic Variables on Agronomic Parameters in Soybean Cultivation Based on Long-Term Data. Plants 2022, 11, 2922. [Google Scholar] [CrossRef] [PubMed]
- Maluk, M.; Giles, M.; Wardell, G.E.; Akramin, A.T.; Ferrando-Molina, F.; Murdoch, A.; Barros, M.; Beukes, C.; Vasconçelos, M.; Harrison, E.; et al. Biological Nitrogen Fixation by Soybean (Glycine max [L.] Merr.), a Novel, High Protein Crop in Scotland, Requires Inoculation with Non-Native Bradyrhizobia. Front. Agron. 2023, 5, 1196873. [Google Scholar] [CrossRef]
- Narożna, D.; Pudełko, K.; Króliczak, J.; Golińska, B.; Sugawara, M.; Mądrzak, C.J.; Sadowsky, M.J. Survival and Competitiveness of Bradyrhizobium Japonicum Strains 20 Years after Introduction into Field Locations in Poland. Appl. Environ. Microbiol. 2015, 81, 5552–5559. [Google Scholar] [CrossRef]
- Sadowsky, M.J.; Tully, R.E.; Cregan, P.B.; Keyser, H.H. Genetic Diversity in Bradyrhizobium japonicum Serogroup 123 and Its Relation to Genotype-Specific Nodulation of Soybean. Appl. Environ. Microbiol. 1987, 53, 2624–2630. [Google Scholar] [CrossRef] [PubMed]
- Richardson, A.E.; Viccars, L.A.; Watson, J.M.; Gibson, A.H. Differentiation of Rhizobium Strains Using the Polymerase Chain Reaction with Random and Directed Primers. Soil Biol. Biochem. 1995, 27, 515–524. [Google Scholar] [CrossRef]
- Shiro, S.; Matsuura, S.; Saiki, R.; Sigua, G.C.; Yamamoto, A.; Umehara, Y.; Hayashi, M.; Saeki, Y. Genetic Diversity and Geographical Distribution of Indigenous Soybean-Nodulating Bradyrhizobia in the United States. Appl. Environ. Microbiol. 2013, 79, 3610–3618. [Google Scholar] [CrossRef]
- Banasiewicz, J.; Granada, C.E.; Lisboa, B.B.; Grzesiuk, M.; Matuśkiewicz, W.; Bałka, M.; Schlindwein, G.; Vargas, L.K.; Passaglia, L.M.P.; Stępkowski, T. Diversity and Phylogenetic Affinities of Bradyrhizobium Isolates from Pampa and Atlantic Forest Biomes. Syst. Appl. Microbiol. 2021, 44, 126203. [Google Scholar] [CrossRef] [PubMed]
- Ferraz Helene, L.C.; O’Hara, G.; Hungria, M. Characterization of Bradyrhizobium Strains Indigenous to Western Australia and South Africa Indicates Remarkable Genetic Diversity and Reveals Putative New Species. Syst. Appl. Microbiol. 2020, 43, 126053. [Google Scholar] [CrossRef]
- Azarias Guimarães, A.; Florentino, L.A.; Alves Almeida, K.; Lebbe, L.; Barroso Silva, K.; Willems, A.; de Souza Moreira, F.M. High Diversity of Bradyrhizobium Strains Isolated from Several Legume Species and Land Uses in Brazilian Tropical Ecosystems. Syst. Appl. Microbiol. 2015, 38, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Degefu, T.; Wolde-meskel, E.; Rasche, F. Genetic Diversity and Symbiotic Effectiveness of Bradyrhizobium Strains Nodulating Selected Annual Grain Legumes Growing in Ethiopia. Int. J. Syst. Evol. Microbiol. 2018, 68, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Ormeño-Orrillo, E.; Martínez-Romero, E. A Genomotaxonomy View of the Bradyrhizobium Genus. Front. Microbiol. 2019, 10, 1334. [Google Scholar] [CrossRef] [PubMed]
- Berrada, H.; Fikri-Benbrahim, K. Taxonomy of the Rhizobia: Current Perspectives. Microbiol. Res. J. Int. 2014, 4, 616–639. [Google Scholar] [CrossRef]
- Liew, M.; Pryor, R.; Palais, R.; Meadows, C.; Erali, M.; Lyon, E.; Wittwer, C. Genotyping of Single-Nucleotide Polymorphisms by High-Resolution Melting of Small Amplicons. Clin. Chem. 2004, 50, 1156–1164. [Google Scholar] [CrossRef] [PubMed]
- Tucker, E.J.; Huynh, B.L. Genotyping by High-Resolution Melting Analysis. In Crop Breeding: Methods and Protocols; Fleury, D., Whitford, R., Eds.; Springer: New York, NY, USA, 2014; pp. 59–66. ISBN 978-1-4939-0446-4. [Google Scholar]
- Wittwer, C.T.; Hemmert, A.C.; Kent, J.O.; Rejali, N.A. DNA Melting Analysis. Mol. Asp. Med. 2024, 97, 101268. [Google Scholar] [CrossRef]
- Simko, I. High-Resolution DNA Melting Analysis in Plant Research. Trends Plant Sci. 2016, 21, 528–537. [Google Scholar] [CrossRef] [PubMed]
- Porcellato, D.; Grønnevik, H.; Rudi, K.; Narvhus, J.; Skeie, S.B. Rapid Lactic Acid Bacteria Identification in Dairy Products by High-resolution Melt Analysis of DGGE Bands. Lett. Appl. Microbiol. 2012, 54, 344–351. [Google Scholar] [CrossRef]
- Martini, M.; Moruzzi, S.; Ermacora, P.; Loi, N.; Firrao, G. Quantitative Real-Time PCR and High-Resolution Melting (HRM) Analysis for Strain-Specific Monitoring of Fluorescent Pseudomonads Used as Biocontrol Agents against Soil-Borne Pathogens of Food Crops. Trends Food Sci. Technol. 2015, 46, 277–285. [Google Scholar] [CrossRef]
- Sakaridis, I.; Ganopoulos, I.; Soultos, N.; Madesis, P.; Tsaftaris, A.; Argiriou, A. Identification of Lactic Acid Bacteria Isolated from Poultry Carcasses by High-Resolution Melting (HRM) Analysis. Eur. Food Res. Technol. 2014, 238, 691–697. [Google Scholar] [CrossRef]
- Parlapani, F.F.; Syropoulou, F.; Tsiartsafis, A.; Ekonomou, S.; Madesis, P.; Exadactylos, A.; Boziaris, I.S. HRM Analysis as a Tool to Facilitate Identification of Bacteria from Mussels during Storage at 4 °C. Food Microbiol. 2020, 85, 103304. [Google Scholar] [CrossRef] [PubMed]
- Iacumin, L.; Ginaldi, F.; Manzano, M.; Anastasi, V.; Reale, A.; Zotta, T.; Rossi, F.; Coppola, R.; Comi, G. High Resolution Melting Analysis (HRM) as a New Tool for the Identification of Species Belonging to the Lactobacillus Casei Group and Comparison with Species-Specific PCRs and Multiplex PCR. Food Microbiol. 2015, 46, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Keikha, M.; Karbalaei, M. High Resolution Melting Assay as a Reliable Method for Diagnosing Drug-Resistant TB Cases: A Systematic Review and Meta-Analysis. BMC Infect. Dis. 2021, 21, 989. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.K.; Anthwal, D.; Bhalla, M.; Tyagi, J.S.; Choudhary, S.; Haldar, S. Direct Detection of Fluoroquinolone Resistance in Sputum Samples from Tuberculosis Patients by High Resolution Melt Curve Analysis. Curr. Microbiol. 2023, 81, 27. [Google Scholar] [CrossRef]
- Delamuta, J.R.M.; Ribeiro, R.A.; Ormeño-Orrillo, E.; Melo, I.S.; Martínez-Romero, E.; Hungria, M. Polyphasic Evidence Supporting the Reclassification of Bradyrhizobium Japonicum Group Ia Strains as Bradyrhizobium Diazoefficiens sp. Nov. Int. J. Syst. Evol. Microbiol. 2013, 63, 3342–3351. [Google Scholar] [CrossRef]
- Somasegaran, P.; Hoben, H.J. Collecting Nodules and Isolating Rhizobia. In Handbook for Rhizobia: Methods in Legume-Rhizobium Technology; Somasegaran, P., Hoben, H.J., Eds.; Springer: New York, NY, USA, 1994; pp. 7–23. ISBN 978-1-4613-8375-8. [Google Scholar]
- Hall, T.; Biosciences, I.; Carlsbad, C. BioEdit: An Important Software for Molecular Biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Kibbe, W.A. OligoCalc: An Online Oligonucleotide Properties Calculator. Nucleic Acids Res. 2007, 35, W43–W46. [Google Scholar] [CrossRef]
- Ramakers, C.; Ruijter, J.M.; Deprez, R.H.L.; Moorman, A.F.M. Assumption-Free Analysis of Quantitative Real-Time Polymerase Chain Reaction (PCR) Data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Herridge, D.F.; Giller, K.E.; Jensen, E.S.; Peoples, M.B. Quantifying Country-to-Global Scale Nitrogen Fixation for Grain Legumes II. Coefficients, Templates and Estimates for Soybean, Groundnut and Pulses. Plant Soil 2022, 474, 1–15. [Google Scholar] [CrossRef]
- de Filippis, I.; de Azevedo, A.C.; Lima, I.d.O.; Ramos, N.F.L.; de Andrade, C.F.; de Almeida, A.E. Accurate, Fast and Cost-Effective Simultaneous Detection of Bacterial Meningitis by Qualitative PCR with High-Resolution Melting. BioTechniques 2023, 74, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Stępkowski, T.; Żak, M.; Moulin, L.; Króliczak, J.; Golińska, B.; Narożna, D.; Safronova, V.I.; Mądrzak, C.J. Bradyrhizobium Canariense and Bradyrhizobium Japonicum Are the Two Dominant Rhizobium Species in Root Nodules of Lupin and Serradella Plants Growing in Europe. Syst. Appl. Microbiol. 2011, 34, 368–375. [Google Scholar] [CrossRef]
- Stępkowski, T.; Watkin, E.; McInnes, A.; Gurda, D.; Gracz, J.; Steenkamp, E.T. Distinct Bradyrhizbium Communities Nodulate Legumes Native to Temperate and Tropical Monsoon Australia. Mol. Phylogenet. Evol. 2012, 63, 265–277. [Google Scholar] [CrossRef]
- van Berkum, P.; Fuhrmann, J.J. Evolutionary Relationships among the Soybean Bradyrhizobia Reconstructed from 16S rRNA Gene and Internally Transcribed Spacer Region Sequence Divergence. Int. J. Syst. Evol. Microbiol. 2000, 50 Pt 6, 2165–2172. [Google Scholar] [CrossRef] [PubMed]
- Osei, O.; Simões Araújo, J.L.; Zilli, J.E.; Boddey, R.M.; Ahiabor, B.D.K.; Abaidoo, R.C.; Rouws, L.F.M. PCR Assay for Direct Specific Detection of Bradyrhizobium Elite Strain BR 3262 in Root Nodule Extracts of Soil-Grown Cowpea. Plant Soil 2017, 417, 535–548. [Google Scholar] [CrossRef]
- Sikora, S.; Redžepović, S. Genotypic Characterisation of Indigenous Soybean Rhizobia by PCR-RFLP of 16S rDNA, Rep-PCR and RAPD Analysis. Food Technol. Biotechnol. 2003, 41, 61–67. [Google Scholar]
- Germano, M.G.; Menna, P.; Mostasso, F.L.; Hungria, M. RFLP Analysis of the rRNA Operon of a Brazilian Collection of Bradyrhizobial Strains from 33 Legume Species. Int. J. Syst. Evol. Microbiol. 2006, 56, 217–229. [Google Scholar] [CrossRef]
- Da Silva, C.G.N.; Vidal, M.S.; Dourado, F.d.S.; Dias, E.S.; Howe, A.C.; Jesus, E.d.C. Specific Primers for the Rapid Detection and Quantitation of Rhizobium Elite Strains of Common Beans in the Plant and Environment. Appl. Soil Ecol. 2024, 193, 105156. [Google Scholar] [CrossRef]
- Vossen, R.H.A.M.; Aten, E.; Roos, A.; den Dunnen, J.T. High-Resolution Melting Analysis (HRMA)—More than Just Sequence Variant Screening. Hum. Mutat. 2009, 30, 860–866. [Google Scholar] [CrossRef]
- Gori, A.; Cerboneschi, M.; Tegli, S. High-Resolution Melting Analysis as a Powerful Tool to Discriminate and Genotype Pseudomonas Savastanoi Pathovars and Strains. PLoS ONE 2012, 7, e30199. [Google Scholar] [CrossRef]
- Landolt, P.; Stephan, R.; Scherrer, S. Development of a New High Resolution Melting (HRM) Assay for Identification and Differentiation of Mycobacterium Tuberculosis Complex Samples. Sci. Rep. 2019, 9, 1850. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.; Huggett, J. qPCR Primer Design Revisited. Biomol. Detect. Quantif. 2017, 14, 19–28. [Google Scholar] [CrossRef]
- Wong, Y.P.; Chua, K.H.; Thong, K.L. One-Step Species-Specific High Resolution Melting Analysis for Nosocomial Bacteria Detection. J. Microbiol. Methods 2014, 107, 133–137. [Google Scholar] [CrossRef]
- Yuan, K.; Reckling, M.; Ramirez, M.D.A.; Djedidi, S.; Fukuhara, I.; Ohyama, T.; Yokoyama, T.; Bellingrath-Kimura, S.D.; Halwani, M.; Egamberdieva, D.; et al. Characterization of Rhizobia for the Improvement of Soybean Cultivation at Cold Conditions in Central Europe. Microbes Environ. 2020, 35, ME19124. [Google Scholar] [CrossRef] [PubMed]
- Delamuta, J.R.M.; Ribeiro, R.A.; Menna, P.; Bangel, E.V.; Hungria, M. Multilocus Sequence Analysis (MLSA) of Bradyrhizobium Strains: Revealing High Diversity of Tropical Diazotrophic Symbiotic Bacteria. Braz. J. Microbiol. 2012, 43, 698–710. [Google Scholar] [CrossRef] [PubMed]
- Benjelloun, I.; Thami Alami, I.; Douira, A.; Udupa, S.M. Phenotypic and Genotypic Diversity Among Symbiotic and Non-Symbiotic Bacteria Present in Chickpea Nodules in Morocco. Front. Microbiol. 2019, 10, 1885. [Google Scholar] [CrossRef]
- Avontuur, J.R.; Palmer, M.; Beukes, C.W.; Chan, W.Y.; Coetzee, M.P.A.; Blom, J.; Stępkowski, T.; Kyrpides, N.C.; Woyke, T.; Shapiro, N.; et al. Genome-Informed Bradyrhizobium Taxonomy: Where to from Here? Syst. Appl. Microbiol. 2019, 42, 427–439. [Google Scholar] [CrossRef]
- Menna, P.; Barcellos, F.G.; Hungria, M. Phylogeny and Taxonomy of a Diverse Collection of Bradyrhizobium Strains Based on Multilocus Sequence Analysis of the 16S rRNA Gene, ITS Region and glnII, recA, atpD and dnaK Genes. Int. J. Syst. Evol. Microbiol. 2009, 59, 2934–2950. [Google Scholar] [CrossRef]
- Tong, S.Y.C.; Giffard, P.M. Microbiological Applications of High-Resolution Melting Analysis. J. Clin. Microbiol. 2012, 50, 3418–3421. [Google Scholar] [CrossRef]
- Borkowska, M.; Celińska, E. Multiple Region High Resolution Melting-Based Method for Accurate Differentiation of Food-Derived Yeasts at Species Level Resolution. Food Microbiol. 2023, 109, 104120. [Google Scholar] [CrossRef] [PubMed]
- Madrzak, C.J.; Golinska, B.; Kroliczak, J.; Pudelko, K.; Lazewska, D.; Lampka, B.; Sadowsky, M.J. Diversity among Field Populations of Bradyrhizobium Japonicum in Poland. Appl. Environ. Microbiol. 1995, 61, 1194. [Google Scholar] [CrossRef] [PubMed]
- Waleron, M.; Waleron, K.; Podhajska, A.J.; Łojkowska, E. Genotyping of Bacteria Belonging to the Former Erwinia Genus by PCR-RFLP Analysis of a recA Gene Fragment. Microbiology 2002, 148, 583–595. [Google Scholar] [CrossRef] [PubMed]
- Costechareyre, D.; Rhouma, A.; Lavire, C.; Portier, P.; Chapulliot, D.; Bertolla, F.; Boubaker, A.; Dessaux, Y.; Nesme, X. Rapid and Efficient Identification of Agrobacterium Species by recA Allele Analysis. Microb. Ecol. 2010, 60, 862–872. [Google Scholar] [CrossRef]
- Blackwood, K.S.; He, C.; Gunton, J.; Turenne, C.Y.; Wolfe, J.; Kabani, A.M. Evaluation of recA Sequences for Identification of Mycobacterium Species. J. Clin. Microbiol. 2000, 38, 2846–2852. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jarzyniak, K.; Narożna, D. Rapid Identification of Rhizobia Nodulating Soybean by a High-Resolution Melting Analysis. Agronomy 2024, 14, 1305. https://doi.org/10.3390/agronomy14061305
Jarzyniak K, Narożna D. Rapid Identification of Rhizobia Nodulating Soybean by a High-Resolution Melting Analysis. Agronomy. 2024; 14(6):1305. https://doi.org/10.3390/agronomy14061305
Chicago/Turabian StyleJarzyniak, Karolina, and Dorota Narożna. 2024. "Rapid Identification of Rhizobia Nodulating Soybean by a High-Resolution Melting Analysis" Agronomy 14, no. 6: 1305. https://doi.org/10.3390/agronomy14061305
APA StyleJarzyniak, K., & Narożna, D. (2024). Rapid Identification of Rhizobia Nodulating Soybean by a High-Resolution Melting Analysis. Agronomy, 14(6), 1305. https://doi.org/10.3390/agronomy14061305




