Alfalfa with Forage Crop Rotation Alleviates Continuous Alfalfa Obstacles through Regulating Soil Enzymes and Bacterial Community Structures
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Soil Sample Collection and Soil Parameter Determination
2.3. Soil DNA Extraction, PCR Amplification and High-Throughput Sequencing
2.4. Processing of Bacterial 16S rRNA Sequencing Data
2.5. Statistical Analysis
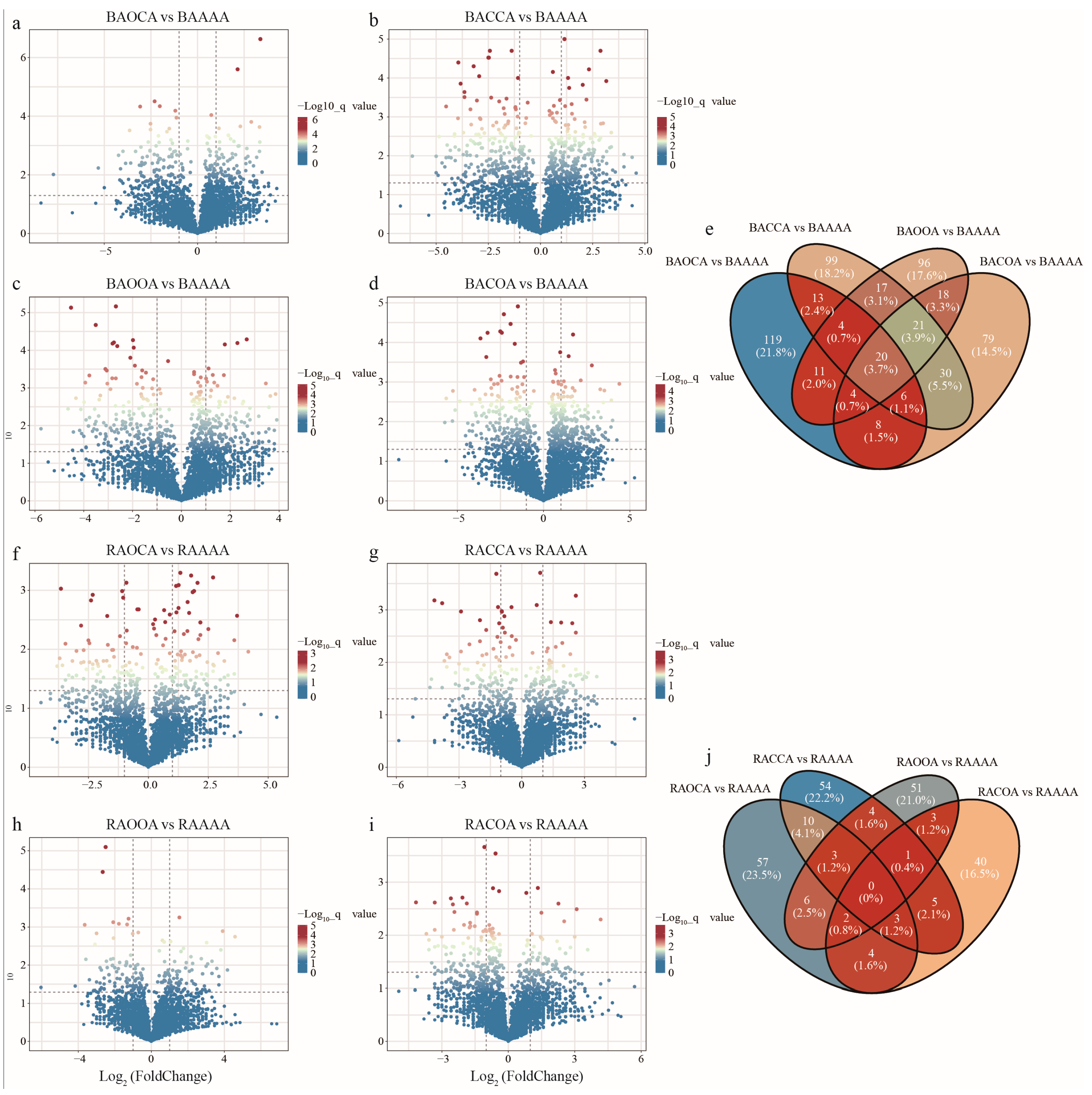
3. Results
3.1. Soil Chemical Properties and Soil Enzyme Activities
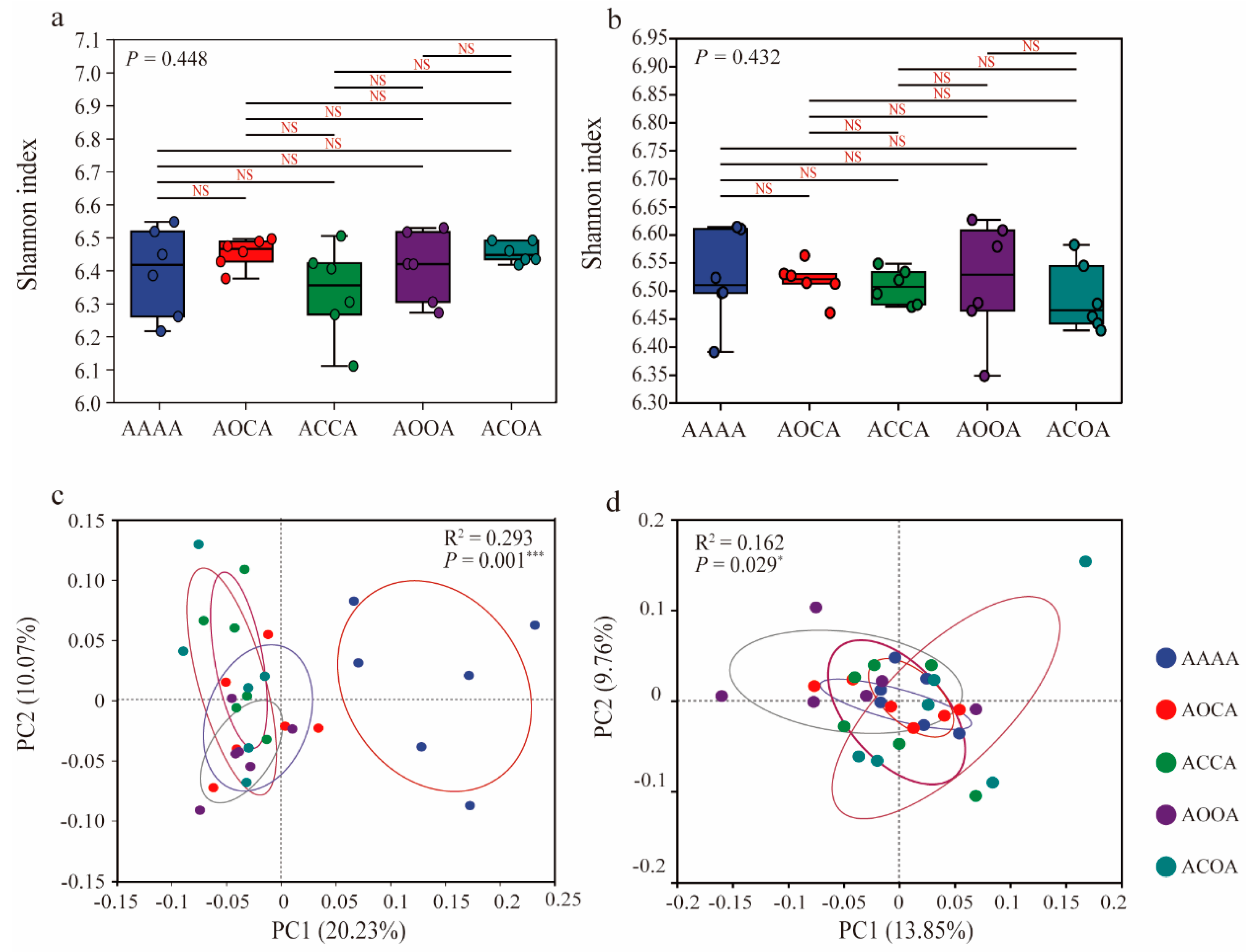
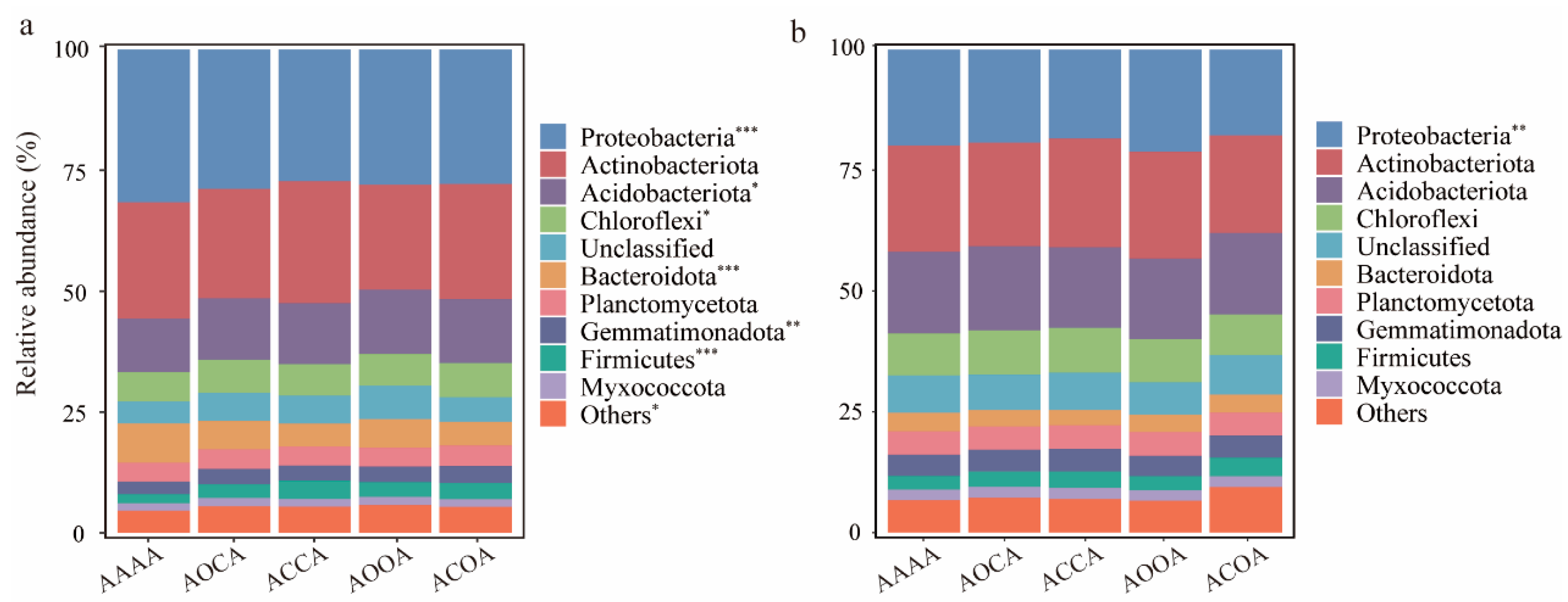
3.2. Soil Bacterial Diversity and Structure
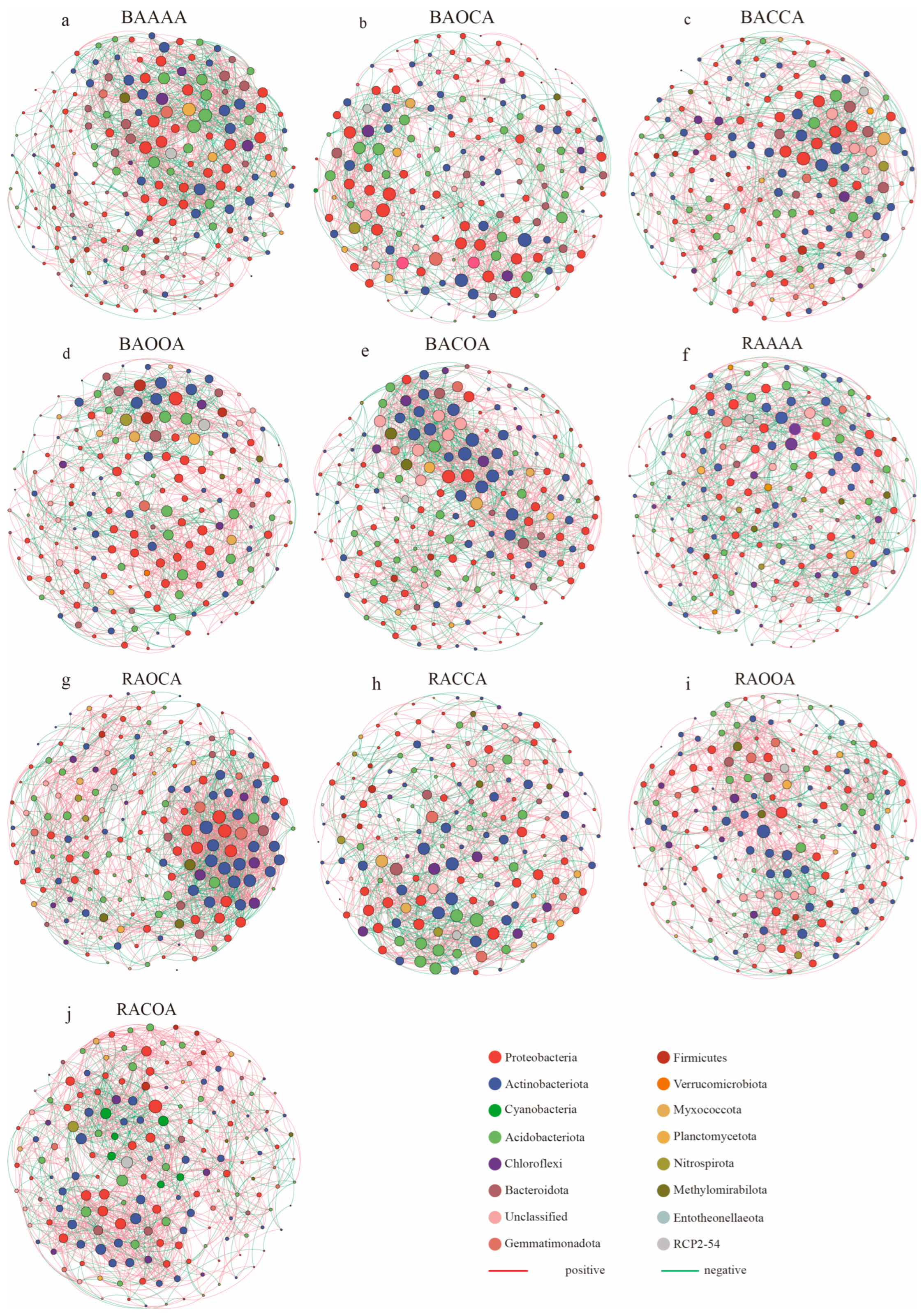
3.3. Soil Bacterial Co-Occurrence Network
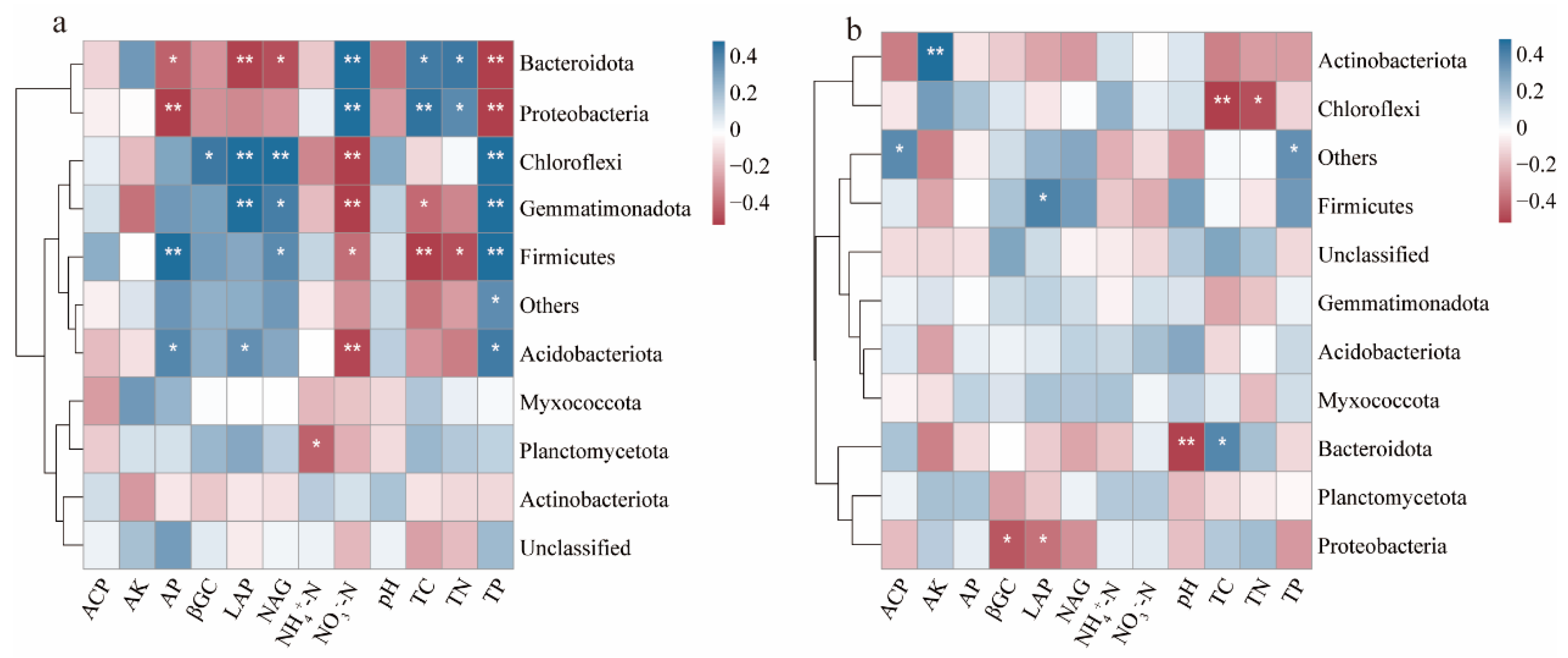
3.4. Correlations between Soil Parameters and Soil Bacterial Community Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liu, X.; Herbert, S.J. Fifteen years of research examining cultivation of continuous soybean in northeast China: A review. Field Crop Res. 2002, 79, 1–7. [Google Scholar] [CrossRef]
- Chen, S.; Qi, G.; Luo, T.; Zhang, H.; Jiang, Q.; Wang, R.; Zhao, X. Continuous-cropping tobacco caused variance of chemical properties and structure of bacterial network in soils. Land. Degrad. Dev. 2018, 29, 4106–4120. [Google Scholar] [CrossRef]
- Pervaiz, Z.H.; Iqbal, J.; Zhang, Q.; Chen, D.; Wei, H.; Saleem, M. Continuous cropping alters multiple biotic and abiotic indicators of soil health. Soil Syst. 2020, 4, 59. [Google Scholar] [CrossRef]
- Liao, J.; Xia, P. Continuous cropping obstacles of medicinal plants: Focus on the plant-soil-microbe interaction system in the rhizosphere. Sci. Hortic. 2024, 328, 112927. [Google Scholar] [CrossRef]
- Zheng, X.; Wei, L.; Lv, W.; Zhang, H.; Zhang, Y.; Zhang, H.; Zhang, H.; Zhu, Z.; Ge, T.; Zhang, W. Long-term bioorganic and organic fertilization improved soil quality and multifunctionality under continuous cropping in watermelon. Agric. Ecosyst. Environ. 2024, 359, 108721. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, W.; Li, Y.; Sun, Y.; Bian, X. Effects of long-term continuous cropping system of cotton on soil physical-chemical properties and activities of soil enzyme in oasis in Xinjiang. Sci. Agric. Sin. 2009, 42, 725–733. [Google Scholar]
- Xu, Y.; Liu, J.; Liu, X.; Li, H.; Yang, Z.; Wang, H.; Huang, X.; Lan, L.; An, Y.; Li, L. Continuous cropping of alfalfa (Medicago sativa L.) reduces bacterial diversity and simplifies cooccurrence networks in aeolian sandy soil. Soil Ecol. Lett. 2022, 4, 113–143. [Google Scholar] [CrossRef]
- Zeeshan Ul Haq, M.; Yu, J.; Yao, G.; Yang, H.; Iqbal, H.A.; Tahir, H.; Cui, H.; Liu, Y.; Wu, Y. A systematic review on the continuous cropping obstacles and control strategies in medicinal plants. Int. J. Mol. Sci. 2023, 24, 12470. [Google Scholar] [CrossRef]
- Pang, Z.; Tayyab, M.; Kong, C.; Liu, Q.; Liu, Y.; Hu, C.; Huang, J.; Weng, P.; Islam, W.; Lin, W. Continuous sugarcane planting negatively impacts soil microbial community structure, soil fertility, and sugarcane agronomic parameters. Microorganisms 2021, 9, 2008. [Google Scholar] [CrossRef]
- Chen, D.M.; Yang, Y.H.; Jin, Y.; Wang, H.B.; Duan, Y.Q.; You, C.H. Constituents of autotoxic chemical from rhizosphere soil under flue-cured tobacco continuous cropping. Pratacultural Sci. 2011, 28, 1766–1769. [Google Scholar]
- Li, H.Q.; Zhang, L.L.; Jiang, X.W.; Liu, Q.Z. Allelopathic effects of phenolic acids on the growth and physiological characteristics of strawberry plants. Allelopathy J. 2015, 35, 61. [Google Scholar]
- Wu, F.; Wang, X.; Xue, C. Effect of cinnamic acid on soil microbial characteristics in the cucumber rhizosphere. Eur. J. Soil Biol. 2009, 45, 356–362. [Google Scholar] [CrossRef]
- Bai, L.; Cui, J.; Jie, W.; Cai, B. Analysis of the community compositions of rhizosphere fungi in soybeans continuous cropping fields. Microbiol. Res. 2015, 180, 49–56. [Google Scholar] [CrossRef]
- Li, M.; Wang, J.; Zhou, Q.; Yasen, M. Effects of continuous melon cropping on rhizospheric fungal communities. Rhizosphere 2023, 27, 100726. [Google Scholar] [CrossRef]
- Strom, N.; Hu, W.; Haarith, D.; Chen, S.; Bushley, K. Interactions between soil properties, fungal communities, the soybean cyst nematode, and crop yield under continuous corn and soybean monoculture. Appl. Soil Ecol. 2020, 147, 103388. [Google Scholar] [CrossRef]
- McDonald, I.; Baral, R.; Min, D. Effects of alfalfa and alfalfa-grass mixtures with nitrogen fertilization on dry matter yield and forage nutritive value. J. Anim. Sci. Technol. 2021, 63, 305. [Google Scholar] [CrossRef]
- Suwignyo, B.; Rini, E.A.; Helmiyati, S. The profile of tropical alfalfa in Indonesia: A review. Saudi J. Biol. Sci. 2023, 30, 103504. [Google Scholar]
- Niu, Y.; Luo, Z.; Cai, L.; Coulter, J.A.; Zhang, Y.; Berti, M. Continuous Monoculture of Alfalfa and Annual Crops Influence Soil Organic Matter and Microbial Communities in the Rainfed Loess Plateau of China. Agronomy 2020, 10, 1054. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, W. Priorities for the development of alfalfa pasture in northern China. Fundam. Res. 2023, 3, 225–228. [Google Scholar] [CrossRef]
- Yang, H.; Feng, X.; Wang, H.; Yan, H.; Zhao, P.; Gao, F.; Guo, X.; Xie, B. Long time-series variation of crop yield under drought stress and drought vulnerability curves in Songnen Plain, Northeast China. Ecol. Indic. 2023, 154, 110624. [Google Scholar] [CrossRef]
- Yang, Z.; Xu, Y.; Li, H.; Li, S.; Wang, X.; Chai, H. Difference of bacterial community structure in the Meadow, Maize, and continuous cropped Alfalfa in Northeast China. Front. Microbiol. 2022, 13, 794848. [Google Scholar] [CrossRef]
- Wang, R.; Liu, J.; Jiang, W.; Ji, P.; Li, Y. Metabolomics and microbiomics reveal impacts of rhizosphere metabolites on alfalfa continuous cropping. Front. Microbiol. 2022, 13, 833968. [Google Scholar]
- Kiani, M.; Hernandez-Ramirez, G.; Quideau, S.; Smith, E.; Janzen, H.; Larney, F.J.; Puurveen, D. Quantifying sensitive soil quality indicators across contrasting long-term land management systems: Crop rotations and nutrient regimes. Agric. Ecosyst. Environ. 2017, 248, 123–135. [Google Scholar] [CrossRef]
- Shah, K.K.; Modi, B.; Pandey, H.P.; Subedi, A.; Aryal, G.; Pandey, M.; Shrestha, J.; Fahad, S. Diversified Crop Rotation: An Approach for Sustainable Agriculture Production. Adv. Agric. 2021, 2021, 8924087. [Google Scholar] [CrossRef]
- Li, A.; Wu, Y.; Tai, X.; Cao, S.; Gao, T. Effects of Alfalfa Crop Rotation on Soil Nutrients and Loss of Soil and Nutrients in Semi-Arid Regions. Sustainability 2023, 15, 15164. [Google Scholar] [CrossRef]
- Singh, A.; Afzal, T.; Woodbury, B.; Wortmann, C.; Iqbal, J. Alfalfa in rotation with annual crops reduced nitrate leaching potential. J. Environ. Qual. 2023, 52, 930–938. [Google Scholar] [CrossRef]
- Liu, Q.; Zhao, Y.; Li, T.; Chen, L.; Chen, Y.; Sui, P. Changes in soil microbial biomass, diversity, and activity with crop rotation in cropping systems: A global synthesis. Appl. Soil Ecol. 2023, 186, 104815. [Google Scholar] [CrossRef]
- Stamenov, D.; Đurić, S.; Hajnal, J.T.; Šeremešić, S. Fertilization and crop rotation effects on the number of different groups of microorganisms. Ratar. I Povrt. 2016, 53, 96–100. [Google Scholar]
- Venter, Z.S.; Jacobs, K.; Hawkins, H. The impact of crop rotation on soil microbial diversity: A meta-analysis. Pedobiologia 2016, 59, 215–223. [Google Scholar] [CrossRef]
- Samaddar, S.; Schmidt, R.; Tautges, N.E.; Scow, K. Adding alfalfa to an annual crop rotation shifts the composition and functional responses of tomato rhizosphere microbial communities. Appl. Soil Ecol. 2021, 167, 104102. [Google Scholar] [CrossRef]
- Brtnicky, M.; Kintl, A.; Hammerschmiedt, T.; Mustafa, A.; Elbl, J.; Kucerik, J.; Vyhnanek, T.; Skladanka, J.; Hunady, I.; Holatko, J. Clover Species Specific Influence on Microbial Abundance and Associated Enzyme Activities in Rhizosphere and Non-Rhizosphere Soils. Agronomy 2021, 11, 2214. [Google Scholar] [CrossRef]
- Jones, D.L.; Willett, V.B. Experimental evaluation of methods to quantify dissolved organic nitrogen (DON) and dissolved organic carbon (DOC) in soil. Soil Biol. Biochem. 2006, 38, 991–999. [Google Scholar] [CrossRef]
- Dospatliev, L.; Ivanova, M. Inductively coupled plasma optical emission spectrometry determination of total phosphorus and sulphur in virginia tobacco leaves. Oxid. Commun. 2022, 45, 115. [Google Scholar]
- Miranda, K.M.; Espey, M.G.; Wink, D.A. A Rapid, Simple Spectrophotometric Method for Simultaneous Detection of Nitrate and Nitrite. Nitric Oxide 2001, 5, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Dahlquist, R.L.; Knoll, J.W. Inductively coupled plasma-atomic emission spectrometry: Analysis of biological materials and soils for major, trace, and ultra-trace elements. Appl. Spectrosc. 1978, 32, 1–30. [Google Scholar] [CrossRef]
- Dick, W.A.; Cheng, L.; Wang, P. Soil acid and alkaline phosphatase activity as pH adjustment indicators. Soil Biol. Biochem. 2000, 32, 1915–1919. [Google Scholar]
- Nidhi, A.; Sneha, S.; Annamma, A.; Arvind, L.; Shams, Y.S. Specific Fusion of β-1,4-Endoglucanase and β-1,4-Glucosidase Enhances Cellulolytic Activity and Helps in Channeling of Intermediates. Appl. Environ. Microb. 2012, 78, 7447–7454. [Google Scholar] [CrossRef]
- Linko-Löppönen, S.; Mäkinen, M. A microtiter plate assay for N-acetyl-β-d-glucosaminidase using a fluorogenic substrate. Anal. Biochem. 1985, 148, 50–53. [Google Scholar] [CrossRef]
- Wang, J.; Huang, S.; He, Q.; Bing, H.; Chen, X.; Zhang, X.; Tian, X.; Zhou, J.; Wilcke, W.; Wu, Y. Microplate fluorimetric assay of soil leucine aminopeptidase activity: Alkalization is not needed before fluorescence reading. Biol. Fert. Soils 2020, 56, 281–285. [Google Scholar] [CrossRef]
- Dineen, S.M.; Aranda, R., IV.; Anders, D.L.; Robertson, J.M. An evaluation of commercial DNA extraction kits for the isolation of bacterial spore DNA from soil. J. Appl. Microbiol. 2010, 109, 1886–1896. [Google Scholar] [CrossRef]
- Chen, L.; Luo, Y.; Xu, J.; Yu, Z.; Zhang, K.; Brookes, P.C. Assessment of bacterial communities and predictive functional profiling in soils subjected to short-term fumigation-incubation. Microb. Ecol. 2016, 72, 240–251. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Jeffrey, I.G. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Amir, A.; McDonald, D.; Navas-Molina, J.A.; Kopylova, E.; Morton, J.T.; Zech Xu, Z.; Kightley, E.P.; Thompson, L.R.; Hyde, E.R.; Gonzalez, A. Deblur rapidly resolves single-nucleotide community sequence patterns. Msystems 2017, 2, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Preheim, S.P.; Perrotta, A.R.; Martin-Platero, A.M.; Gupta, A.; Alm, E.J. Distribution-based clustering: Using ecology to refine the operational taxonomic unit. Appl. Environ. Microb. 2013, 79, 6593–6603. [Google Scholar] [CrossRef] [PubMed]
- Almeida, A.; Mitchell, A.L.; Tarkowska, A.; Finn, R.D. Benchmarking taxonomic assignments based on 16S rRNA gene profiling of the microbiota from commonly sampled environments. Gigascience 2018, 7, giy054. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Lkhagva, E.; Chung, H.; Hong, J.; Tang, W.H.W.; Lee, S.; Hong, S.; Lee, S. The regional diversity of gut microbiome along the GI tract of male C57BL/6 mice. BMC Microbiol. 2021, 21, 1–13. [Google Scholar] [CrossRef]
- Team, R. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2009; Volume 14, pp. 12–21. [Google Scholar]
- Shi, Z.; Shi, S.; Li, W.; Wang, C.; Hu, F. Bacterial community characteristics in epigeic and anecic earthworm vermicompartments within soil-earthworm systems. Pedosphere 2023, 9, 1–21. [Google Scholar] [CrossRef]
- Rojas-Jimenez, K.; Grossart, H.; Cordes, E.; Cortés, J. Fungal communities in sediments along a depth gradient in the Eastern Tropical Pacific. Front. Microbiol. 2020, 11, 575207. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Yang, T.; Bao, Y.; He, P.; Yang, K.; Mei, X.; Wei, Z.; Xu, Y.; Shen, Q.; Banerjee, S. Network analysis and subsequent culturing reveal keystone taxa involved in microbial litter decomposition dynamics. Soil Biol. Biochem. 2021, 157, 108230. [Google Scholar] [CrossRef]
- Cherven, K. Mastering Gephi Network Visualization; Packt Publishing Ltd.: Birmingham, UK, 2015. [Google Scholar]
- Böer, S.I.; Hedtkamp, S.I.; Van Beusekom, J.E.; Fuhrman, J.A.; Boetius, A.; Ramette, A. Time-and sediment depth-related variations in bacterial diversity and community structure in subtidal sands. ISME J. 2009, 3, 780–791. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Singh, U.B.; Sahu, P.K.; Paul, S.; Kumar, A.; Malviya, D.; Singh, S.; Kuppusamy, P.; Singh, P.; Paul, D. Linking soil microbial diversity to modern agriculture practices: A review. Int. J. Environ. Res. Public Health 2022, 19, 3141. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Plaza-Bonilla, D.; Coulter, J.A.; Kutcher, H.R.; Beckie, H.J.; Wang, L.; Floc’H, J.; Hamel, C.; Siddique, K.H.; Li, L. Diversifying crop rotations enhances agroecosystem services and resilience. Adv. Agron. 2022, 173, 299–335. [Google Scholar]
- Stirling, G.; Hayden, H.; Pattison, T.; Stirling, M. Soil Health, Soil Biology, Soilborne Diseases and Sustainable Agriculture: A Guide; Csiro Publishing: Clayton, Australia, 2016. [Google Scholar]
- Chen, T.; Li, J.; Wu, L.K.; Lin, S.; Wang, J.H.; Li, Z.F.; Zhang, Z.Y.; Lin, W.X. Effects of continuous monoculture of Achyranthes bidentata on microbial community structure and functional diversity in soil. Allelopath. J. 2015, 36, 197. [Google Scholar]
- Wang, J.Y.; Xu, J.H.; Wu, L.K.; Wu, H.M.; Zhu, Q.; Kong, L.F.; Lin, W.X. Analysis of physicochemical properties and microbial diversity in rhizosphere soil of Achyranthes bidentata under different cropping years. Acta Ecol. Sin. 2017, 37, 5621–5629. [Google Scholar]
- Liu, Z.; Liu, J.; Yu, Z.; Yao, Q.; Li, Y.; Liang, A.; Zhang, W.; Mi, G.; Jin, J.; Liu, X. Long-term continuous cropping of soybean is comparable to crop rotation in mediating microbial abundance, diversity and community composition. Soil Tillage Res. 2020, 197, 104503. [Google Scholar] [CrossRef]
- Lian, T.; Cheng, L.; Liu, Q.; Yu, T.; Cai, Z.; Nian, H.; Hartmann, M. Potential relevance between soybean nitrogen uptake and rhizosphere prokaryotic communities under waterlogging stress. ISME Commun. 2023, 3, 71. [Google Scholar] [CrossRef]
- Lian, T.; Ma, Q.; Shi, Q.; Cai, Z.; Zhang, Y.; Cheng, Y.; Nian, H. High aluminum stress drives different rhizosphere soil enzyme activities and bacterial community structure between aluminum-tolerant and aluminum-sensitive soybean genotypes. Plant Soil 2019, 440, 409–425. [Google Scholar] [CrossRef]
- Fan, K.; Weisenhorn, P.; Gilbert, J.A.; Chu, H. Wheat rhizosphere harbors a less complex and more stable microbial co-occurrence pattern than bulk soil. Soil Biol. Biochem. 2018, 125, 251–260. [Google Scholar] [CrossRef]
- Bag, S.; Mondal, A.; Banik, A. Exploring tea (Camellia sinensis) microbiome: Insights into the functional characteristics and their impact on tea growth promotion. Microbiol. Res. 2022, 254, 126890. [Google Scholar] [CrossRef] [PubMed]
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z. Current knowledge and perspectives of Paenibacillus: A review. Microb. Cell Fact. 2016, 15, 203. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Yu, T.; Shi, Q.; Han, D.; Yu, K.; Wang, L.; Wang, S.; Xiang, H.; Wen, R.; Nian, H. Rhizosphere soil bacterial communities of continuous cropping-tolerant and sensitive soybean genotypes respond differently to long-term continuous cropping in mollisols. Front. Microbiol. 2021, 12, 729047. [Google Scholar] [CrossRef]
- Rastogi, M.; Verma, S.; Kumar, S.; Bharti, S.; Kumar, G.; Azam, K.; Singh, V. Soil health and sustainability in the age of organic amendments: A review. Int. J. Environ. Clim. Chang. 2023, 13, 2088–2102. [Google Scholar] [CrossRef]


| Treatments | pH | TC | TN | TP | NH4+-N | NO3−-N | AP | AK |
|---|---|---|---|---|---|---|---|---|
| AAAA | 7.49 ± 0.017 a | 19.9 ± 0.372 a | 1.39 ± 0.031 a | 0.461 ± 0.023 c | 105 ± 2.72 ab | 6.50 ± 0.116 a | 22.2 ± 1.41 b | 218 ± 14.7 a |
| ACCA | 7.53 ± 0.014 a | 18.6 ± 0.198 b | 1.27 ± 0.007 b | 0.599 ± 0.041 b | 109 ± 3.37 ab | 4.87 ± 0.362 b | 29.4 ± 1.99 a | 223 ± 15.0 a |
| ACOA | 7.51 ± 0.025 a | 19.0 ± 0.189 b | 1.32 ± 0.009 ab | 0.695 ± 0.018 a | 99.0 ± 2.67 b | 4.78 ± 0.095 b | 28.0 ± 1.04 a | 195 ± 5.39 a |
| AOCA | 7.53 ± 0.011 a | 18.8 ± 0.300 b | 1.31 ± 0.034 ab | 0.617 ± 0.009 b | 104 ± 3.18 ab | 5.05 ± 0.223 b | 29.4 ± 1.93 a | 200 ± 4.92 a |
| AOOA | 7.52 ± 0.018 a | 18.7 ± 0.349 b | 1.26 ± 0.040 b | 0.622 ± 0.009 b | 112 ± 4.15 a | 5.00 ± 0.522 b | 29.5 ± 0.915 a | 207 ± 6.77 a |
| P | 0.466 | 0.032 * | 0.023 * | 0.0001 *** | 0.101 | 0.003 ** | 0.0092 ** | 0.2780 |
| Treatments | ACP | βGC | LAP | NAG |
|---|---|---|---|---|
| AAAA | 6.43 ± 0.103 b | 27.2 ± 1.22 c | 2.79 ± 0.136 d | 2.70 ± 0.353 c |
| AOCA | 7.80 ± 0.326 a | 41.9 ± 1.72 a | 6.31 ± 0.184 a | 5.28 ± 0.258 a |
| ACCA | 6.65 ± 0.153 b | 33.2 ± 2.71 b | 4.56 ± 0.326 b | 3.86 ± 0.202 b |
| AOOA | 6.74 ± 0.182 b | 29.1 ± 1.08 bc | 3.65 ± 0.226 c | 3.57 ± 0.247 b |
| ACOA | 8.36 ± 0.626 a | 42.5 ± 2.17 a | 6.59 ± 0.437 a | 5.25 ± 0.181 a |
| P | 0.0013 | 0.0001 | 0.0001 | 0.0001 |
| Network Indicator | BAAAA | BAOCA | BACCA | BAOOA | BACOA | RAAAA | RAOCA | RACCA | RAOOA | RACOA |
|---|---|---|---|---|---|---|---|---|---|---|
| Node | 188 | 178 | 177 | 176 | 183 | 181 | 180 | 172 | 182 | 179 |
| Edge | 1699 | 1326 | 1372 | 1343 | 1538 | 1363 | 1854 | 1335 | 1345 | 1580 |
| Positive correlation | 958 | 780 | 835 | 862 | 913 | 791 | 1194 | 796 | 893 | 1079 |
| Negative correlation | 741 | 546 | 537 | 481 | 625 | 572 | 660 | 539 | 452 | 501 |
| Average degree | 18.1 | 14.9 | 15.5 | 15.3 | 16.8 | 15.1 | 20.6 | 15.8 | 14.8 | 17.7 |
| Average path length | 2.92 | 3.05 | 3.01 | 3.00 | 2.96 | 3.01 | 2.89 | 3.00 | 3.07 | 2.88 |
| Graph density | 0.097 | 0.084 | 0.088 | 0.087 | 0.092 | 0.084 | 0.115 | 0.092 | 0.082 | 0.099 |
| Network diameter | 6 | 6 | 6 | 6 | 6 | 7 | 6 | 7 | 6 | 6 |
| Average clustering coefficient | 0.490 | 0.515 | 0.543 | 0.530 | 0.499 | 0.498 | 0.487 | 0.507 | 0.507 | 0.480 |
| Average weighted degree | 2.59 | 3.15 | 4.40 | 6.10 | 3.98 | 2.71 | 8.97 | 3.91 | 7.07 | 9.83 |
| Connecting components | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 |
| Modularity | 0.423 | 0.467 | 0.468 | 0.493 | 0.455 | 0.437 | 0.335 | 0.446 | 0.473 | 0.421 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Liu, Z.; Shen, Z.; Yang, Z.; Fu, X.; Wang, X.; Li, S.; Chai, H.; Wang, R.; Liu, X.; et al. Alfalfa with Forage Crop Rotation Alleviates Continuous Alfalfa Obstacles through Regulating Soil Enzymes and Bacterial Community Structures. Agronomy 2024, 14, 1349. https://doi.org/10.3390/agronomy14071349
Xu Y, Liu Z, Shen Z, Yang Z, Fu X, Wang X, Li S, Chai H, Wang R, Liu X, et al. Alfalfa with Forage Crop Rotation Alleviates Continuous Alfalfa Obstacles through Regulating Soil Enzymes and Bacterial Community Structures. Agronomy. 2024; 14(7):1349. https://doi.org/10.3390/agronomy14071349
Chicago/Turabian StyleXu, Yanxia, Zhuxiu Liu, Zhongbao Shen, Zhao Yang, Xuepeng Fu, Xiaolong Wang, Shasha Li, Hua Chai, Ruoding Wang, Xiaobing Liu, and et al. 2024. "Alfalfa with Forage Crop Rotation Alleviates Continuous Alfalfa Obstacles through Regulating Soil Enzymes and Bacterial Community Structures" Agronomy 14, no. 7: 1349. https://doi.org/10.3390/agronomy14071349
APA StyleXu, Y., Liu, Z., Shen, Z., Yang, Z., Fu, X., Wang, X., Li, S., Chai, H., Wang, R., Liu, X., & Liu, J. (2024). Alfalfa with Forage Crop Rotation Alleviates Continuous Alfalfa Obstacles through Regulating Soil Enzymes and Bacterial Community Structures. Agronomy, 14(7), 1349. https://doi.org/10.3390/agronomy14071349








