Identification of miRNAs Interacting with Abscisic Acid to Regulate Fatty Acid Metabolism
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Treatment Procedures
2.2. RNA Extraction and Real-Time Quantitative PCR (RT-qPCR)
2.3. Small RNA Sequencing Data Analysis and miRNA Identification
2.4. Target Gene Prediction for miRNAs
2.5. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analysis
2.6. Determination of Fatty Acid
3. Results
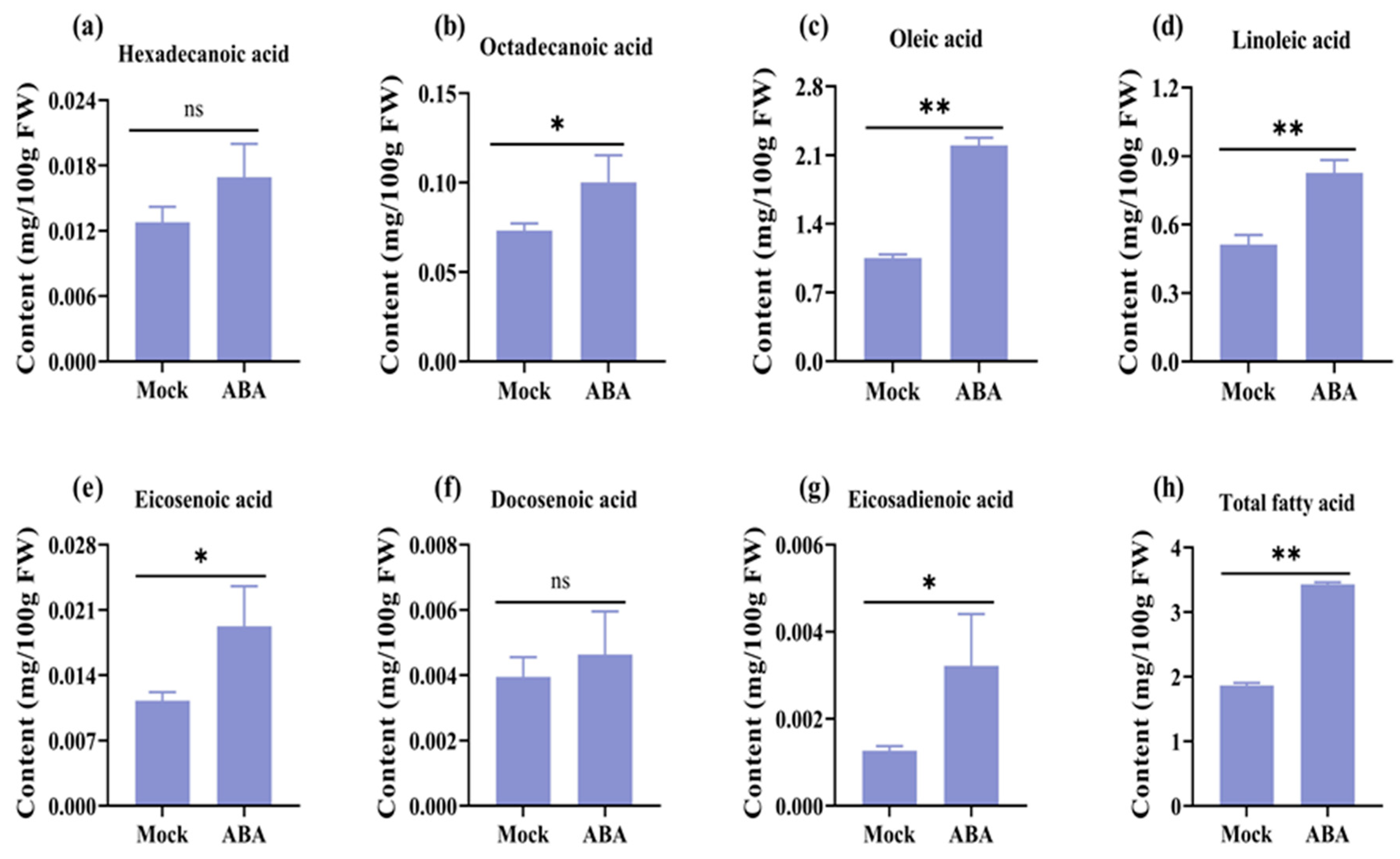
3.1. Exogenous Application of ABA Can Efficiently Improve Unsaturated Fatty Acids Content in Rapeseeds
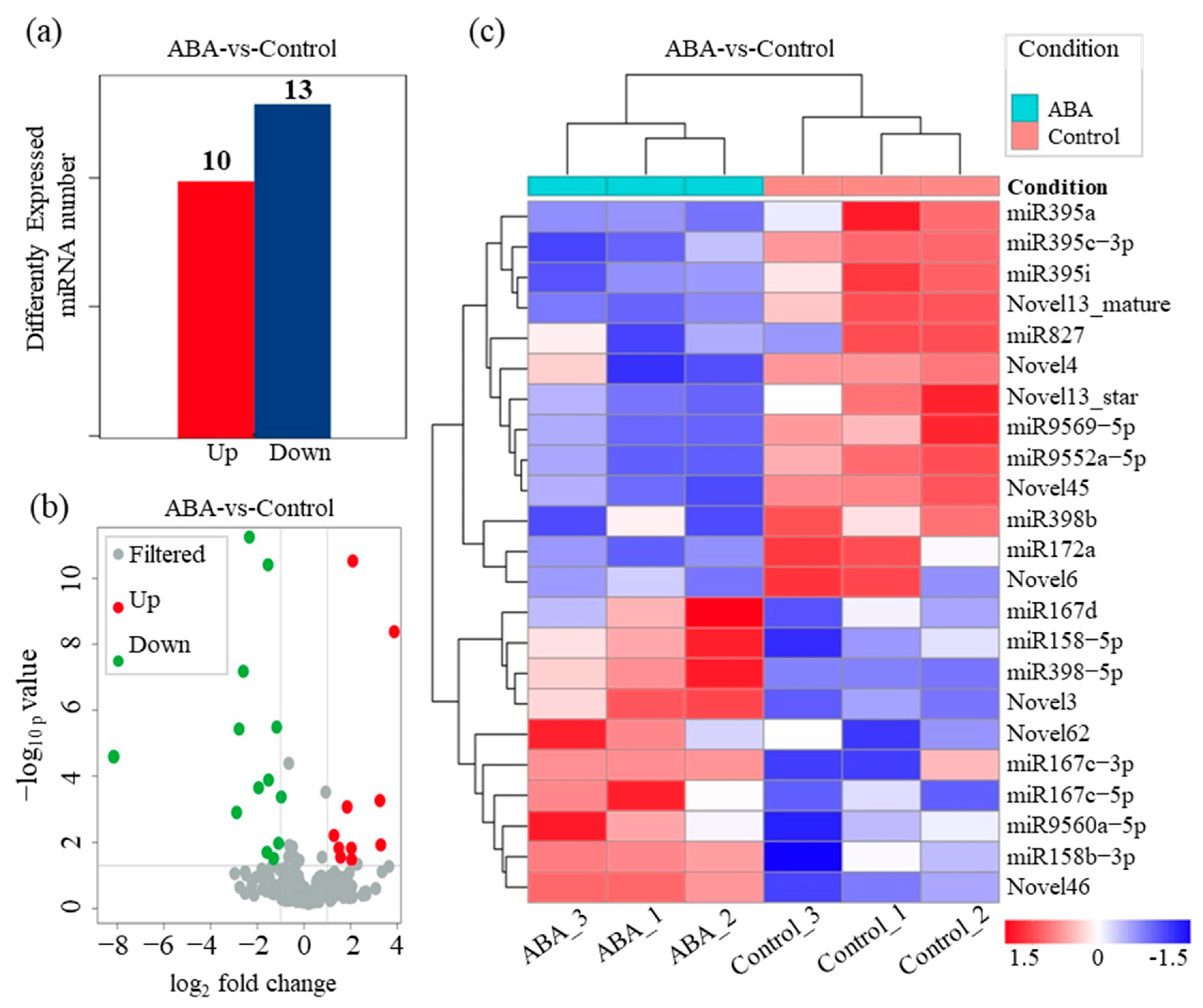
3.2. Overview of Small RNA Library Sequencing for Rapeseeds
3.3. Identification of ABA-Interacting miRNAs Involved in Fatty Acid Metabolism in B. napus
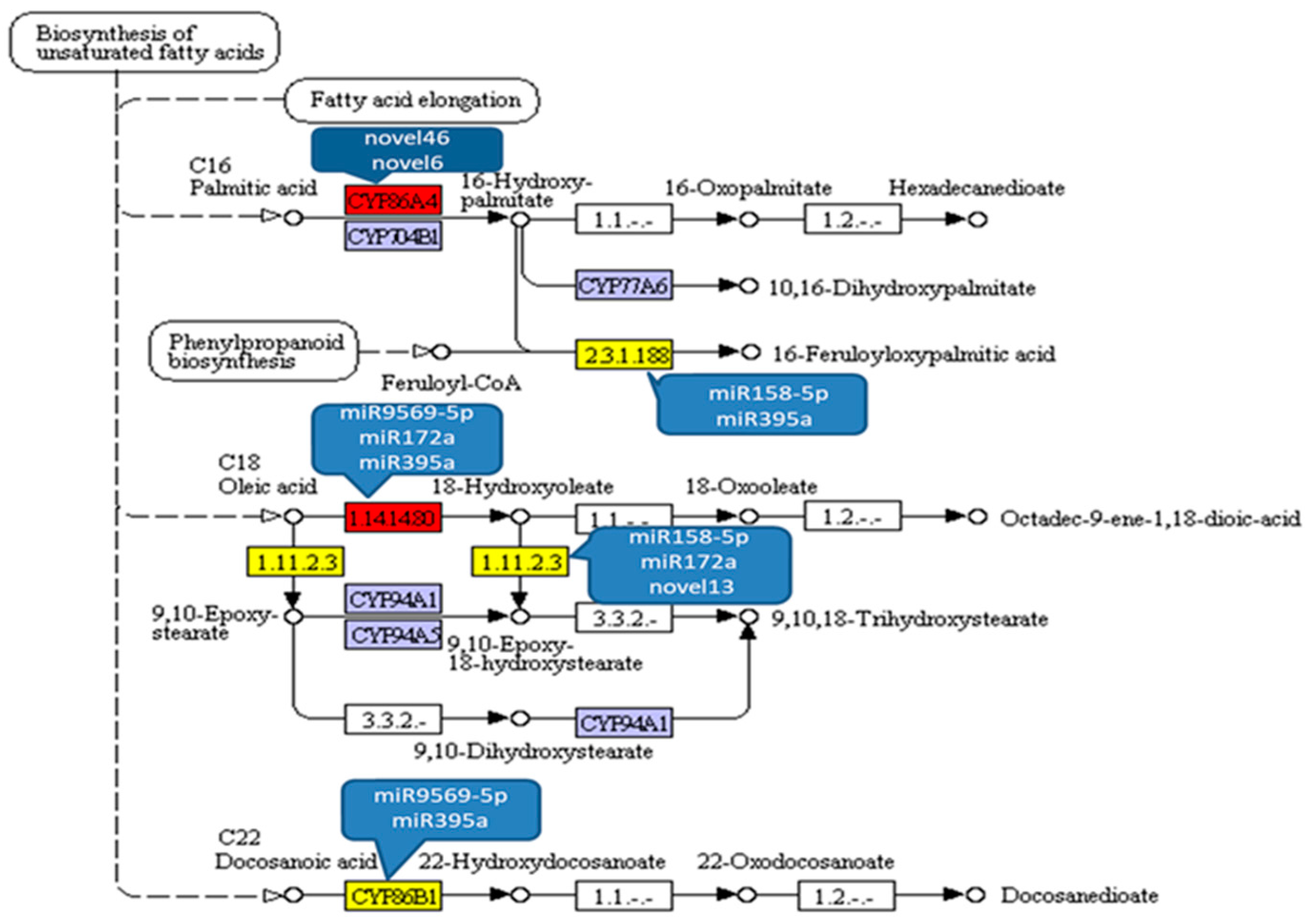
3.4. Prediction of Target Genes by miRNAs Interacting with ABA and Functional Analysis
3.5. Expression Profiles of miRNAs Interacting with ABA Involved in Fatty Acids Biosynthesis
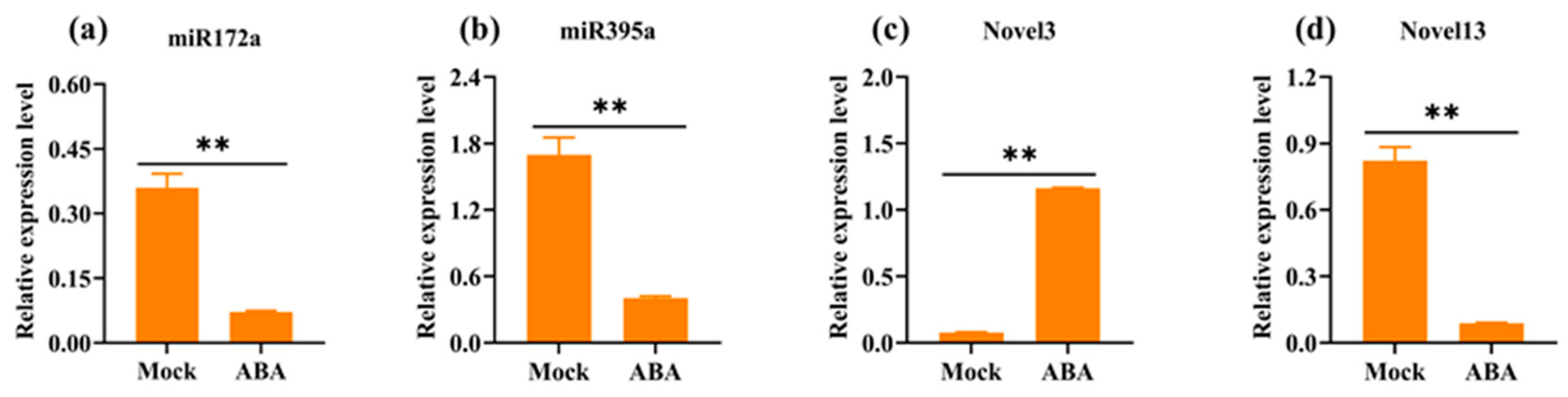
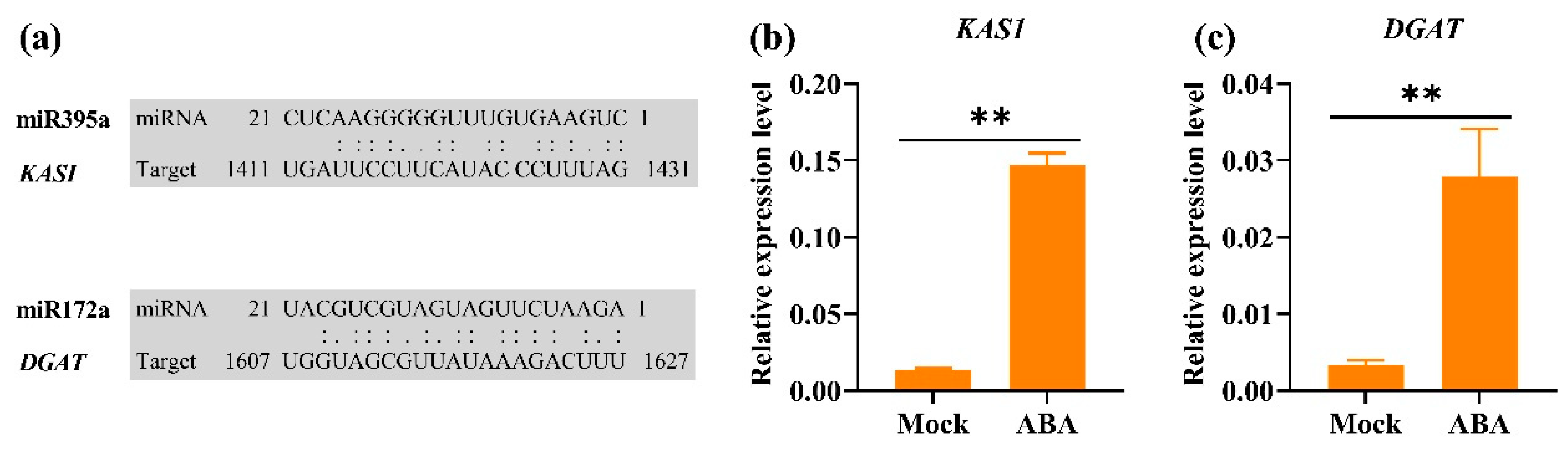
3.6. RT-qPCR Validation of miRNAs and Corresponding Target Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Petrie, J.R.; Zhou, X.R.; Leonforte, A.; McAllister, J.; Shrestha, P.; Kennedy, Y.; Belide, S.; Buzza, G.; Gororo, N.; Gao, W.; et al. Development of a Brassica napus (Canola) Crop Containing Fish Oil-like Levels of DHA in the Seed Oil. Front. Plant Sci. 2020, 11, 727. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, F.; Yu, Z.; Shi, X.; Zhou, X.; Wang, P.; Song, Y.; Hong, D.; Yang, G. Pyramiding of multiple genes generates rapeseed introgression lines with clubroot and herbicide resistance, high oleic acid content, and early maturity. Crop J. 2023, 11, 895–903. [Google Scholar] [CrossRef]
- Wang, H.; Wang, Q.; Pak, H.; Yan, T.; Chen, M.; Chen, X.; Wu, D.; Jiang, L. Genome-wide association study reveals a patatin-like lipase relating to the reduction of seed oil content in Brassica napus. BMC Plant Biol. 2021, 21, 6. [Google Scholar] [CrossRef]
- He, M.; Qin, C.X.; Wang, X.; Ding, N.-Z. Plant Unsaturated Fatty Acids: Biosynthesis and Regulation. Front. Plant Sci. 2020, 11, 390. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Zhan, J.; Meyers, B.C. Plant Small RNAs: Their Biogenesis, Regulatory Roles, and Functions. Annu. Rev. Plant Biol. 2023, 74, 21–51. [Google Scholar] [CrossRef]
- Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef]
- Llave, C.; Xie, Z.; Kasschau, K.D.; Carrington, J.C. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 2002, 297, 2053–2056. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. Plant microRNA: A small regulatory molecule with big impact. Dev. Biol. 2006, 289, 3–16. [Google Scholar] [CrossRef]
- Li, J.; Reichel, M.; Li, Y.; Millar, A.A. The functional scope of plant microRNA-mediated silencing. Trends Plant Sci. 2014, 19, 750–756. [Google Scholar] [CrossRef]
- Yuan, S.; Zhao, J.; Li, Z.; Hu, Q.; Yuan, N.; Zhou, M.; Xia, X.; Noorai, R.; Saski, C.; Li, S.; et al. MicroRNA396-mediated alteration in plant development and salinity stress response in creeping bentgrass. Hortic. Res. 2019, 6, 48. [Google Scholar] [CrossRef]
- Iliopoulos, D.; Drosatos, K.; Hiyama, Y.; Goldberg, I.J.; Zannis, V.I. MicroRNA-370 controls the expression of MicroRNA-122 and Cpt1α and affects lipid metabolism. J. Lipid Res. 2010, 51, 1513–1523. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, N.; Nakagawa, Y.; Tokushige, N.; Aoki, N.; Matsuzaka, T.; Ishii, K.; Yahagi, N.; Kobayashi, K.; Yatoh, S.; Takahashi, A.; et al. The up-regulation of microRNA-335 is associated with lipid metabolism in liver and white adipose tissue of genetically obese mice. Biochem. Biophys. Res. Commun. 2009, 385, 492–496. [Google Scholar] [CrossRef]
- Körbes, A.P.; Machado, R.D.; Guzman, F.; Almerão, M.P.; de Oliveira, L.F.; Loss-Morais, G.; Turchetto-Zolet, A.C.; Cagliari, A.; dos Santos Maraschin, F.; Margis-Pinheiro, M.; et al. Identifying conserved and novel microRNAs in developing seeds of Brassica napus using deep sequencing. PLoS ONE 2012, 7, e50663. [Google Scholar] [CrossRef]
- Wang, J.; Jian, H.; Wang, T.; Wei, L.; Li, J.; Li, C.; Liu, L. Identification of microRNAs Actively Involved in Fatty Acid Biosynthesis in Developing Brassica napus Seeds Using High-Throughput Sequencing. Front. Plant Sci. 2016, 7, 1570. [Google Scholar] [CrossRef]
- Chen, L.; Chen, L.; Zhang, X.; Liu, T.; Niu, S.; Wen, J.; Yi, B.; Ma, C.; Tu, J.; Fu, T.; et al. Identification of miRNAs that regulate silique development in Brassica napus. Plant Sci. 2018, 269, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Qiao, Y.; Zhang, J.; Shi, W.; Zhang, J. Genome wide identification of microRNAs involved in fatty acid and lipid metabolism of Brassica napus by small RNA and degradome sequencing. Gene 2017, 619, 61–70. [Google Scholar] [CrossRef]
- Zhao, Y.-T.; Wang, M.; Fu, S.X.; Yang, W.C.; Qi, C.K.; Wang, X.-J. Small RNA Profiling in Two Brassica napus Cultivars Identifies MicroRNAs with Oil Production- and Development-Correlated Expression and New Small RNA Classes. Plant Physiol. 2012, 158, 813–823. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Li, G.; Jiang, X.; Wang, Y.; Ma, Z.; Niu, Z.; Wang, Z.; Geng, X. Small RNA and degradome profiling involved in seed development and oil synthesis of Brassica napus. PLoS ONE 2018, 13, e0204998. [Google Scholar] [CrossRef]
- Zhang, C.; Chang, W.; Li, X.; Yang, B.; Zhang, L.; Xiao, Z.; Li, J.; Lu, K. Transcriptome and Small RNA Sequencing Reveal the Mechanisms Regulating Harvest Index in Brassica napus. Front. Plant Sci. 2022, 13, 855486. [Google Scholar] [CrossRef]
- Zou, X.; Zhang, K.; Wu, D.; Lu, M.; Wang, H.; Shen, Q. Integrated analysis of miRNA, transcriptome, and degradome sequencing provides new insights into lipid metabolism in perilla seed. Gene 2024, 895, 147953. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ruan, C.; Bao, A.; Li, H. Small RNA profiling for identification of microRNAs involved in regulation of seed development and lipid biosynthesis in yellowhorn. BMC Plant Biol. 2021, 21, 464. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Ruan, C.; Shah, A.H.; Li, D.; Li, H.; Ding, J.; Li, J.; Du, W. Identification of miRNA-mRNA Regulatory Modules Involved in Lipid Metabolism and Seed Development in a Woody Oil Tree (Camellia oleifera). Cells 2021, 11, 71. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhu, Y.; Li, S.; Zhang, W.; Yin, C.; Lin, Y. Regulation of Phytohormones on the Growth and Development of Plant Root Hair. Front. Plant Sci. 2022, 13, 865302. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, L.; Bussell, J.D.; Păcurar, D.I.; Schwambach, J.l.; Păcurar, M.; Bellini, C. Phenotypic Plasticity of Adventitious Rooting in Arabidopsis Is Controlled by Complex Regulation of AUXIN RESPONSE FACTOR Transcripts and MicroRNA Abundance. Plant Cell 2009, 21, 3119–3132. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.-S.; Xie, Q.; Fei, J.F.; Chua, N.-H. MicroRNA Directs mRNA Cleavage of the Transcription Factor NAC1 to Downregulate Auxin Signals for Arabidopsis Lateral Root Development. Plant Cell 2005, 17, 1376–1386. [Google Scholar] [CrossRef]
- Kinoshita, N.; Wang, H.; Kasahara, H.; Liu, J.; MacPherson, C.; Machida, Y.; Kamiya, Y.; Hannah, M.A.; Chua, N.-H. IAA-Ala Resistant3, an Evolutionarily Conserved Target of miR167, MediatesArabidopsisRoot Architecture Changes during High Osmotic Stress. Plant Cell 2012, 24, 3590–3602. [Google Scholar] [CrossRef] [PubMed]
- Yoon, E.K.; Yang, J.H.; Lim, J.; Kim, S.H.; Kim, S.K.; Lee, W.S. Auxin regulation of the microRNA390-dependent transacting small interfering RNA pathway in Arabidopsis lateral root development. Nucleic Acids Res. 2010, 38, 1382–1391. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Zhang, Y.; Moss, B.L.; Bargmann, B.O.R.; Wang, R.; Prigge, M.; Nemhauser, J.L.; Estelle, M. Untethering the TIR1 auxin receptor from the SCF complex increases its stability and inhibits auxin response. Nat. Plants 2015, 1, 14030. [Google Scholar] [CrossRef]
- Jodder, J. miRNA-mediated regulation of auxin signaling pathway during plant development and stress responses. J Biosci. 2020, 45, 91. [Google Scholar] [CrossRef]
- Yan, A.; Chen, Z. The pivotal role of abscisic acid signaling during transition from seed maturation to germination. Plant Cell Rep. 2017, 36, 689–703. [Google Scholar] [CrossRef] [PubMed]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed]
- Brookbank, B.P.; Patel, J.; Gazzarrini, S.; Nambara, E. Role of Basal ABA in Plant Growth and Development. Genes 2021, 12, 1936. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.W.; Baek, W.; Jung, J.; Kim, J.H.; Lee, S.C. Function of ABA in Stomatal Defense against Biotic and Drought Stresses. Int. J. Mol. Sci. 2015, 16, 15251–15270. [Google Scholar] [CrossRef] [PubMed]
- Sano, N.; Marion-Poll, A. ABA Metabolism and Homeostasis in Seed Dormancy and Germination. Int. J. Mol. Sci. 2021, 22, 5069. [Google Scholar] [CrossRef] [PubMed]
- Wilmer, J.A.; Helsper, J.P.F.G.; van der Plas, L.H.W. Effects of abscisic acid and temperature on erucic acid accumulation in oilseed rape (Brassica napus L.). J. Plant Physiol. 1997, 150, 414–419. [Google Scholar] [CrossRef]
- Ma, H.; Wang, S. Histidine Regulates Seed Oil Deposition through Abscisic Acid Biosynthesis and β-oxidation. Plant Physiol. 2016, 172, 848–857. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef] [PubMed]
- Abel, Y.; Rederstorff, M. Stem-Loop qRT-PCR-Based Quantification of miRNAs. Methods Mol. Biol. 2021, 2300, 59–64. [Google Scholar]
- Huang, D.; Koh, C.; Feurtado, J.A.; Tsang, E.W.; Cutler, A.J. MicroRNAs and their putative targets in Brassica napusseed maturation. BMC Genom. 2013, 14, 1–25. [Google Scholar] [CrossRef]
- GB/T17376–2008; Animal and vegetable fats and oils-Preparation of methyl esters of fatty acids. Standards Press of China: Beijing, China, 2008.
- Axtell, M.J.; Meyers, B.C. Revisiting Criteria for Plant MicroRNA Annotation in the Era of Big Data. Plant Cell 2018, 30, 272–284. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Xie, M.; Liang, L.; Yang, L.; Han, H.; Qin, X.; Zhao, J.; Hou, Y.; Dai, W.; Du, C.; et al. Genome-Wide Association Analysis Combined With Quantitative Trait Loci Mapping and Dynamic Transcriptome Unveil the Genetic Control of Seed Oil Content in Brassica napus L. Front. Plant Sci. 2022, 13, 929197. [Google Scholar] [CrossRef]
- Boulard, C.; Bardet, M.; Chardot, T.; Dubreucq, B.; Gromova, M.; Guillermo, A.; Miquel, M.; Nesi, N.; Yen-Nicolaÿ, S.; Jolivet, P. The structural organization of seed oil bodies could explain the contrasted oil extractability observed in two rapeseed genotypes. Planta 2015, 242, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Yu, L.; Wei, L.; Yu, P.; Wang, J.; Zhao, H.; Zhang, Y.; Zhang, S.; Yang, Z.; Chen, G.; et al. BnTIR: An online transcriptome platform for exploring RNA-seq libraries for oil crop Brassica napus. Plant Biotechnol. J. 2021, 19, 1895–1897. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R.; Somerville, C. Abscisic acid or high osmoticum promote accumulation of long-chain fatty acids in developing embryos of Brassica napus. Plant Sci. 1989, 61, 213–217. [Google Scholar] [CrossRef]
- Yu, S.; Galvão, V.C.; Zhang, Y.C.; Horrer, D.; Zhang, T.Q.; Hao, Y.H.; Feng, Y.Q.; Wang, S.; Schmid, M.; Wang, J.-W. Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell 2012, 24, 3320–3332. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, S.; Hou, Q.; Zhao, J.; Fang, C.; An, X.; Wan, X. Fatty acid de novo biosynthesis in plastids: Key enzymes and their critical roles for male reproduction and other processes in plants. Plant Physiol. Biochem. 2024, 210, 108654. [Google Scholar] [CrossRef] [PubMed]
- Ke, J.; Behal, R.H.; Back, S.L.; Nikolau, B.J.; Wurtele, E.S.; Oliver, D.J. The Role of Pyruvate Dehydrogenase and Acetyl-Coenzyme A Synthetase in Fatty Acid Synthesis in Developing Arabidopsis Seeds. Plant Physiol. 2000, 123, 497–508. [Google Scholar] [CrossRef]
- Meï, C.; Michaud, M.; Cussac, M.; Albrieux, C.; Gros, V.; Maréchal, E.; Block, M.A.; Jouhet, J.; Rébeillé, F. Levels of polyunsaturated fatty acids correlate with growth rate in plant cell cultures. Sci. Rep. 2015, 5, 15207. [Google Scholar] [CrossRef]
- Wang, S.H.; Pan, Y.; Li, J.; Chen, H.-q.; Zhang, H.; Chen, W.; Gu, Z.N.; Chen, Y.Q. Endogenous omega-3 long-chain fatty acid biosynthesis from alpha-linolenic acid is affected by substrate levels, gene expression, and product inhibition. RSC Adv. 2017, 7, 40946–40951. [Google Scholar] [CrossRef]
- Perez-Arcoiza, A.; Luisa Hernández, M.; Dolores Sicardo, M.; Hernandez Santana, V.; Diaz Espejo, A.; Martinez Rivas, J.M. Carbon supply and water status regulate fatty acid and triacylglycerol biosynthesis at transcriptional level in the olive mesocarp. Plant Cell Environ. 2022, 45, 2366–2380. [Google Scholar] [CrossRef]
- Liang, G.; Yu, D. Reciprocal regulation among miR395, APS and SULTR2;1 in Arabidopsis thaliana. Plant Signal Behav. 2010, 5, 1257–1259. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Yang, F.; Yu, D. MicroRNA395 mediates regulation of sulfate accumulation and allocation in Arabidopsis thaliana. Plant J. 2010, 62, 1046–1057. [Google Scholar] [CrossRef]
- Chung, M.Y.; Nath, U.K.; Vrebalov, J.; Gapper, N.; Lee, J.M.; Lee, D.J.; Kim, C.K.; Giovannoni, J. Ectopic expression of miRNA172 in tomato (Solanum lycopersicum) reveals novel function in fruit development through regulation of an AP2 transcription factor. BMC Plant Biol. 2020, 20, 283. [Google Scholar] [CrossRef]
- Nowak, K.; Morończyk, J.; Grzyb, M.; Szczygieł-Sommer, A.; Gaj, M.D. miR172 Regulates WUS during Somatic Embryogenesis in Arabidopsis via AP2. Cells 2022, 11, 718. [Google Scholar] [CrossRef] [PubMed]
- Zeng, V.; Uauy, C.; Chen, Y. Identification of a novel SNP in the miR172 binding site of Q homoeolog AP2L-D5 is associated with spike compactness and agronomic traits in wheat (Triticum aestivum L.). Theor. Appl. Genet. 2023, 137, 13. [Google Scholar] [CrossRef]
- Jung, J.H.; Lee, S.; Yun, J.; Lee, M.; Park, C.M. The miR172 target TOE3 represses AGAMOUS expression during Arabidopsis floral patterning. Plant Sci. 2014, 215–216, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, Q.; Zhu, H.; Song, M.; Zhang, K.; Ge, W. Molecular mechanisms of miR172a and its target gene LbrTOE3 regulating maturation in Lilium. Planta 2023, 258, 53. [Google Scholar] [CrossRef]
- Liu, J.; Liu, X.; Zhang, S.; Liang, S.; Luan, W.; Ma, X. TarDB: An online database for plant miRNA targets and miRNA-triggered phased siRNAs. BMC Genom. 2021, 22, 348. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Z.; Yi, Z.; Zhao, J. Identification of miRNAs Interacting with Abscisic Acid to Regulate Fatty Acid Metabolism. Agronomy 2024, 14, 1358. https://doi.org/10.3390/agronomy14071358
Xu Z, Yi Z, Zhao J. Identification of miRNAs Interacting with Abscisic Acid to Regulate Fatty Acid Metabolism. Agronomy. 2024; 14(7):1358. https://doi.org/10.3390/agronomy14071358
Chicago/Turabian StyleXu, Zhijun, Zhenxie Yi, and Jing Zhao. 2024. "Identification of miRNAs Interacting with Abscisic Acid to Regulate Fatty Acid Metabolism" Agronomy 14, no. 7: 1358. https://doi.org/10.3390/agronomy14071358



