Integrated Analysis of the lncRNA-miRNA-mRNA Expression Profiles in Response to Meloidogyne incognita in Radish (Raphanus sativus L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Growth Condition
2.2. Measurement of Endogenous Jasmonic Acid (JA) and Abscisic Acid (ABA) Content
2.3. Strand-Specific RNA Sequencing and LncRNA Identification
2.4. Label-Free Proteomics Analysis
2.5. Small RNA Sequencing, miRNA Identification and Targets’ Prediction
2.6. Quantitative Reverse Transcription-PCR (qRT-PCR)
3. Results
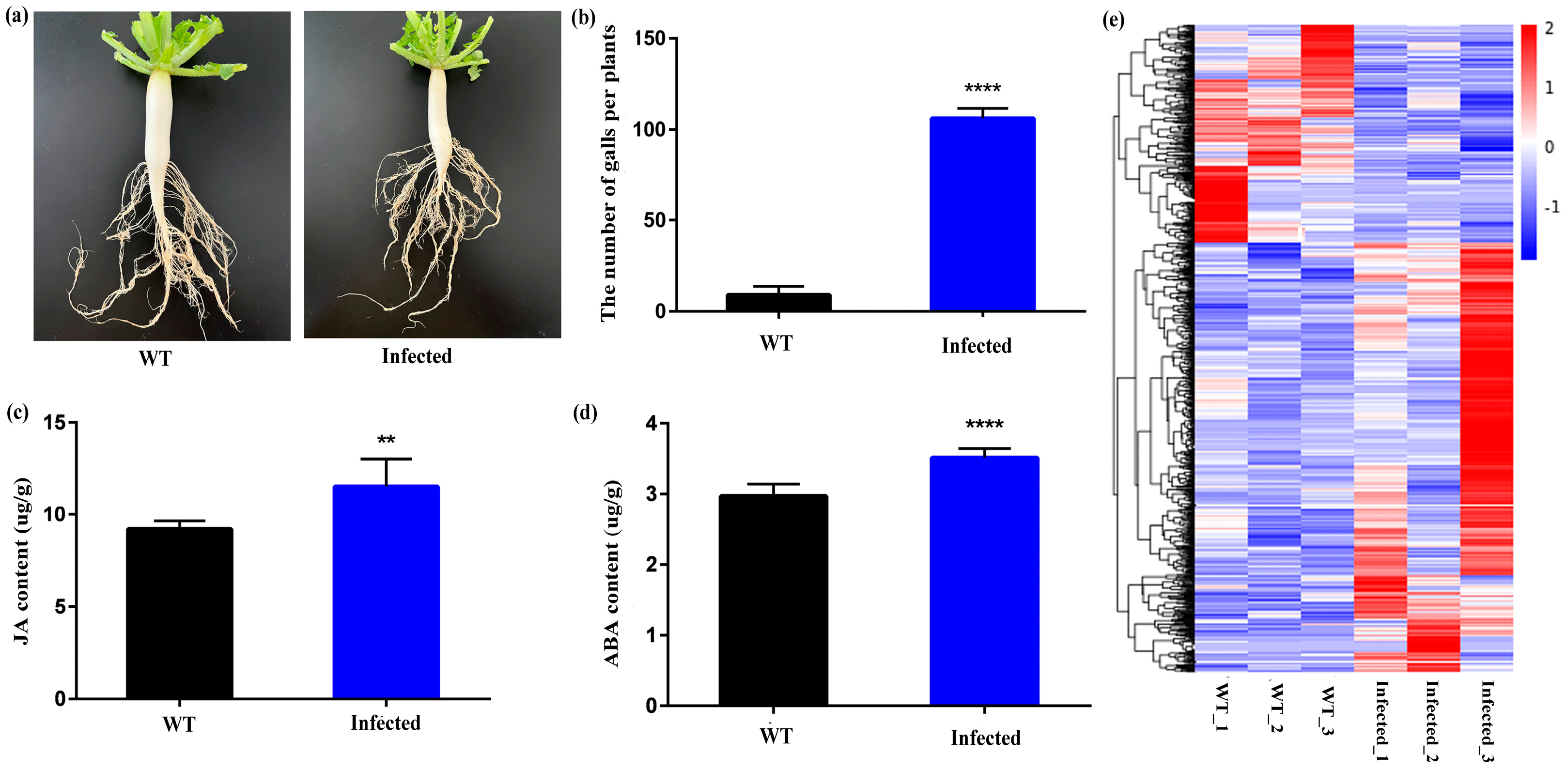
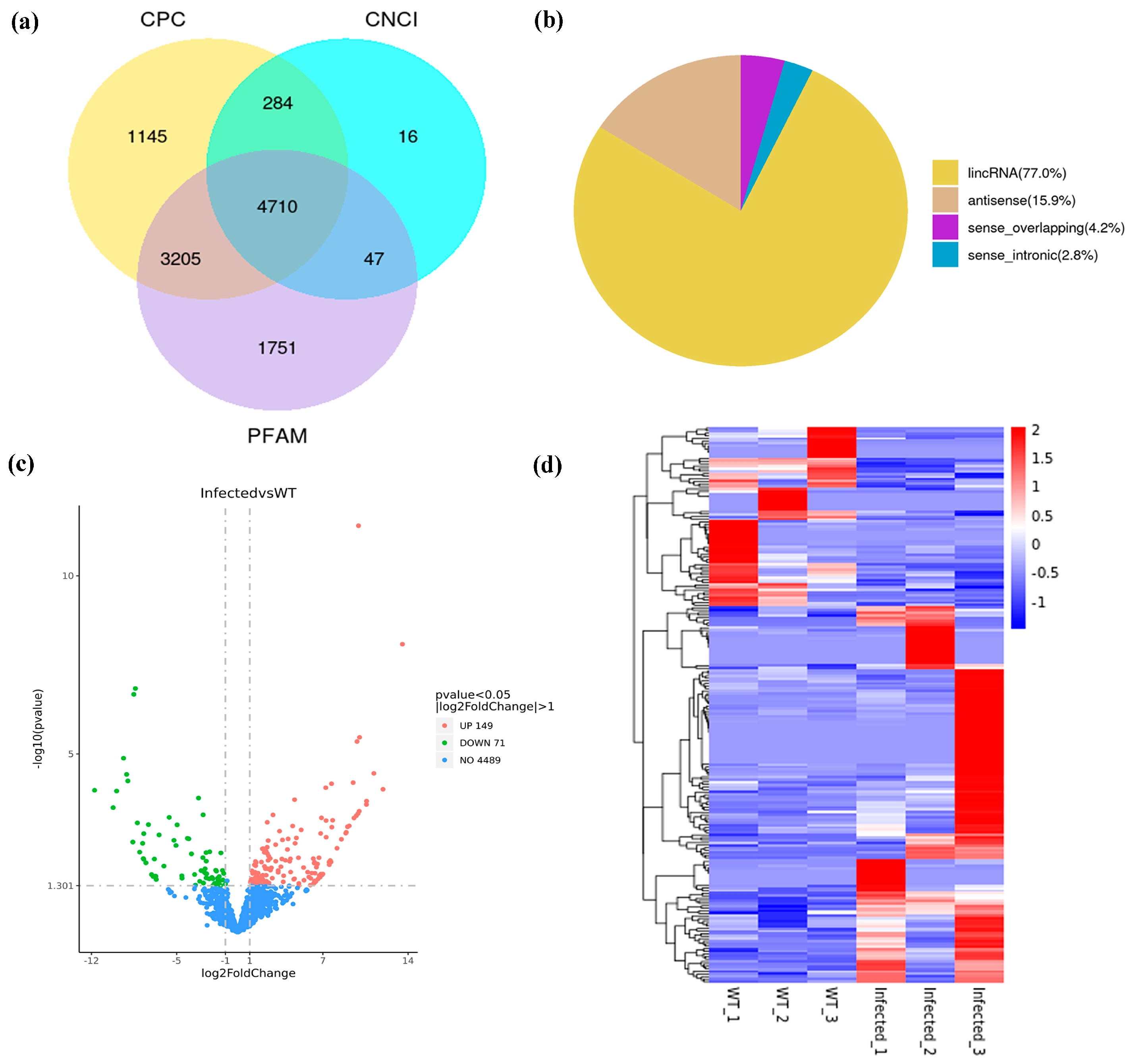
3.1. Identification and Characterization of DElncRNAs and DEGs
3.2. The Differential Expression Genes Involved in RKN Defense Response
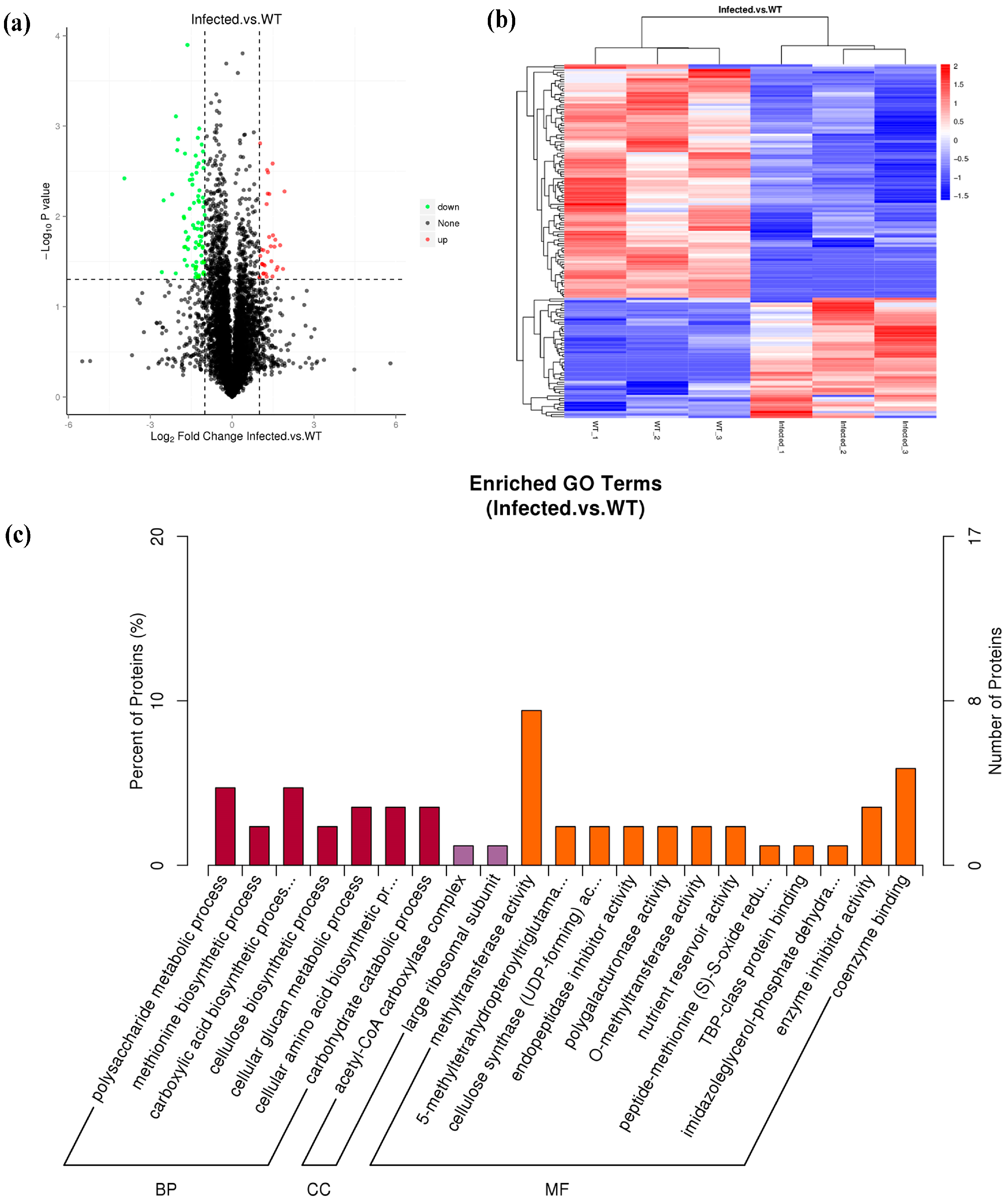
3.3. Proteome-Wide Analysis of the Radish Root in Response to RKN Infection
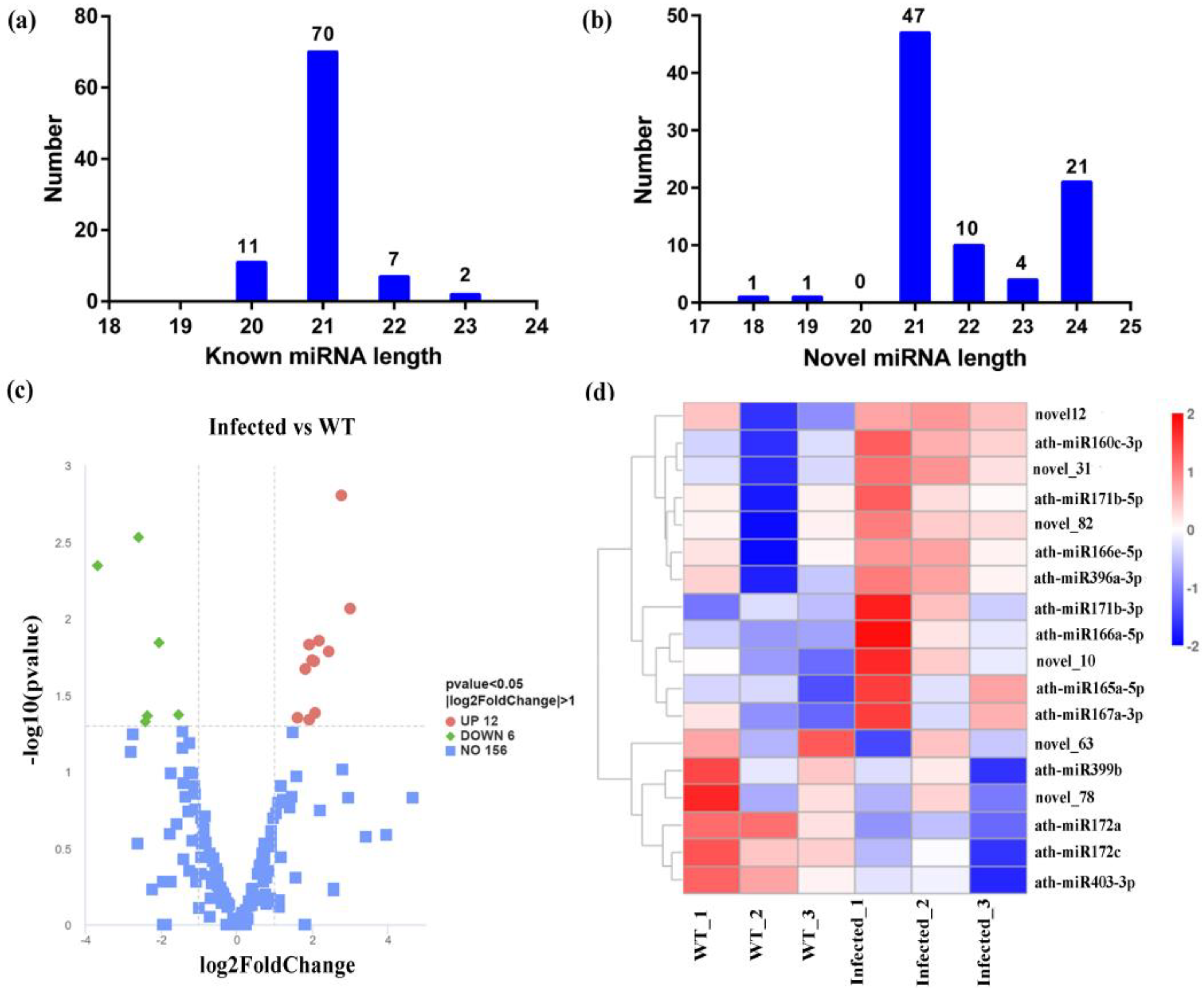
3.4. The sRNA Sequencing Data, miRNA Identification and Target Genes Analysis
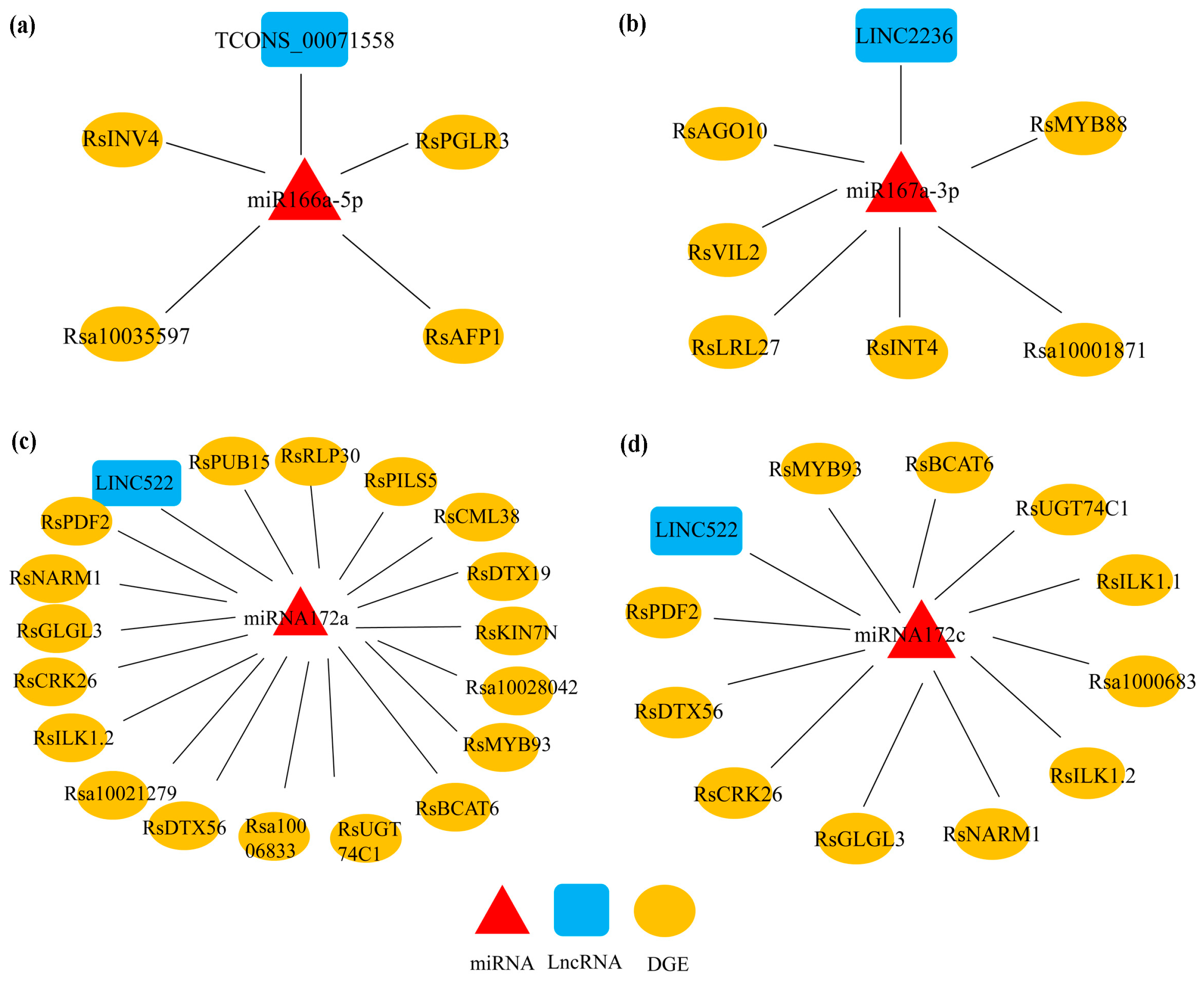
3.5. The Regulatory Network of lncRNA-miRNA-mRNA in Response to RKN Infection
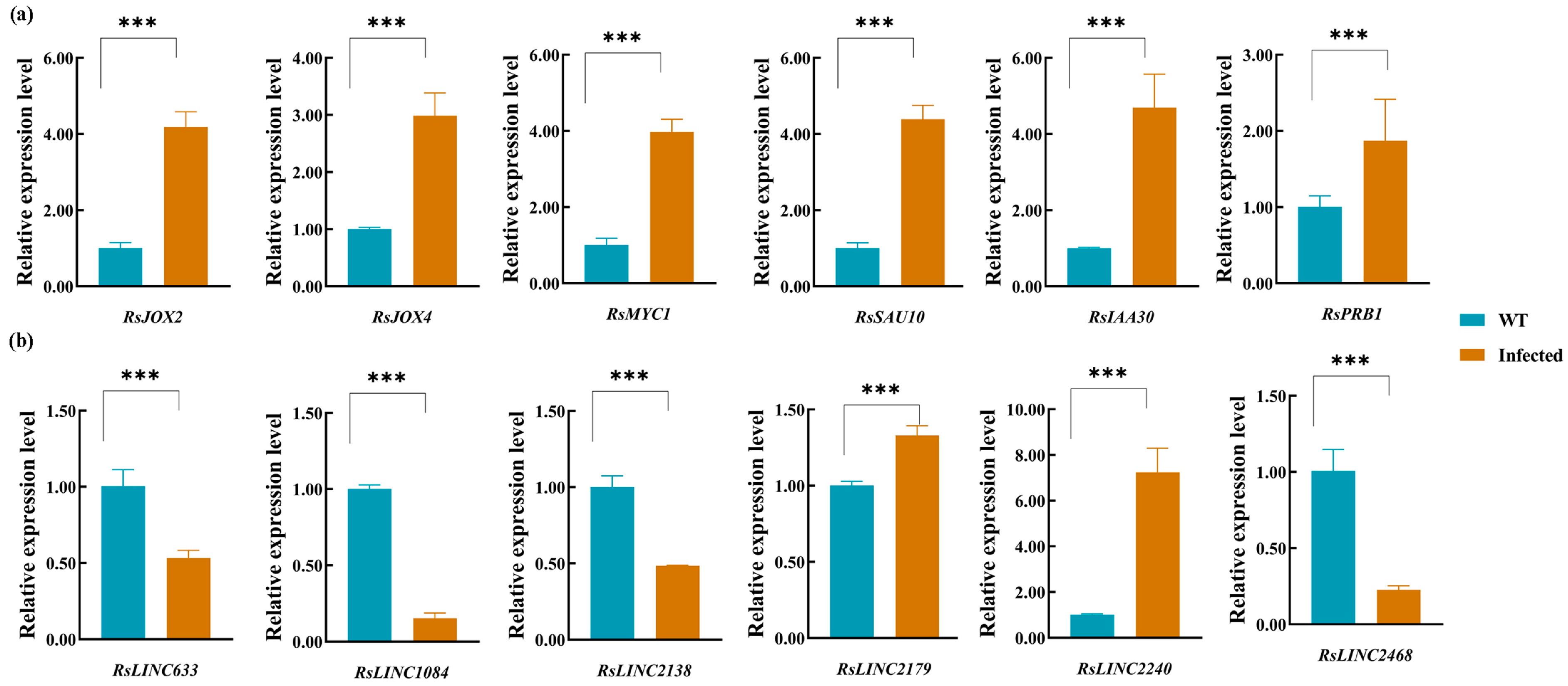
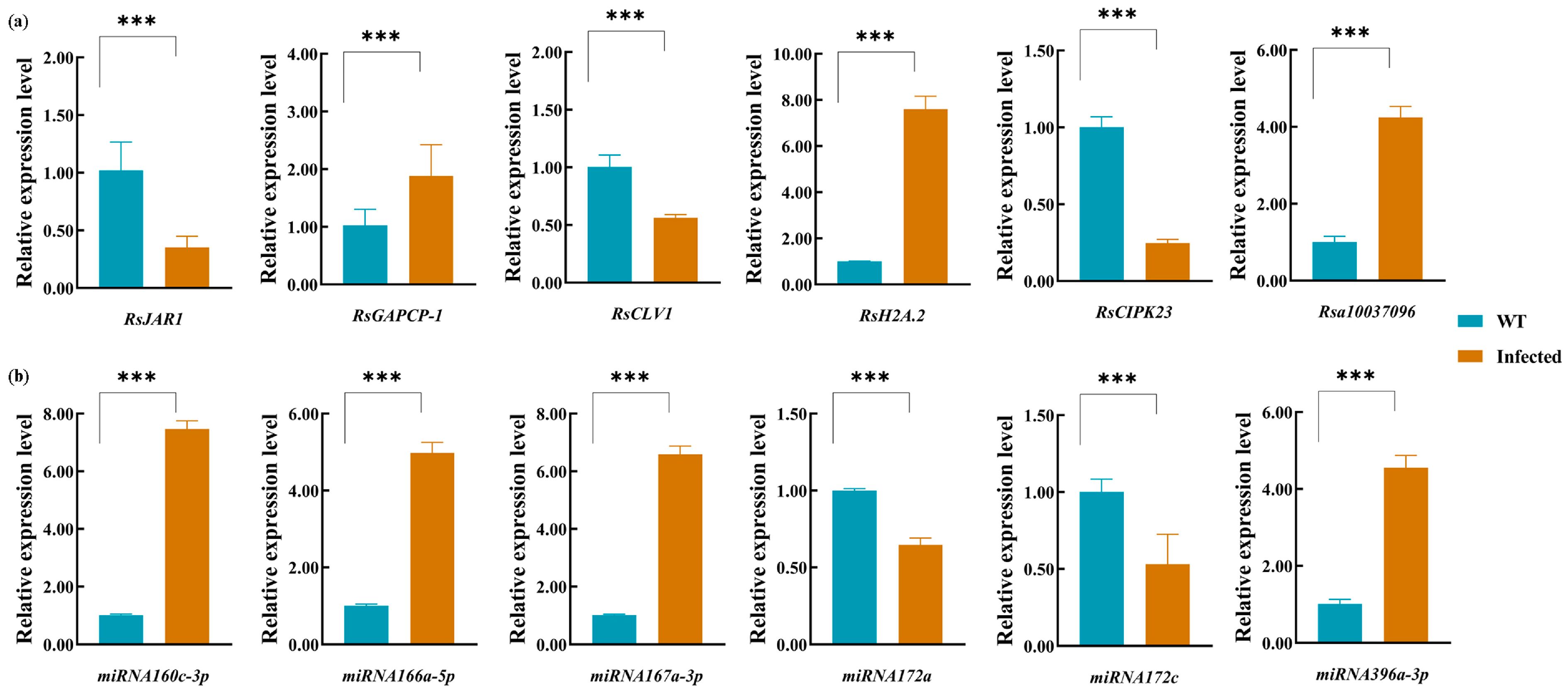
3.6. Validation of Gene Expression of mRNAs, lncRNAs and miRNAs
4. Discussion
4.1. miRNA Response to Root-Knot Nematode Infection in Radish
4.2. RKN Infection Is Associated to Changes in Hormone Homeostasis in Roots
4.3. The Important DGEs Involved in RKN Infections
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abad, P.; Gouzy, J.; Aury, J.M.; Castagnone-Sereno, P.; Danchin, E.G.; Deleury, E.; Perfus-Barbeoch, L.; Anthouard, V.; Artiguenave, F.; Blok, V.C.; et al. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 2008, 26, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Mejias, J.; Quentin, M.; Chen, Y.; Almeida-Engler, J.; Mao, Z.; Sun, Q.; Liu, Q.; Xie, B.; Abad, P.; et al. The root-knot nematode effector MiPDI1 targets a stress-associated protein (SAP) to establish disease in Solanaceae and Arabidopsis. New Phytol. 2020, 228, 1417–1430. [Google Scholar] [CrossRef] [PubMed]
- Goodey, B.; Franklin, M.F.; Hooper, D.J. The Nematode Parasites of Plants Classified under Their Hosts, 3rd ed.; CAB International: Farnahm Royal, UK, 1965. [Google Scholar]
- Warmerdam, S.; Sterken, M.G.; Schaik, C.V.; Oortwijn, M.E.P.; Sukarta, O.C.A.; Lozano-Torres, J.L.; Dicke, M.; Helder, J.; Kammenga, J.E.; Goverse, A.; et al. Genome-wide association mapping of the architecture of susceptibility to the root-knot nematode Meloidogyne incognita in Arabidopsis thaliana. New Phytol. 2018, 218, 724–737. [Google Scholar] [CrossRef] [PubMed]
- Oloka, B.M.; Pereira, G.D.S.; Amankwaah, V.A.; Mollinari, M.A.; Pecota, K.V.; Yada, B.; Olukolu, B.A.; Zeng, G.B.; Yencho, G. Discovery of a major QTL for root-knot nematode (Meloidogyne incognita) resistance in cultivated sweetpotato (Ipomoea batatas). Theor. Appl. Genet. 2021, 134, 1945–1955. [Google Scholar] [CrossRef] [PubMed]
- Alekcevetch, J.C.; Passianotto, A.L.L.; Ferreira, E.G.C.; Santos, A.B.; Silva, D.C.G.; Dias, W.P.; Belzile, F.; Ricardo Vilela Abdelnoor, R.V.; Marcelino-Guimarães, F.C. Genome-wide association study for resistance to the Meloidogyne javanica causing root-knot nematode in soybean. Theor. Appl. Genet. 2021, 134, 777–792. [Google Scholar] [PubMed]
- Mejias, J.; Bazin, J.; Truong, N.M.; Chen, Y.; Marteu, N.; Bouteiller, N.; Sawa, S.; Crespi, M.; Vaucheret, H.; Abad, P.; et al. The root-knot nematode effector MiEFF18 interacts with the plant core spliceosomal protein SmD1 required for giant cell formation. New Phytol. 2021, 229, 3408–3423. [Google Scholar] [PubMed]
- Zhao, W.; Liang, J.; Huang, H.; Yang, J.; Feng, J.; Sun, L.; Wang, S. Tomato defence against Meloidogyne incognita by jasmonic acid-mediated fine-tuning of kaempferol homeostasis. New Phytol. 2023, 238, 1651. [Google Scholar]
- Huang, L.; Dong, H.; Zhou, D.; Li, M.; Liu, Y.; Zhang, F.; Feng, Y.; Yu, D.; Lin, S.; Cao, J. Systematic identification of long non-coding RNA s during pollen development and fertilization in Brassica rapa. Plant J. 2018, 96, 203–222. [Google Scholar] [PubMed]
- Yang, F.; Zhao, D.; Fan, H.; Zhu, X.; Wang, Y.; Liu, X.; Duan, Y.; Xuan, Y.; Chen, L. Functional analysis of long non-coding RNAs reveal their novel roles in biocontrol of bacteria-induced tomato resistance to Meloidogyne incognita. Int. J. Mol. Sci. 2020, 21, 911. [Google Scholar] [CrossRef]
- Khoei, M.A.; Karimi, M.; Karamian, R.; Amini, S.; Soorni, A. Identification of the complex interplay between nematode-related lncRNAs and their target genes in Glycine max L. Front. Plant Sci. 2021, 12, 779597. [Google Scholar] [CrossRef]
- Xu, P.; Li, H.; Wang, X.; Zhao, G.; Lu, X.; Dai, S.; Cui, X.; Mei, Y.; Liu, Z. Integrated analysis of the lncRNA/circRNA-miRNA-mRNA expression profiles reveals novel insights into potential mechanisms in response to root-knot nematodes in peanut. BMC Genom. 2022, 23, 239. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Yang, T.; Wang, B.; Han, B.; Zhou, H.; Wang, Y.; Li, D.; Liu, A. Differential expression networks and inheritance patterns of long non-coding RNA s in castor bean seeds. Plant J. 2018, 95, 324–340. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.B.; Sung, S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 2011, 331, 76. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Fu, D.; Zhu, B.; Luo, Y.; Zhu, H. CRISPR/Cas9-mediated mutagenesis of lncRNA1459 alters tomato fruit ripening. Plant J. 2018, 94, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Martínez, D.; Barrero-Gil, J.; Tranque, E.; Ruiz, M.F.; Catalá, R.; Salinas, J. SVALKA-POLYCOMB REPRESSIVE COMPLEX2 module controls C-REPEAT BINDING FACTOR3 induction during cold acclimation. Plant Physiol. 2024, 195, 1152–1160. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.; Li, C.; Sun, R.; Zhang, B.; Nichols, R.L.; Hake, K.D.; Pan, X. Small RNA and degradome deep sequencing reveals important roles of microRNAs in cotton (Gossypium hirsutum L.) response to root-knot nematode Meloidogyne incognita infection. Genomics 2021, 113, 1146–1156. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.W.; Czech, B.; Weigel, D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 2009, 138, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Xue, T.; Liu, Z.; Dai, X.; Xiang, F. Primary root growth in Arabidopsis thaliana is inhibited by the miR159 mediated repression of MYB33, MYB65 and MYB101. Plant Sci. 2017, 262, 182–189. [Google Scholar] [CrossRef]
- Barciszewska-Pacak, M.; Milanowska, K.; Knop, K.; Bielewicz, D.; Nuc, P.; Plewka, P.; Pacak, A.M.; Vazquez, F.; Karlowski, W.; Jarmolowski, A.; et al. Arabidopsis microRNA expression regulation in a wide range of abiotic stress responses. Front. Plant Sci. 2015, 6, 410. [Google Scholar] [CrossRef]
- Zhao, W.; Li, Z.; Fan, J.; Hu, C.; Yang, R.; Qi, X.; Chen, H.; Zhao, F.; Wang, S. Identification of jasmonic acid-associated microRNAs and characterization of the regulatory roles of the miR319/TCP4 module under root-knot nematode stress in tomato. J. Exp. Bot. 2015, 66, 4653–4667. [Google Scholar] [CrossRef]
- Medina, C.; Rocha, M.; Magliano, M.; Ratpopoulo, A.; Revel, B.; Marteu, N.; Magnone, V.; Lebrigand, K.; Cabrera, J.; Barcala, M.; et al. Characterization of microRNAs from Arabidopsis galls highlights a role for miR159 in the plant response to the root-knot nematode Meloidogyne incognita. New Phytol. 2017, 216, 882–896. [Google Scholar] [CrossRef] [PubMed]
- Noureddine, Y.; Mejias, J.; Rocha, M.; Thomine, S.; Quentin, M.; Abad, P.; Favery, B.; Jaubert-Possamai, S. Copper microRNAs modulate the formation of giant feeding cells induced by the root knot nematode Meloidogyne incognita in Arabidopsis thaliana. New Phytol. 2022, 236, 283–295. [Google Scholar] [PubMed]
- Noureddine, Y.; da Rocha, M.; An, J.; Médina, C.; Mejias, J.; Mulet, K.; Quentin, M.; Abad, P.; Zouine, M.; Favery, B.; et al. AUXIN RESPONSIVE FACTOR8 regulates development of the feeding site induced by root-knot nematodes in tomato. J. Exp. Bot. 2023, 74, 5752–5766. [Google Scholar] [CrossRef] [PubMed]
- Bünte, R.; Müller, J.; Friedt, W. Genetic variation and response to selection for resistance to root-knot nematodes in oil radish (Raphanus sativus ssp. oleiferus). Plant Breed. 1997, 116, 263–266. [Google Scholar] [CrossRef]
- Min, Y.Y.; Toyota, K.; Sato, E.; Takada, A. Effects of anaerobically digested slurry on Meloidogyne incognita and Pratylenchus penetrans in tomato and radish production. Appl. Environ. Soil Sci. 2011, 2011, 1687–7667. [Google Scholar]
- Luo, X.; Xu, L.; Wang, Y.; Dong, J.; Chen, Y.; Tang, M.; Fan, L.; Zhu, Y.; Liu, L. An ultra-high-density genetic map provides insights into genome synteny, recombination landscape and taproot skin colour in radish (Raphanus sativus L.). Plant Biotechnol. J. 2020, 18, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Luo, X.; Li, Y.; Peng, X.; Wu, L.; Yang, G.; Xu, X.; Pei, Y.; Li, W.; Zhang, W. Fine mapping and analysis of candidate genes for qBT2 and qBT7.2 locus controlling bolting time in radish (Raphanus sativus L.). Theor. Appl. Genet. 2024, 137, 4. [Google Scholar]
- Wang, X.; Li, Q.; Xie, J.; Huang, M.; Cai, J.; Zhou, Q.; Dai, T.; Jiang, D. Abscisic acid and jasmonic acid are involved in drought priming-induced tolerance to drought in wheat. Crop J. 2021, 9, 120–132. [Google Scholar]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNASeq. Nat. Med. 2008, 5, 621–628. [Google Scholar]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.P.; Li, W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhang, Y.; Ye, Z.; Liu, X.; Zhao, S.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 36, W345–W349. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Jing, F.; Zhou, Y.; Zhang, P.; Huang, R.; Fan, W.; Li, B.; Li, G.; Song, X.; Pei, D. Identification of Key Lipogenesis Stages and Proteins Involved in Walnut Kernel Development. J. Agric. Food Chem. 2023, 71, 4306–4318. [Google Scholar]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast andmemory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Wen, M.; Shen, Y.; Shi, S.; Tang, T. miREvo: An integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinform. 2012, 13, 140. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhuang, Z.; Zhao, P. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Xu, X.; Fang, P.; Zhang, H.; Chi, C.; Song, L.; Xia, X.; Shi, K.; Zhou, Y.; Jie Zhou, J.; Yu, J. Strigolactones positively regulate defense against root-knot nematodes in tomato. J. Exp. Bot. 2019, 70, 1325–1337. [Google Scholar] [CrossRef]
- Zou, J.; Chen, X.; Liu, C.; Guo, M.; Kanwar, M.K.; Qi, Z.; Yang, P.; Wang, G.; Bao, Y.; Bassham, D.C.; et al. Autophagy promotes jasmonate-mediated defense against nematodes. Nat. Commun. 2023, 14, 4769. [Google Scholar] [CrossRef]
- Cabrera, J.; Barcala, M.; García, A.; Ana Rio-Machín, A.; Medina, C.; Jaubert-Possamai, S.; Favery, B.; Maizel, A.; Ruiz-Ferrer, V.; Fenoll, C.; et al. Differentially expressed small RNAs in arabidopsis galls formed by meloidogyne javanica: A functional role for miR390 and its TAS3-derived tasiRNAs. New Phytol. 2015, 209, 1625–1640. [Google Scholar] [CrossRef]
- Díaz-Manzano, F.E.; Cabrera, J.; Ripoll, J.; Olmo, I.; Andrés, M.; Silva, A.; Barcala, M.; Sánchez, M.; Ruíz-Ferrer, V.; Almeida-Engler, J.; et al. A role for the gene regulatory module microRNA172/TARGET OF EARLY ACTIVATION TAGGED 1/FLOWERING LOCUS T (miRNA172/TOE 1/FT) in the feeding sites induced by Meloidogyne javanica in Arabidopsis thaliana. New Phytol. 2018, 217, 813–827. [Google Scholar] [CrossRef]
- Noon, J.B.; Hewezi, T.; Baum, T.J. Homeostasis in the soybean miRNA396-GRF network is essential for productive soybean cyst nematode infections. J. Exp. Bot. 2019, 70, 1653–1668. [Google Scholar] [CrossRef]
- Pan, X.; Nichols, R.L.; Li, C.; Zhang, B. MicroRNA-target gene responses to root knot nematode (Meloidogyne incognita) infection in cotton (Gossypium hirsutum L.). Genomics 2019, 111, 383–390. [Google Scholar] [CrossRef]
- Nahar, K.; Kyndt, T.; Nzogela, Y.B.; Gheysen, G. Abscisic acid interacts antagonistically with classical defense pathways in rice–migratory nematode interaction. New Phytol. 2012, 196, 901–913. [Google Scholar] [CrossRef]
- Grunewald, W.; Cannoot, B.; Friml, J.; Gheysen, G. Parasitic nematodes modulate PIN-mediated auxin transport to facilitate infection. PLoS Pathog. 2009, 5, e1000266. [Google Scholar] [CrossRef]
- Caarls, L.; Elberse, J.; Awwanah, M.; Ludwig, N.R.; Vries, M.; Zeilmaker, T.; Van Wees, S.C.M.; Schuurink, R.C.; Ackerveken, G. Arabidopsis JASMONATE-INDUCED OXYGENASES down-regulate plant immunity by hydroxylation and inactivation of the hormone jasmonic acid. Proc. Natl. Acad. Sci. USA 2017, 114, 6388–6393. [Google Scholar] [CrossRef]
- Nahar, K.; Kyndt, T.; De Vleesschauwer, D.; Höfte, M.; Gheysen, G. The jasmonate pathway is a key player in systemically induced defense against root knot nematodes in rice. Plant Physiol. 2011, 157, 305–316. [Google Scholar] [CrossRef]
- Shukla, N.; Yadav, R.; Kaur, P.; Rasmussen, S.; Goel, S.; Agarwal, M.; Jagannath, A.; Gupta, R.; Kumar, A. Transcriptome analysis of root-knot nematode (Meloidogyne incognita)-infected tomato (Solanum lycopersicum) roots reveals complex gene expression profiles and metabolic networks of both host and nematode during susceptible and resistance responses. Mol. Plant Pathol. 2018, 19, 615–633. [Google Scholar] [CrossRef]
- Martínez-Medina, A.; Fernandez, I.; Lok, G.B.; Pozo, M.J.; Pieterse, C.M.; Van Wees, S.C. Shifting from priming of salicylic acid-to jasmonic acid-regulated defences by Trichoderma protects tomato against the root knot nematode Meloidogyne incognita. New Phytol. 2017, 213, 1363–1377. [Google Scholar] [CrossRef] [PubMed]
- Chinnapandi, B.; Bucki, P.; Braun Miyara, S. SlWRKY45, nematode-responsive tomato WRKY gene, enhances susceptibility to the root knot nematode; M. javanica infection. Plant Signal. Behav. 2017, 12, e1356530. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, H.; Tan, J.; Huang, S.; Chen, X.; Jiang, D.; Xiao, X. Transcriptome analysis of eggplant root in response to root-knot nematode infection. Pathogens 2021, 10, 470. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Zhao, W.; Qiao, H.; Li, C.; Sun, L.; Yang, R.; Ma, X.; Ma, J.; Song, S.; Wang, S. SlWRKY45 interacts with jasmonate-ZIM domain proteins to negatively regulate defense against the root-knot nematode Meloidogyne incognita in tomato. Hortic. Res. 2022, 9, uhac197. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, C.; de Melo, B.P.; Lourenço-Tessutti, I.T.; Ballesteros, H.F.; Ribeiro, K.V.G.; Menuet, K.; Heyman, J.; Hemerly, A.; de Sá, M.; Veylder, L.; et al. The regeneration conferring transcription factor complex ERF115-PAT1 coordinates a wound-induced response in root-knot nematode induced galls. New Phytol. 2024, 241, 878–895. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Xu, X.; Wang, F.; Wang, Y.; Xia, X.J.; Shi, K.; Zhou, Y.; Zhou, J.; Yu, J. Brassinosteroids act as a positive regulator for resistance against root-knot nematode involving RESPIRATORY BURST OXIDASE HOMOLOG-dependent activation of MAPKs in tomato. Plant Cell Environ. 2018, 41, 1113–1125. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Hu, C.; Zhou, J.; Liu, Y.; Cai, J.; Pan, C.; Wang, Y.; Wu, X.; Shi, K.; Xia, X.; et al. Systemic root-shoot signaling drives jasmonate-based root defense against nematodes. Curr. Biol. 2019, 29, 3430–3438. [Google Scholar] [CrossRef] [PubMed]
- Hamamouch, N.; Li, C.; Seo, P.J.; Park, C.M.; Davis, E.L. Expression of Arabidopsis pathogenesis-related genes during nematode infection. Mol. Plant Pathol. 2011, 12, 355–364. [Google Scholar] [CrossRef]
- Truong, N.M.; Chen, Y.; Mejias, J.; Soulé, S.; Mulet, K.; Jaouannet, M.; Jaubert-Possamai, S.; Sawa, S.; Abad, P.; Favery, B.; et al. The Meloidogyne incognita nuclear effector MiEFF1 interacts with Arabidopsis cytosolic glyceraldehyde-3-phosphate dehydrogenases to promote parasitism. Front. Plant Sci. 2021, 12, 641480. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, X.; Jin, Y.; Shen, F.; Zhang, W. Integrated Analysis of the lncRNA-miRNA-mRNA Expression Profiles in Response to Meloidogyne incognita in Radish (Raphanus sativus L.). Agronomy 2024, 14, 1603. https://doi.org/10.3390/agronomy14081603
Luo X, Jin Y, Shen F, Zhang W. Integrated Analysis of the lncRNA-miRNA-mRNA Expression Profiles in Response to Meloidogyne incognita in Radish (Raphanus sativus L.). Agronomy. 2024; 14(8):1603. https://doi.org/10.3390/agronomy14081603
Chicago/Turabian StyleLuo, Xiaobo, Yueyue Jin, Feng Shen, and Wanping Zhang. 2024. "Integrated Analysis of the lncRNA-miRNA-mRNA Expression Profiles in Response to Meloidogyne incognita in Radish (Raphanus sativus L.)" Agronomy 14, no. 8: 1603. https://doi.org/10.3390/agronomy14081603
APA StyleLuo, X., Jin, Y., Shen, F., & Zhang, W. (2024). Integrated Analysis of the lncRNA-miRNA-mRNA Expression Profiles in Response to Meloidogyne incognita in Radish (Raphanus sativus L.). Agronomy, 14(8), 1603. https://doi.org/10.3390/agronomy14081603





