Performance of Machine Learning Models in Predicting Common Bean (Phaseolus vulgaris L.) Crop Nitrogen Using NIR Spectroscopy
Abstract
1. Introduction
2. Materials and Methods
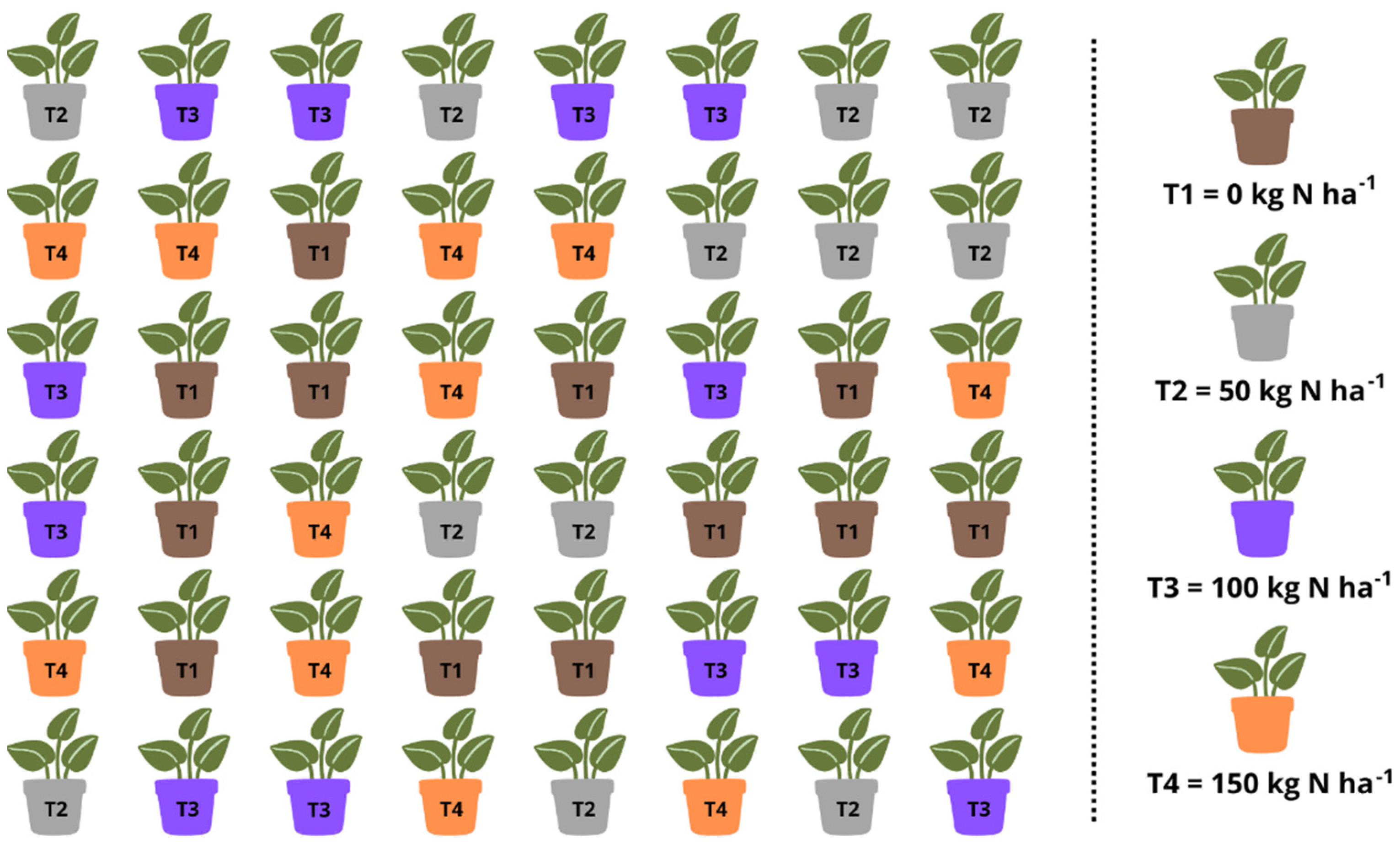
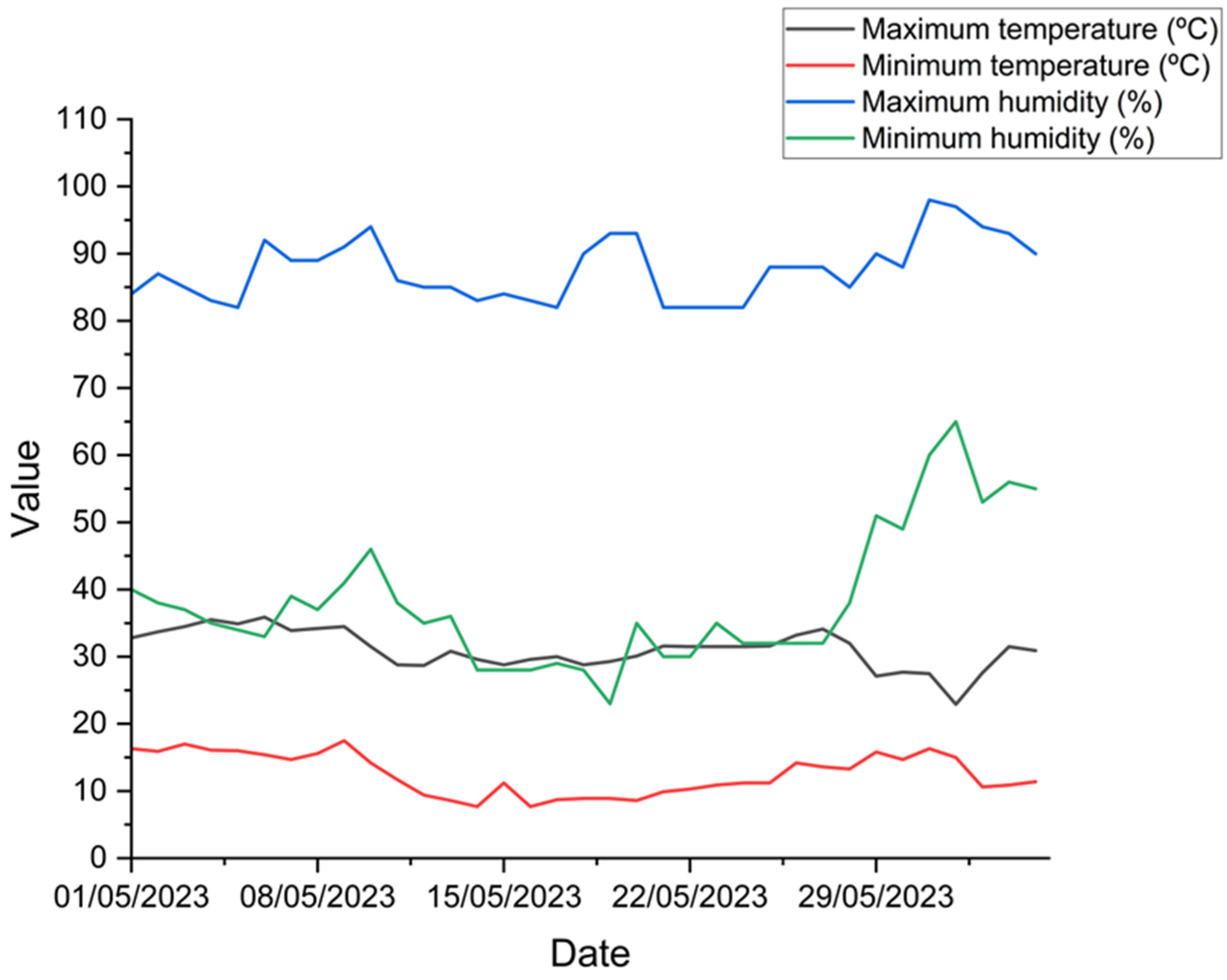
2.1. Study Place and Experimental Design
2.2. Foliar Collection, Hyperespectral Data Acquisition, and Quantification of Leaf Nitrogen
2.3. Model Description and Performance Analysis
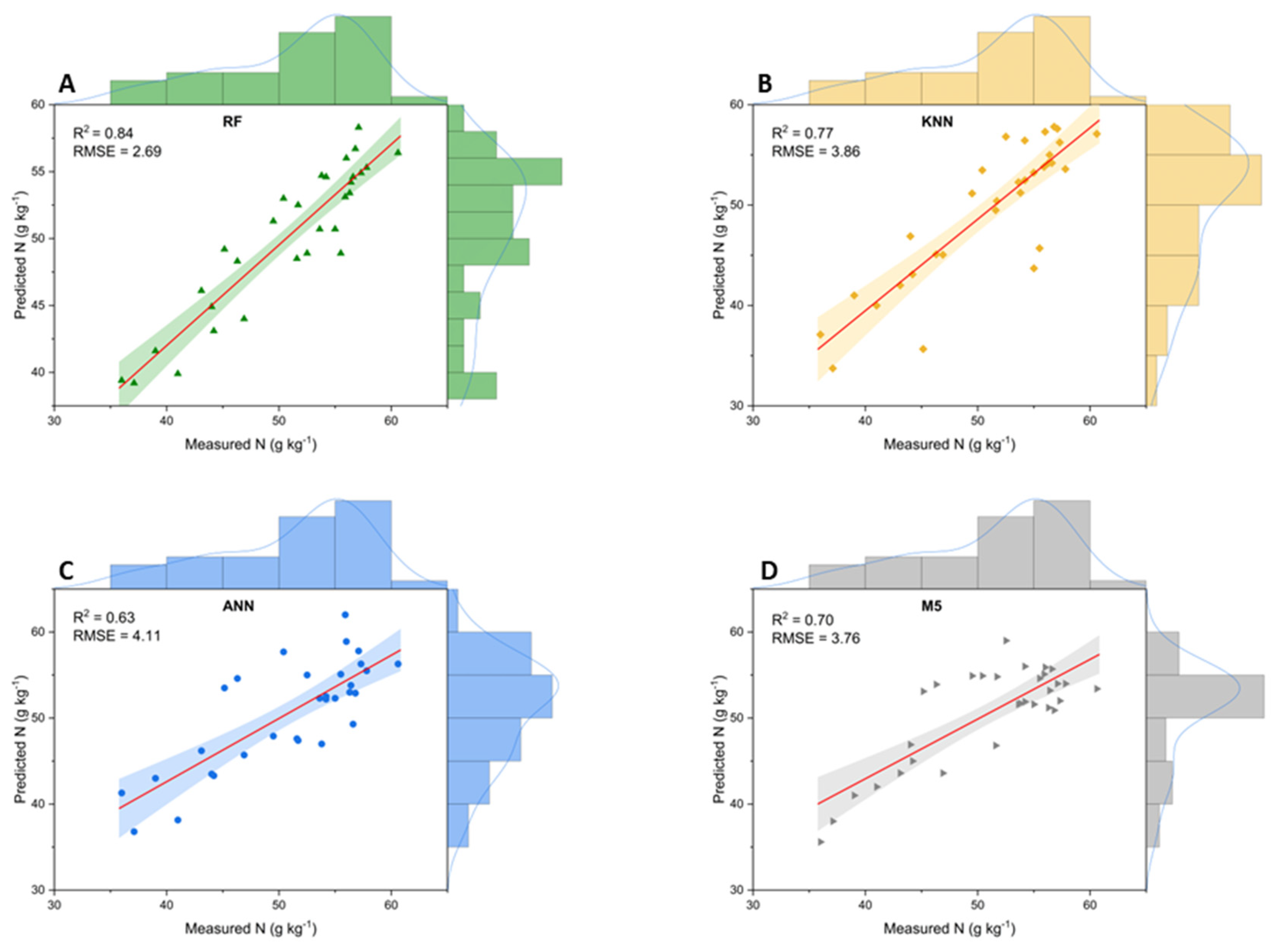
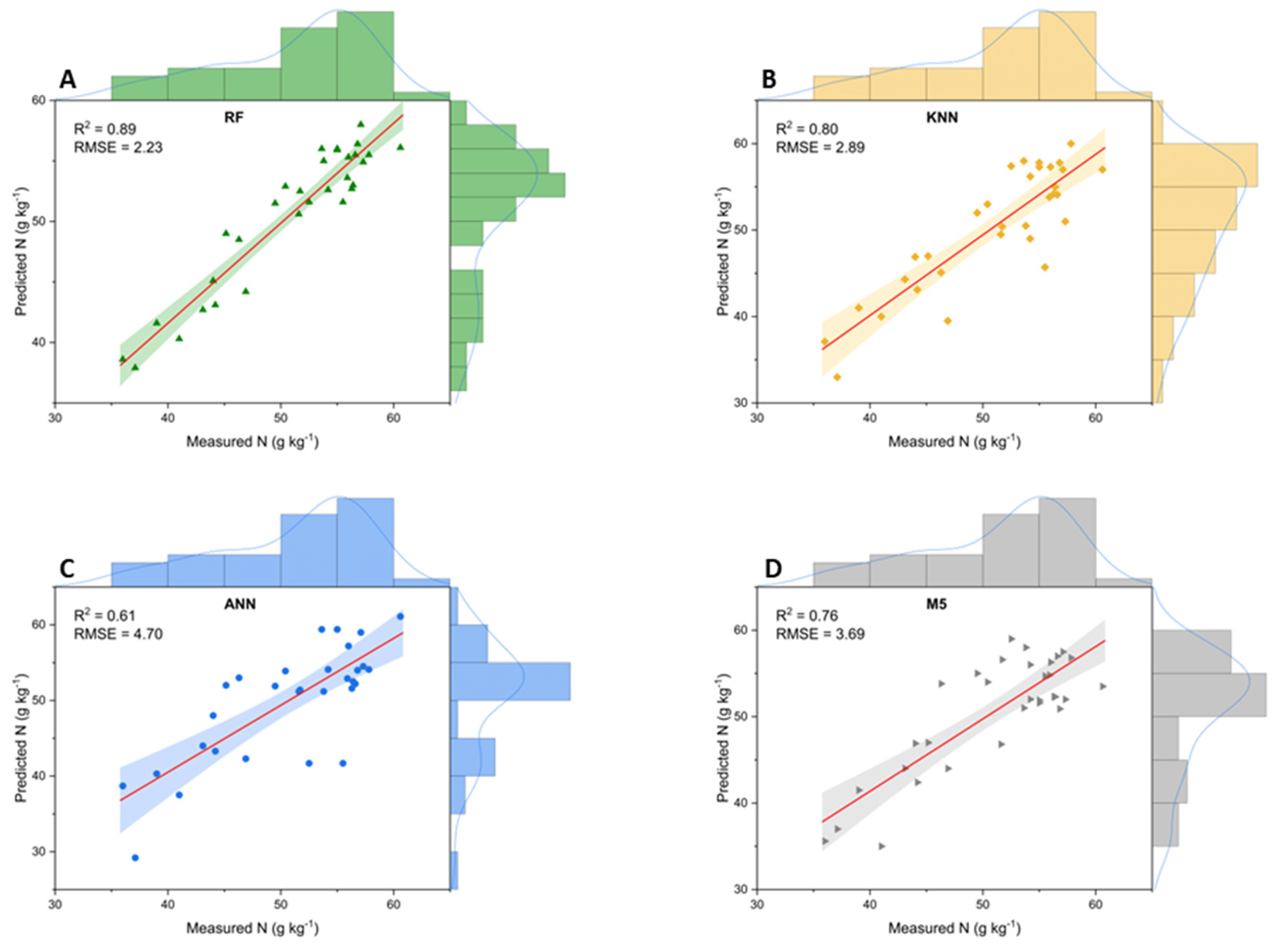
3. Results
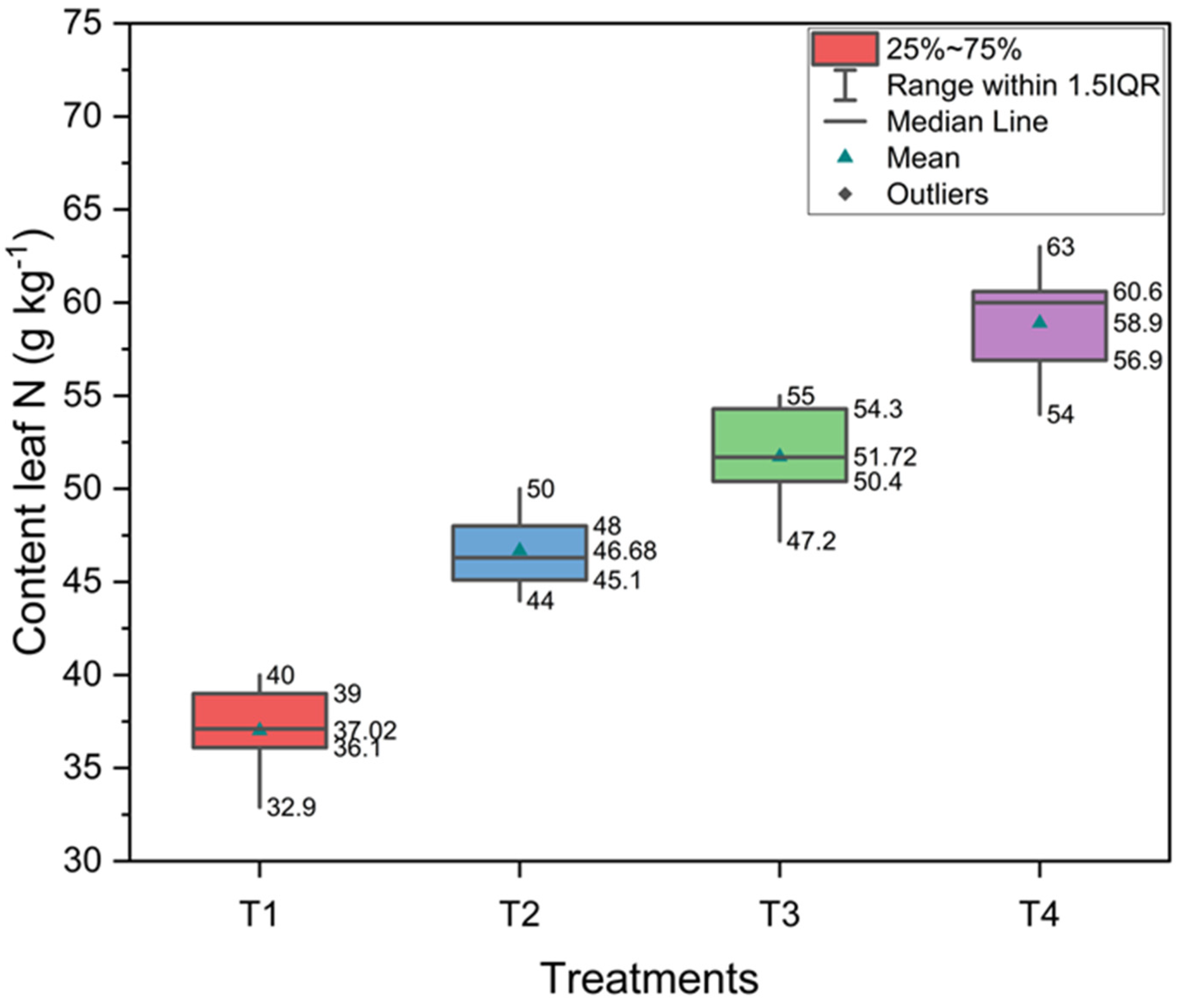
3.1. Foliar Nitrogen Content
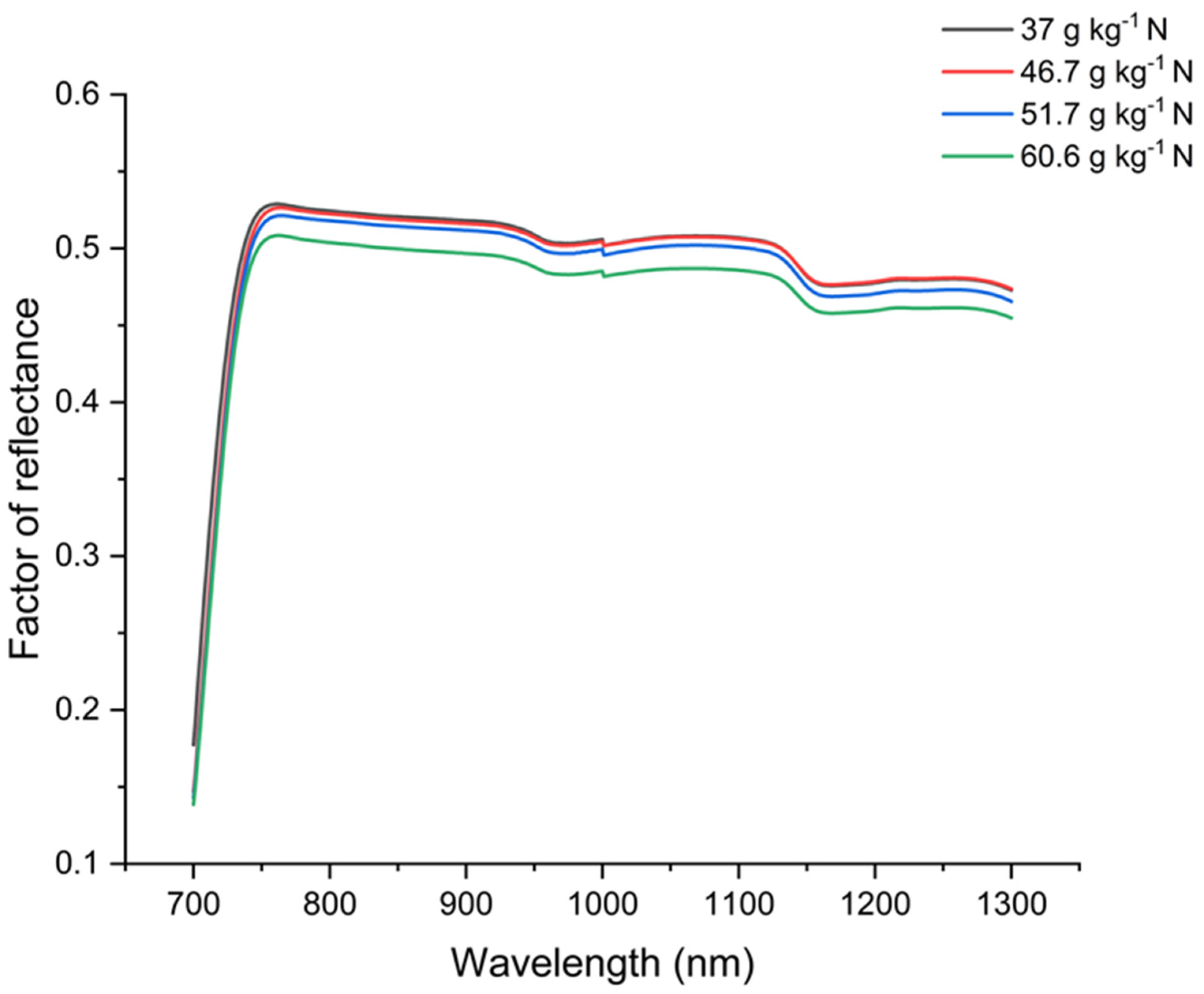
3.2. Leaf Spectral Analysis
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A

References
- Antolin, L.A.S.; Heinemann, A.B.; Marin, F.R. Impact Assessment of Common Bean Availability in Brazil under Climate Change Scenarios. Agric. Syst. 2021, 191, 103174. [Google Scholar] [CrossRef]
- Heinemann, A.B.; Costa-Neto, G.; Fritsche-Neto, R.; da Matta, D.H.; Fernandes, I.K. Enviromic Prediction Is Useful to Define the Limits of Climate Adaptation: A Case Study of Common Bean in Brazil. Field Crops Res. 2022, 286, 108628. [Google Scholar] [CrossRef]
- FAO. FAOSTAT Statistical Database; Food and Agriculture Organisation of the United Nations: Rome, Italy, 2020. [Google Scholar]
- Shumi, D. Response of Common Bean (Phaseolus vulgaris L.) Varieties to Rates of Blended NPS Fertilizer in Adola District, Southern Ethiopia. Afr. J. Plant Sci. 2018, 12, 164–179. [Google Scholar] [CrossRef]
- Araujo Robusti, E.; Godoy Androcioli, H.; Ventura, M.U.; Hata, F.T.; Soares Júnior, D.; Menezes Júnior, A.d.O. Integrated Pest Management versus Conventional System in the Common Bean Crop in Brazil: Insecticide Reduction and Financial Maximization. Int. J. Pest Manag. 2023, 1–11. [Google Scholar] [CrossRef]
- da Silva Borges, M.P.; Trezzi, M.M.; Mendes, K.F.; Fuzinatto, E.; Pilatti, G.; da Silva, A.A. Tolerance of Brazilian Bean Cultivars to S-Metolachlor and Poaceae Weed Control in Two Agricultural Soils. Agronomy 2023, 13, 2919. [Google Scholar] [CrossRef]
- Xie, K.; Ren, Y.; Chen, A.; Yang, C.; Zheng, Q.; Chen, J.; Wang, D.; Li, Y.; Hu, S.; Xu, G. Plant Nitrogen Nutrition: The Roles of Arbuscular Mycorrhizal Fungi. J. Plant Physiol. 2022, 269, 153591. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Jia, X.; Hu, M.; Zhou, M.; Li, D.; Liu, W.; Wang, R.; Zhang, J.; Xie, C.; Liu, L.; et al. An Effective Data Augmentation Strategy for CNN-Based Pest Localization and Recognition in the Field. IEEE Access 2019, 7, 160274–160283. [Google Scholar] [CrossRef]
- Silva, B.C.d.; Prado, R.d.M.; Baio, F.H.R.; Campos, C.N.S.; Teodoro, L.P.R.; Teodoro, P.E.; Santana, D.C.; Fernandes, T.F.S.; Silva Junior, C.A.d.; Loureiro, E.d.S. New Approach for Predicting Nitrogen and Pigments in Maize from Hyperspectral Data and Machine Learning Models. Remote Sens. Appl. 2024, 33, 101110. [Google Scholar] [CrossRef]
- Fu, Y.; Yang, G.; Pu, R.; Li, Z.; Li, H.; Xu, X.; Song, X.; Yang, X.; Zhao, C. An Overview of Crop Nitrogen Status Assessment Using Hyperspectral Remote Sensing: Current Status and Perspectives. Eur. J. Agron. 2021, 124, 126241. [Google Scholar] [CrossRef]
- Acosta, M.; Quiñones, A.; Munera, S.; de Paz, J.M.; Blasco, J. Rapid Prediction of Nutrient Concentration in Citrus Leaves Using Vis-NIR Spectroscopy. Sensors 2023, 23, 6530. [Google Scholar] [CrossRef]
- Fiorio, P.R.; Silva, C.A.A.C.; Rizzo, R.; Demattê, J.A.M.; dos Santos Luciano, A.C.; da Silva, M.A. Prediction of Leaf Nitrogen in Sugarcane (Saccharum spp.) by Vis-NIR-SWIR Spectroradiometry. Heliyon 2024, 10, e26819. [Google Scholar] [CrossRef]
- Sanaeifar, A.; Yang, C.; de la Guardia, M.; Zhang, W.; Li, X.; He, Y. Proximal Hyperspectral Sensing of Abiotic Stresses in Plants. Sci. Total Environ. 2023, 861, 160652. [Google Scholar] [CrossRef]
- Azadnia, R.; Rajabipour, A.; Jamshidi, B.; Omid, M. New Approach for Rapid Estimation of Leaf Nitrogen, Phosphorus, and Potassium Contents in Apple-Trees Using Vis/NIR Spectroscopy Based on Wavelength Selection Coupled with Machine Learning. Comput. Electron. Agric. 2023, 207, 107746. [Google Scholar] [CrossRef]
- Amaral, J.B.C.; Lopes, F.B.; Magalhães, A.C.M.d.; Kujawa, S.; Taniguchi, C.A.K.; Teixeira, A.d.S.; Lacerda, C.F.d.; Queiroz, T.R.G.; Andrade, E.M.d.; Araújo, I.C.d.S.; et al. Quantifying Nutrient Content in the Leaves of Cowpea Using Remote Sensing. Appl. Sci. 2022, 12, 458. [Google Scholar] [CrossRef]
- Ji, F.; Li, F.; Hao, D.; Shiklomanov, A.N.; Yang, X.; Townsend, P.A.; Dashti, H.; Nakaji, T.; Kovach, K.R.; Liu, H.; et al. Unveiling the Transferability of PLSR Models for Leaf Trait Estimation: Lessons from a Comprehensive Analysis with a Novel Global Dataset. New Phytol. 2024, 16, 243. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Lu, X.; Yu, S.; Gu, S.; Huang, G.; Guo, X.; Zhao, C. The Application of Machine Learning Models Based on Leaf Spectral Reflectance for Estimating the Nitrogen Nutrient Index in Maize. Agriculture 2022, 12, 1839. [Google Scholar] [CrossRef]
- Mustaqimah; Devianti; Munawar, A.A.; Sufardi, S. Capability of Short Vis-NIR Band Tandem with Machine Learning to Rapidly Predict NPK Content in Tropical Farmland: A Case Study of Aceh Province Agricultural Soil Dry Land, Indonesia. Case Stud. Chem. Environ. Eng. 2024, 9, 100711. [Google Scholar] [CrossRef]
- Osco, L.P.; Ramos, A.P.M.; Faita Pinheiro, M.M.; Moriya, É.A.S.; Imai, N.N.; Estrabis, N.; Ianczyk, F.; Araújo, F.F.d.; Liesenberg, V.; Jorge, L.A.d.C.; et al. A Machine Learning Framework to Predict Nutrient Content in Valencia-Orange Leaf Hyperspectral Measurements. Remote Sens. 2020, 12, 906. [Google Scholar] [CrossRef]
- Barcala, V.; Rozemeijer, J.; Ouwerkerk, K.; Gerner, L.; Osté, L. Value and Limitations of Machine Learning in High-Frequency Nutrient Data for Gap-Filling, Forecasting, and Transport Process Interpretation. Environ. Monit. Assess 2023, 195, 892. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Hu, Q.; Ruan, S.; Liu, J.; Zhang, J.; Hu, C.; Liu, Y.; Dian, Y.; Zhou, J. Utilizing Hyperspectral Reflectance and Machine Learning Algorithms for Non-Destructive Estimation of Chlorophyll Content in Citrus Leaves. Remote Sens. 2023, 15, 4934. [Google Scholar] [CrossRef]
- Alvares, C.A.; Stape, J.L.; Sentelhas, P.C.; de Moraes Gonçalves, J.L.; Sparovek, G. Köppen’s Climate Classification Map for Brazil. Meteorol. Z. 2013, 22, 711–728. [Google Scholar] [CrossRef] [PubMed]
- Cantarella, H.; Quaggio, J.A.; Júnior, D.M.; Boaretto, R.M.; Raij, B.v. Boletim 100: Recomendações de Adubação e Calagem Para o Estado de São Paulo; Elsiver: Singapore, 2022. [Google Scholar]
- Patel, M.K.; Padarian, J.; Western, A.W.; Fitzgerald, G.J.; McBratney, A.B.; Perry, E.M.; Suter, H.; Ryu, D. Retrieving Canopy Nitrogen Concentration and Aboveground Biomass with Deep Learning for Ryegrass and Barley: Comparing Models and Determining Waveband Contribution. Field Crops Res. 2023, 294, 108859. [Google Scholar] [CrossRef]
- Falcioni, R.; Oliveira, R.B.d.; Chicati, M.L.; Antunes, W.C.; Demattê, J.A.M.; Nanni, M.R. Estimation of Biochemical Compounds in Tradescantia Leaves Using VIS-NIR-SWIR Hyperspectral and Chlorophyll a Fluorescence Sensors. Remote Sens. 2024, 16, 1910. [Google Scholar] [CrossRef]
- Miao, X.; Miao, Y.; Liu, Y.; Tao, S.; Zheng, H.; Wang, J.; Wang, W.; Tang, Q. Measurement of Nitrogen Content in Rice Plant Using near Infrared Spectroscopy Combined with Different PLS Algorithms. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2023, 284, 121733. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, M.; Zhang, S.; Ma, H.; Wang, J.; Lu, H.; Wu, Y. Rapid Determination of Geniposide in the Extraction and Concentration Processes of Lanqin Oral Solution by Near-Infrared Spectroscopy Coupled with Chemometric Algorithms. Vib. Spectrosc. 2020, 107, 103023. [Google Scholar] [CrossRef]
- Lynch, J.M.; Barbano, D.M. Kjeldahl Nitrogen Analysis as a Reference Method for Protein Determination in Dairy Products. J. AOAC Int. 1999, 82, 1389–1398. [Google Scholar] [CrossRef] [PubMed]
- Bakthavatchalam, K.; Karthik, B.; Thiruvengadam, V.; Muthal, S.; Jose, D.; Kotecha, K.; Varadarajan, V. IoT Framework for Measurement and Precision Agriculture: Predicting the Crop Using Machine Learning Algorithms. Technologies 2022, 10, 13. [Google Scholar] [CrossRef]
- Shi, M.; Hu, W.; Li, M.; Zhang, J.; Song, X.; Sun, W. Ensemble Regression Based on Polynomial Regression-Based Decision Tree and Its Application in the in-Situ Data of Tunnel Boring Machine. Mech. Syst. Signal Process. 2023, 188, 110022. [Google Scholar] [CrossRef]
- Sha’abani, M.N.A.H.; Fuad, N.; Jamal, N.; Ismail, M.F. KNN and SVM Classification for EEG: A Review; Elisiver: Singapore, 2020; pp. 555–565. [Google Scholar] [CrossRef]
- Roopashree, S.; Anitha, J.; Mahesh, T.R.; Vinoth Kumar, V.; Viriyasitavat, W.; Kaur, A. An IoT Based Authentication System for Therapeutic Herbs Measured by Local Descriptors Using Machine Learning Approach. Measurement 2022, 200, 111484. [Google Scholar] [CrossRef]
- Wang, Y.; Ian, H. Induzindo Árvores Modelo Para Classes Contínuas. In Proceedings of the Induzindo Árvores Modelo para Classes Contínuas. In Proceedings of the 9th Conferência Europeia sobre Aprendizado de Máquina, Prague, Czech Republic, 23–25 April 1997. [Google Scholar]
- Thai, T.H.; Omari, R.A.; Barkusky, D.; Bellingrath-Kimura, S.D. Statistical Analysis versus the M5P Machine Learning Algorithm to Analyze the Yield of Winter Wheat in a Long-Term Fertilizer Experiment. Agronomy 2020, 10, 1779. [Google Scholar] [CrossRef]
- Afzal, S.; Ziapour, B.M.; Shokri, A.; Shakibi, H.; Sobhani, B. Building Energy Consumption Prediction Using Multilayer Perceptron Neural Network-Assisted Models; Comparison of Different Optimization Algorithms. Energy 2023, 282, 128446. [Google Scholar] [CrossRef]
- Harsányi, E.; Bashir, B.; Arshad, S.; Ocwa, A.; Vad, A.; Alsalman, A.; Bácskai, I.; Rátonyi, T.; Hijazi, O.; Széles, A.; et al. Data Mining and Machine Learning Algorithms for Optimizing Maize Yield Forecasting in Central Europe. Agronomy 2023, 13, 1297. [Google Scholar] [CrossRef]
- Zovko, M.; Žibrat, U.; Knapič, M.; Kovačić, M.B.; Romić, D. Hyperspectral Remote Sensing of Grapevine Drought Stress. Precis. Agric. 2019, 20, 335–347. [Google Scholar] [CrossRef]
- Nunes, H.D.; Leal, F.T.; Mingotte, F.L.C.; Damião, V.D.; Junior, P.A.C.; Lemos, L.B. Agronomic Performance, Quality and Nitrogen Use Efficiency by Common Bean Cultivars. J. Plant Nutr. 2021, 44, 995–1009. [Google Scholar] [CrossRef]
- Falcioni, R.; Moriwaki, T.; Pattaro, M.; Herrig Furlanetto, R.; Nanni, M.R.; Camargos Antunes, W. High Resolution Leaf Spectral Signature as a Tool for Foliar Pigment Estimation Displaying Potential for Species Differentiation. J. Plant Physiol. 2020, 249, 153161. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, S.; Zhang, B. Evaluation of Hyperspectral Indices for Retrieval of Canopy Equivalent Water Thickness and Gravimetric Water Content. Int. J. Remote Sens. 2016, 37, 3384–3399. [Google Scholar] [CrossRef]
- Machado, M.L.; Pinto, F.d.A.C.; Paula Junior, T.J.d.; Queiroz, D.M.d.; Cerqueira, O.d.A.T. White Mold Detection in Common Beans through Leaf Reflectance Spectroscopy. Eng. Agrícola 2015, 35, 1117–1126. [Google Scholar] [CrossRef]
- Liang, L.; Qin, Z.; Zhao, S.; Di, L.; Zhang, C.; Deng, M.; Lin, H.; Zhang, L.; Wang, L.; Liu, Z. Estimating Crop Chlorophyll Content with Hyperspectral Vegetation Indices and the Hybrid Inversion Method. Int. J. Remote Sens. 2016, 37, 2923–2949. [Google Scholar] [CrossRef]
- Crusiol, L.G.T.; Sun, L.; Sun, Z.; Chen, R.; Wu, Y.; Ma, J.; Song, C. In-Season Monitoring of Maize Leaf Water Content Using Ground-Based and UAV-Based Hyperspectral Data. Sustainability 2022, 14, 9039. [Google Scholar] [CrossRef]
- Falcioni, R.; Antunes, W.C.; Demattê, J.A.M.; Nanni, M.R. Biophysical, Biochemical, and Photochemical Analyses Using Reflectance Hyperspectroscopy and Chlorophyll a Fluorescence Kinetics in Variegated Leaves. Biology 2023, 12, 704. [Google Scholar] [CrossRef]
- Maimaitijiang, M.; Sagan, V.; Sidike, P.; Daloye, A.M.; Erkbol, H.; Fritschi, F.B. Crop Monitoring Using Satellite/UAV Data Fusion and Machine Learning. Remote Sens. 2020, 12, 1357. [Google Scholar] [CrossRef]
- Flynn, K.C.; Baath, G.; Lee, T.O.; Gowda, P.; Northup, B. Hyperspectral Reflectance and Machine Learning to Monitor Legume Biomass and Nitrogen Accumulation. Comput. Electron. Agric. 2023, 211, 107991. [Google Scholar] [CrossRef]
- Khan, M.; Ullah, Z.; Mašek, O.; Raza Naqvi, S.; Nouman Aslam Khan, M. Artificial Neural Networks for the Prediction of Biochar Yield: A Comparative Study of Metaheuristic Algorithms. Bioresour. Technol. 2022, 355, 127215. [Google Scholar] [CrossRef] [PubMed]
- Sreedhara, B.M.; Rao, M.; Mandal, S. Application of an Evolutionary Technique (PSO–SVM) and ANFIS in Clear-Water Scour Depth Prediction around Bridge Piers. Neural Comput. Appl. 2019, 31, 7335–7349. [Google Scholar] [CrossRef]
- Narmilan, A.; Gonzalez, F.; Salgadoe, A.S.A.; Kumarasiri, U.W.L.M.; Weerasinghe, H.A.S.; Kulasekara, B.R. Predicting Canopy Chlorophyll Content in Sugarcane Crops Using Machine Learning Algorithms and Spectral Vegetation Indices Derived from UAV Multispectral Imagery. Remote Sens. 2022, 14, 1140. [Google Scholar] [CrossRef]
- Yoosefzadeh-Najafabadi, M.; Earl, H.J.; Tulpan, D.; Sulik, J.; Eskandari, M. Application of Machine Learning Algorithms in Plant Breeding: Predicting Yield from Hyperspectral Reflectance in Soybean. Front. Plant Sci. 2021, 11, 624273. [Google Scholar] [CrossRef]

| Soil (Quartzarenic Neosol): 0–20 cm | ||||||||
|---|---|---|---|---|---|---|---|---|
| pH (CaCl2) | P (res) mg. dm−3 | S (PPM) | K (res) mmolc. dm−3 | Ca mmolc. dm−3 | Mg mmolc. dm−3 | Al mmolc. dm−3 | H + Al mmolc. dm−3 | C.T. (g/kg) |
| 4.4 | 8 | 4 | 0.3 | 16 | 4 | 4.1 | 30 | 9.3 |
| Tanned Cattle Manure | ||||||||
| pH | MS total (%) | M.O (%) | Gray (%) | C Total (%) | C org (%) | N (g/kg) | P2O5 (g/kg) | Relation C/N |
| 6.6 | 63.5 | 17.6 | 80 | 10.2 | 6.8 | 10.2 | 5.9 | 10:1 |
| Crushed Sugarcane Straw | ||||||||
| pH | MS total (%) | M.O (%) | Gray (%) | C Total (%) | C org (%) | N (g/kg) | P2O5 (g/kg) | Relation C/N |
| 4.9 | 7.6 | 50.6 | 28.4 | 29.4 | 12.8 | 9.1 | 4.8 | 32:1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tavares, M.S.; Silva, C.A.A.C.; Regazzo, J.R.; Sardinha, E.J.d.S.; da Silva, T.L.; Fiorio, P.R.; Baesso, M.M. Performance of Machine Learning Models in Predicting Common Bean (Phaseolus vulgaris L.) Crop Nitrogen Using NIR Spectroscopy. Agronomy 2024, 14, 1634. https://doi.org/10.3390/agronomy14081634
Tavares MS, Silva CAAC, Regazzo JR, Sardinha EJdS, da Silva TL, Fiorio PR, Baesso MM. Performance of Machine Learning Models in Predicting Common Bean (Phaseolus vulgaris L.) Crop Nitrogen Using NIR Spectroscopy. Agronomy. 2024; 14(8):1634. https://doi.org/10.3390/agronomy14081634
Chicago/Turabian StyleTavares, Marcos Silva, Carlos Augusto Alves Cardoso Silva, Jamile Raquel Regazzo, Edson José de Souza Sardinha, Thiago Lima da Silva, Peterson Ricardo Fiorio, and Murilo Mesquita Baesso. 2024. "Performance of Machine Learning Models in Predicting Common Bean (Phaseolus vulgaris L.) Crop Nitrogen Using NIR Spectroscopy" Agronomy 14, no. 8: 1634. https://doi.org/10.3390/agronomy14081634
APA StyleTavares, M. S., Silva, C. A. A. C., Regazzo, J. R., Sardinha, E. J. d. S., da Silva, T. L., Fiorio, P. R., & Baesso, M. M. (2024). Performance of Machine Learning Models in Predicting Common Bean (Phaseolus vulgaris L.) Crop Nitrogen Using NIR Spectroscopy. Agronomy, 14(8), 1634. https://doi.org/10.3390/agronomy14081634






