Drought Stress Increases the Complexity of the Bacterial Network in the Rhizosphere and Endosphere of Rice (Oryza sativa L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Soil Sampling and Basic Soil Properties
2.2. Pot Experiment and Sample Collection
2.3. DNA Extraction and PCR Amplification
2.4. Bioinformatic Analysis
2.5. Statistical Analysis
3. Results
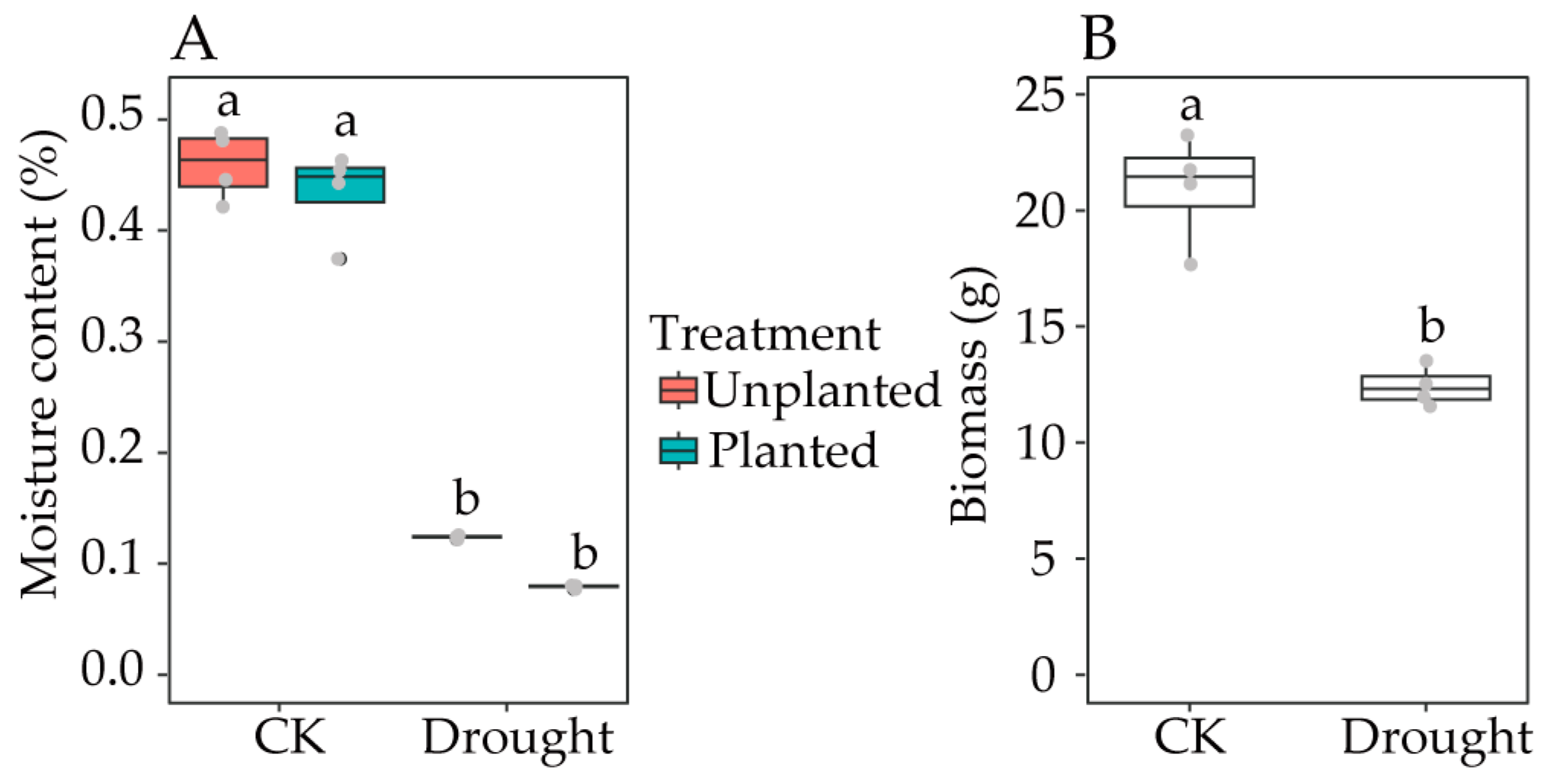
3.1. Influences of Drought Stress on Soil Moisture Content and Rice Biomass
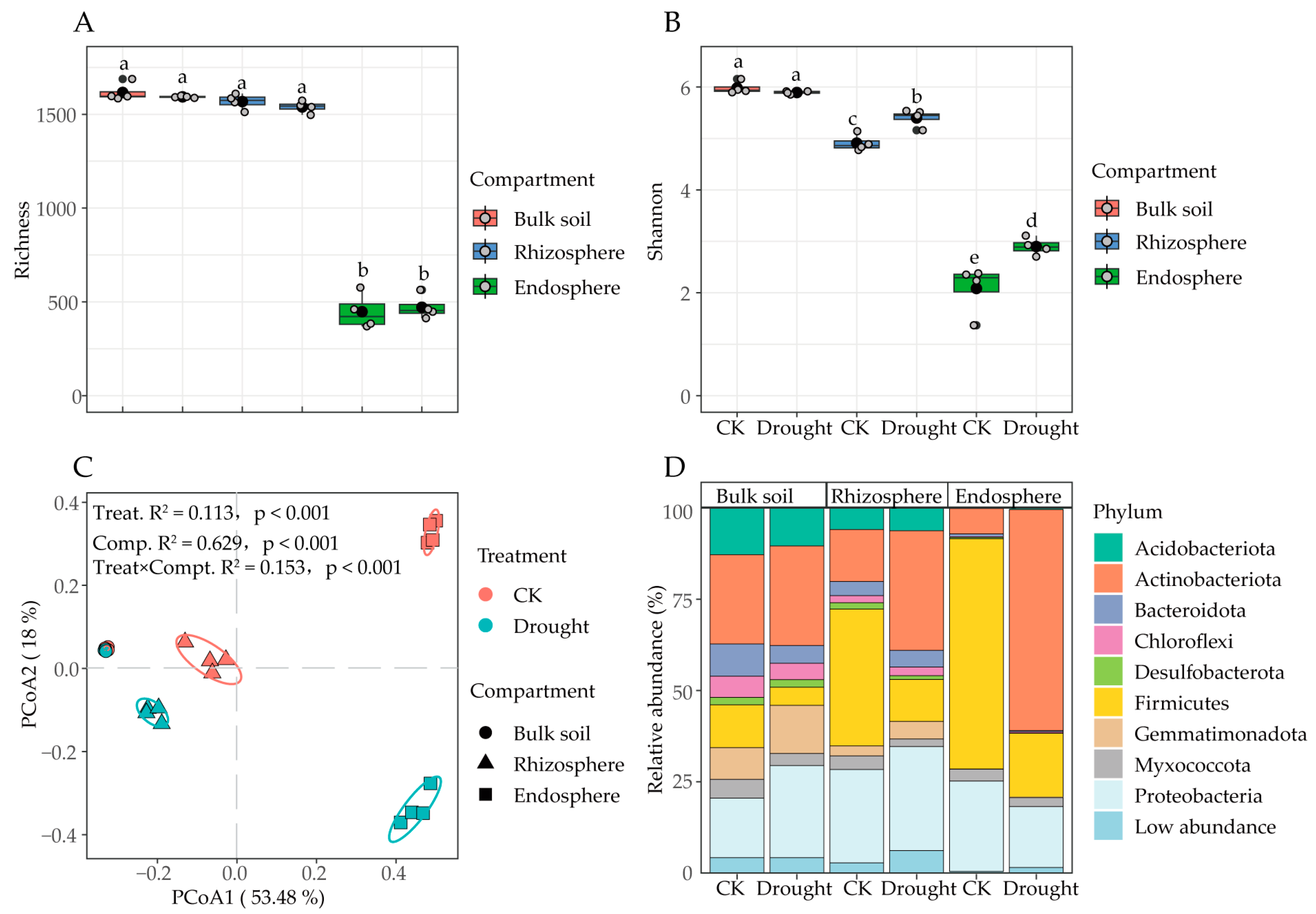
3.2. Influences of Drought Stress on α- and β-Diversity as Well as Community Assemblage of Bacteria
3.3. Community Composition of Bacteria at Genus Level in Different Compartments
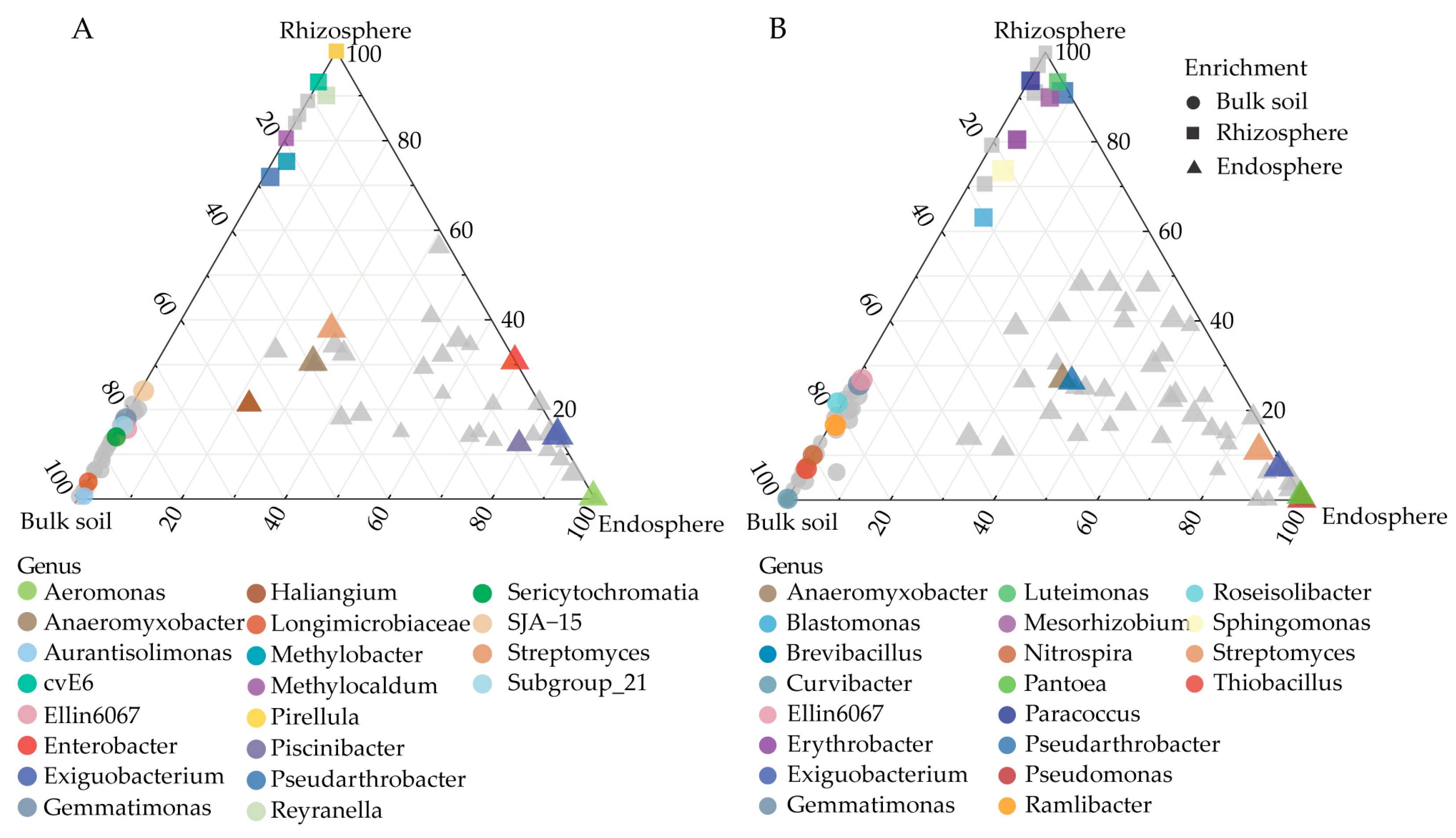
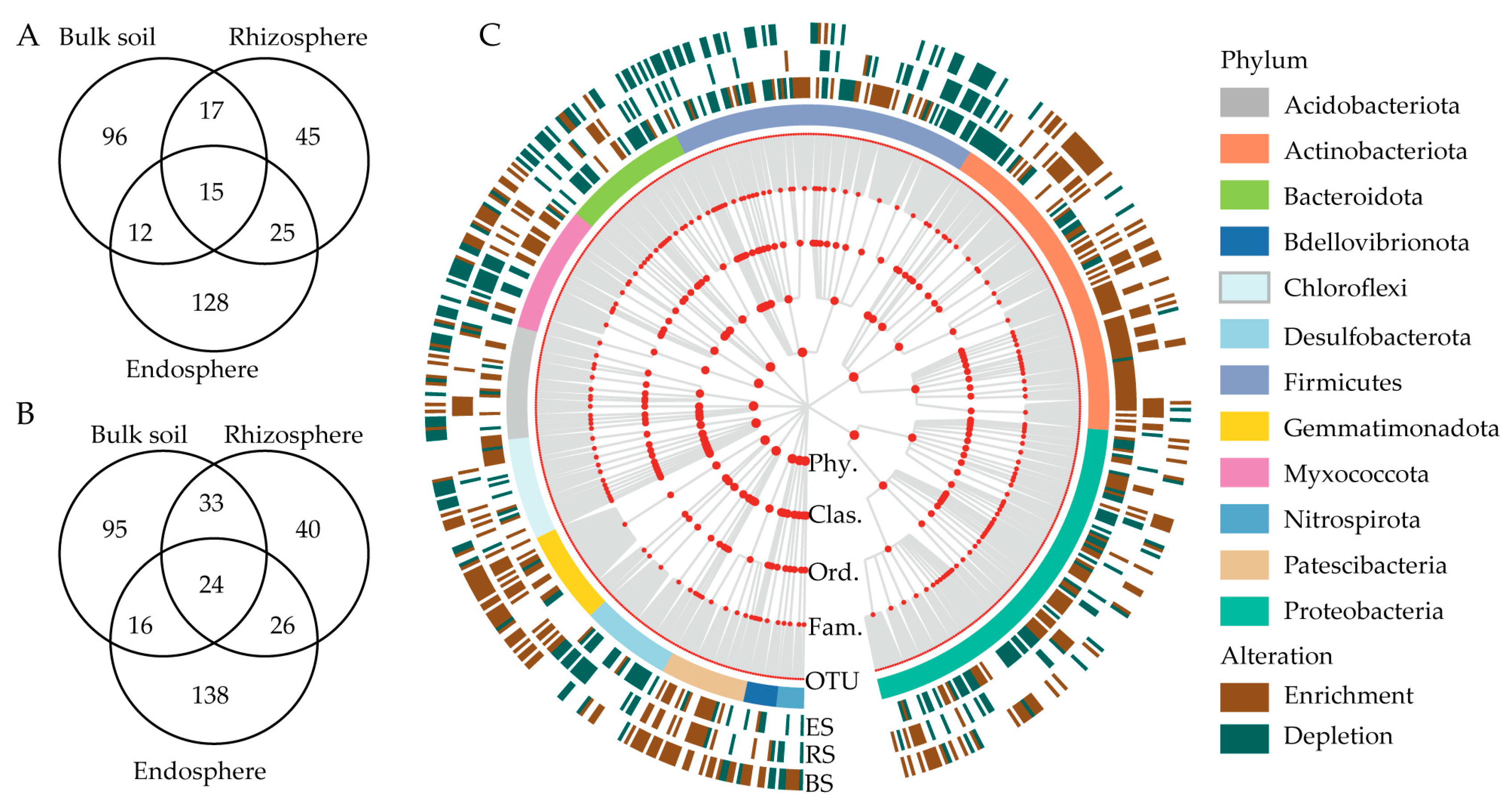
3.4. OTUs Significantly Changed under Drought Treatment
3.5. Network Analysis
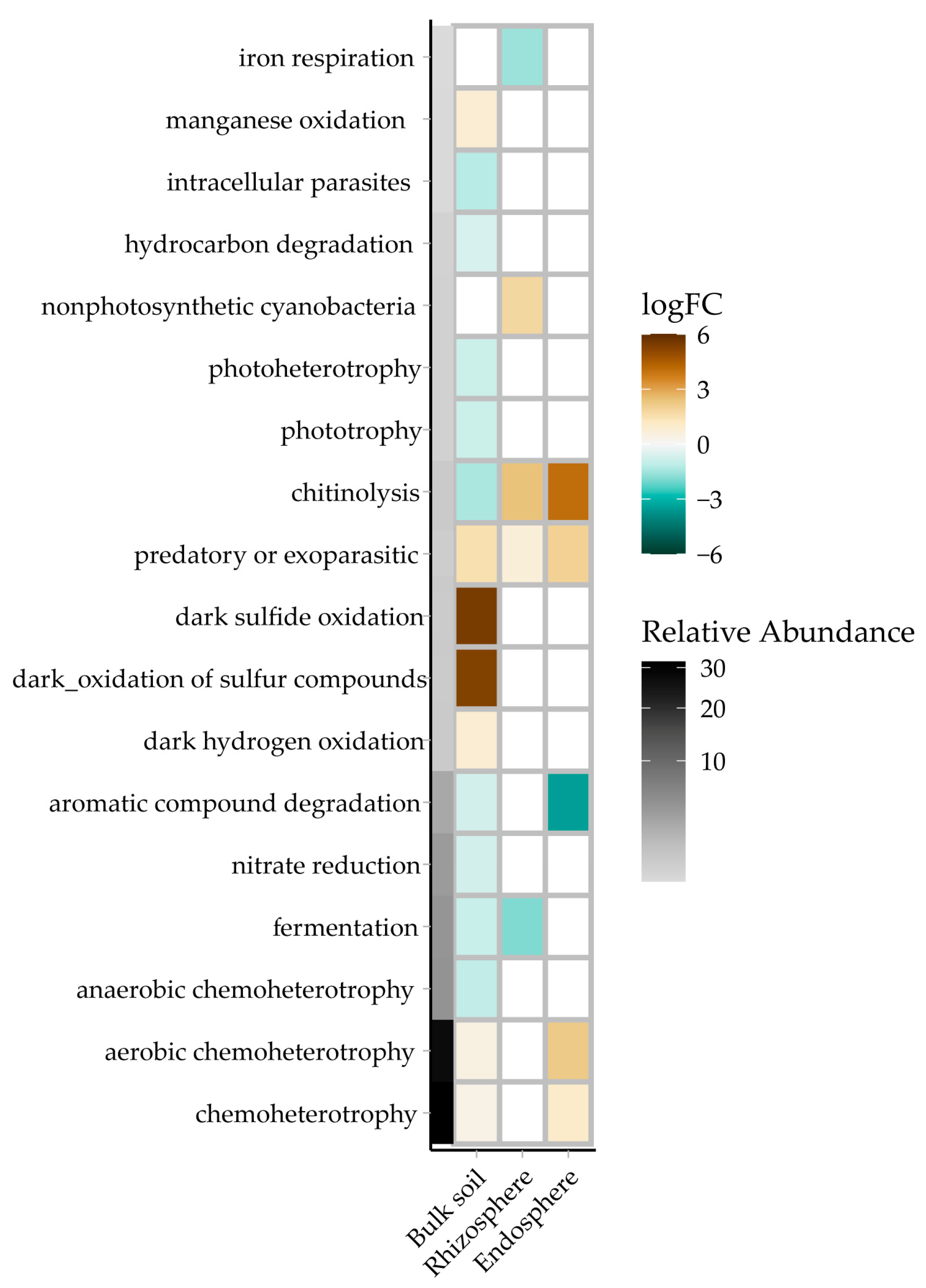
3.6. Effect of Drought on Ecological Functional Potentials
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lesk, C.; Rowhani, P.; Ramankutty, N. Influence of extreme weather disasters on global crop production. Nature 2016, 529, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Tyagi, A.; Park, S.; Mir, R.A.; Mushtaq, M.; Bhat, B.; Mahmoudi, H.; Bae, H. Deciphering the plant microbiome to improve drought tolerance: Mechanisms and perspectives. Environ. Exp. Bot. 2022, 201, 104933. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant–microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef] [PubMed]
- de Vries, F.T.; Griffiths, R.I.; Knight, C.G.; Nicolitch, O.; Williams, A. Harnessing rhizosphere microbiomes for drought-resilient crop production. Science 2020, 368, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Coleman-Derr, D. Causes and consequences of a conserved bacterial root microbiome response to drought stress. Curr. Opin. Microbiol. 2019, 49, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Naylor, D.; Coleman-Derr, D. Drought stress and root-associated bacterial communities. Front. Plant Sci. 2018, 8, 2223. [Google Scholar] [CrossRef] [PubMed]
- Naylor, D.; DeGraaf, S.; Purdom, E.; Coleman-Derr, D. Drought and host selection influence bacterial community dynamics in the grass root microbiome. ISME J. 2017, 11, 2691–2704. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Naylor, D.; Dong, Z.; Simmons, T.; Pierroz, G.; Hixson, K.K.; Kim, Y.-M.; Zink, E.M.; Engbrecht, K.M.; Wang, Y.; et al. Drought delays development of the sorghum root microbiome and enriches for monoderm bacteria. Proc. Natl. Acad. Sci. USA 2018, 115, E4284–E4293. [Google Scholar] [CrossRef] [PubMed]
- Santos-Medellin, C.; Edwards, J.; Liechty, Z.; Nguyen, B.; Sundaresan, V. Drought stress results in a compartment-specific restructuring of the rice root-associated microbiomes. mBio 2017, 8, e00764-17. [Google Scholar] [CrossRef] [PubMed]
- Santos-Medellin, C.; Liechty, Z.; Edwards, J.; Nguyen, B.; Huang, B.; Weimer, B.C.; Sundaresan, V. Prolonged drought imparts lasting compositional changes to the rice root microbiome. Nat. Plants 2021, 7, 1065–1077. [Google Scholar] [CrossRef]
- Fitzpatrick, C.R.; Copeland, J.; Wang, P.W.; Guttman, D.S.; Kotanen, P.M.; Johnson, M.T.J. Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl. Acad. Sci. USA 2018, 115, E1157. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Nuccio, E.E.; Shi, Z.J.; He, Z.; Zhou, J.; Firestone, M.K. The interconnected rhizosphere: High network complexity dominates rhizosphere assemblages. Ecol. Lett. 2016, 19, 926–936. [Google Scholar] [CrossRef] [PubMed]
- Durán, P.; Thiergart, T.; Garrido-Oter, R.; Agler, M.; Kemen, E.; Schulze-Lefert, P.; Hacquard, S. Microbial interkingdom interactions in roots pomote Arabidopsis survival. Cell 2018, 175, 973–983.e14. [Google Scholar] [CrossRef] [PubMed]
- Michalska-Smith, M.; Song, Z.; Spawn-Lee, S.A.; Hansen, Z.A.; Johnson, M.; May, G.; Borer, E.T.; Seabloom, E.W.; Kinkel, L.L. Network structure of resource use and niche overlap within the endophytic microbiome. ISME J. 2022, 16, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Vikram, P.; Swamy, B.P.M.; Dixit, S.; Singh, R.; Singh, B.P.; Miro, B.; Kohli, A.; Henry, A.; Singh, N.K.; Kumar, A. Drought susceptibility of modern rice varieties: An effect of linkage of drought tolerance with undesirable traits. Sci. Rep. 2015, 5, 14799. [Google Scholar] [CrossRef]
- Edwards, J.; Johnson, C.; Santos-Medellin, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, M.; Liu, D.; Sun, H.; Wu, J.; Huo, Y.; Chen, X.; Fang, R.; Zhang, L. Designing specific bacterial 16S primers to sequence and quantitate plant endo-bacteriome. Sci. China Life Sci. 2022, 65, 1000–1013. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, Z.D.; Müller, C.L.; Miraldi, E.R.; Littman, D.R.; Blaser, M.J.; Bonneau, R.A. Sparse and Compositionally Robust Inference of Microbial Ecological Networks. PLoS Comput. Biol. 2015, 11, e1004226. [Google Scholar] [CrossRef]
- Liu, C.; Cui, Y.; Li, X.; Yao, M. microeco: An R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 2020, 97, fiaa255. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 18 September 2021).
- Oksanen, J.; Simpson, G.L.; Blanchet, G.F.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. Vegan: Community Ecology Package. R Package Version 2.6-4. 2022. Available online: https://CRAN.R-project.org/package=vegan (accessed on 6 April 2023).
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going back to the roots themcirobial ecology of the rhzosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; del Rio, T.G.; et al. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef]
- Li, G.; Wang, K.; Qin, Q.; Li, Q.; Mo, F.; Nangia, V.; Liu, Y. Integrated microbiome and metabolomic analysis reveal responses of rhizosphere bacterial communities and root exudate composition to drought and genotype in rice (Oryza sativa L.). Rice 2023, 16, 19. [Google Scholar] [CrossRef]
- Williams, A.; de Vries, F.T. Plant root exudation under drought: Implications for ecosystem functioning. New Phytol. 2020, 225, 1899–1905. [Google Scholar] [CrossRef]
- Preece, C.; Peñuelas, J. Rhizodeposition under drought and consequences for soil communities and ecosystem resilience. Plant Soil 2016, 409, 1–17. [Google Scholar] [CrossRef]
- Jennifer, M.D.; Lauren, T.N.; Mariam, N.F.; Amy, M.J.; Mark, R. Global biogeography and quantitative seasonal dynamics of Gemmatimonadetes in soil. Appl. Environ. Microbiol. 2011, 77, 6295–6300. [Google Scholar]
- van Bergeijk, D.A.; Terlouw, B.R.; Medema, M.H.; van Wezel, G.P. Ecology and genomics of Actinobacteria: New concepts for natural product discovery. Nat. Rev. Microbiol. 2020, 18, 546–558. [Google Scholar] [CrossRef]
- Lei, Z.; Ding, Y.; Xu, W.; Zhang, Y. Microbial community structure in rice rhizosheaths under drought stress. J. Plant Ecol. 2023, 16, rtad012. [Google Scholar] [CrossRef]
- Bai, B.; Liu, W.; Qiu, X.; Zhang, J.; Zhang, J.; Bai, Y. The root microbiome: Community assembly and its contributions to plant fitness. J. Integr. Plant Biol. 2022, 64, 230–243. [Google Scholar] [CrossRef]
- Ding, X.; Peng, X.; Chen, Z.; Li, Y.; Mao, L.; Xie, H.; Cai, Y. Microbiota community assembly in wild rice (Oryza longistaminata) in response to drought stress shows phylogenetic conservation, stochasticity, and aboveground-belowground patterns. Res. Sq. 2020. [Google Scholar] [CrossRef]
- Yuan, M.M.; Guo, X.; Wu, L.; Zhang, Y.; Xiao, N.; Ning, D.; Shi, Z.; Zhou, X.; Wu, L.; Yang, Y.; et al. Climate warming enhances microbial network complexity and stability. Nat. Clim. Chang. 2021, 11, 343–348. [Google Scholar] [CrossRef]
- Hernandez, D.J.; David, A.S.; Menges, E.S.; Searcy, C.A.; Afkhami, M.E. Environmental stress destabilizes microbial networks. ISME J. 2021, 15, 1722–1734. [Google Scholar] [CrossRef]
- Bertness, M.D.; Callaway, R. Positive interactions in communities. Trends Ecol. Evol. 1994, 9, 191–193. [Google Scholar] [CrossRef]
- Gao, C.; Xu, L.; Montoya, L.; Madera, M.; Hollingsworth, J.; Chen, L.; Purdom, E.; Singan, V.; Vogel, J.; Hutmacher, R.B.; et al. Co-occurrence networks reveal more complexity than community composition in resistance and resilience of microbial communities. Nat. Commun. 2022, 13, 3867. [Google Scholar] [CrossRef]
- Connell, J.H. Diversity in tropical rain forests and coral reefs. Science 1978, 199, 1302–1310. [Google Scholar] [CrossRef]
- Conrad, R. Soil microorganisms as controllers of atmospheric trace gases(H2, CO, CH4, OCS, N2O, NO). Microbiol. Rev. 1996, 60, 609–640. [Google Scholar] [CrossRef]
- Burgin, A.J.; Yang, W.H.; Hamilton, S.K.; Silver, W.L. Beyond carbon and nitrogen: How the microbial energy ecocnomgy couples elemental cycles in diverse ecosystems. Front. Ecol. Environ. 2011, 1, 44–52. [Google Scholar] [CrossRef]
- da Silva, E.C.; Nogueira, R.; da Silva, M.A.; de Albuquerque, M.B. Drought stress and plant nutrition. Plant Stress 2011, 5, 32–41. [Google Scholar]
- Bouskill, N.J.; Lim, H.C.; Borglin, S.; Salve, R.; Wood, T.E.; Silver, W.L.; Brodie, E.L. Pre-exposure to drought increases the resistance of tropical forest soil bacterial communities to extended drought. ISME J. 2013, 7, 384–394. [Google Scholar] [CrossRef]
- Xu, L.; Dong, Z.; Chiniquy, D.; Pierroz, G.; Deng, S.; Gao, C.; Diamond, S.; Simmons, T.; Wipf, H.M.L.; Caddell, D.; et al. Genome-resolved metagenomics reveals role of iron metabolism in drought-induced rhizosphere microbiome dynamics. Nat. Commun. 2021, 12, 3209. [Google Scholar] [CrossRef]
- Zhang, L.; Zeng, Q.; Liu, X.; Chen, P.; Guo, X.; Ma, L.Z.; Dong, H.; Huang, Y. Iron reduction by diverse actinobacteria under oxic and pH-neutral conditions and the formation of secondary minerals. Chem. Geol. 2019, 525, 390–399. [Google Scholar] [CrossRef]

| Topological Attributes | Bulk Soil | Rhizosphere | Endosphere | |||
|---|---|---|---|---|---|---|
| CK | Drought | CK | Drought | CK | Drought | |
| No. of nodes | 592.000 | 616.000 | 382.000 | 544.000 | 35.000 | 53.000 |
| No. of edges | 1213.000 | 1302.000 | 663.000 | 1095.000 | 28.000 | 36.000 |
| Average degree | 4.098 | 4.227 | 3.471 | 4.026 | 1.600 | 1.359 |
| Average path length | 6.897 | 6.906 | 6.007 | 7.200 | 2.578 | 1.529 |
| Diameter | 18.000 | 18.000 | 16.000 | 19.000 | 7.000 | 4.000 |
| Clustering coefficient | 0.456 | 0.459 | 0.503 | 0.438 | 0.182 | 0.261 |
| Density | 0.007 | 0.007 | 0.009 | 0.007 | 0.047 | 0.026 |
| Centralization coefficient | 0.022 | 0.021 | 0.030 | 0.020 | 0.071 | 0.032 |
| Modularity | 0.790 | 0.791 | 0.803 | 0.794 | 0.752 | 0.921 |
| Proportion of negative edges | 0.525 | 0.489 | 0.425 | 0.258 | 0.464 | 0.361 |
| No. of module hubs | 4.000 | 1.000 | - | 4.000 | - | - |
| No. of connectors | 6.000 | 13.000 | 4.000 | 7.000 | - | - |
| No. of network hubs | - | - | - | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, C.; Zhang, X.; Liu, Y.; Tang, X.; Li, Y.; Sun, T.; Yan, G.; Yin, C. Drought Stress Increases the Complexity of the Bacterial Network in the Rhizosphere and Endosphere of Rice (Oryza sativa L.). Agronomy 2024, 14, 1662. https://doi.org/10.3390/agronomy14081662
Wu C, Zhang X, Liu Y, Tang X, Li Y, Sun T, Yan G, Yin C. Drought Stress Increases the Complexity of the Bacterial Network in the Rhizosphere and Endosphere of Rice (Oryza sativa L.). Agronomy. 2024; 14(8):1662. https://doi.org/10.3390/agronomy14081662
Chicago/Turabian StyleWu, Chunyan, Xiaoqin Zhang, Yinxiu Liu, Xu Tang, Yan Li, Tao Sun, Guochao Yan, and Chang Yin. 2024. "Drought Stress Increases the Complexity of the Bacterial Network in the Rhizosphere and Endosphere of Rice (Oryza sativa L.)" Agronomy 14, no. 8: 1662. https://doi.org/10.3390/agronomy14081662





