Characterization of Improved Barley Germplasm under Desert Environments Using Agro-Morphological and SSR Markers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Data Collection and Phenotyping
2.3. Genotyping Using SSR Markers
2.4. Statistical Analysis
2.4.1. Morphological Diversity Analysis
2.4.2. Molecular Analyses
3. Results
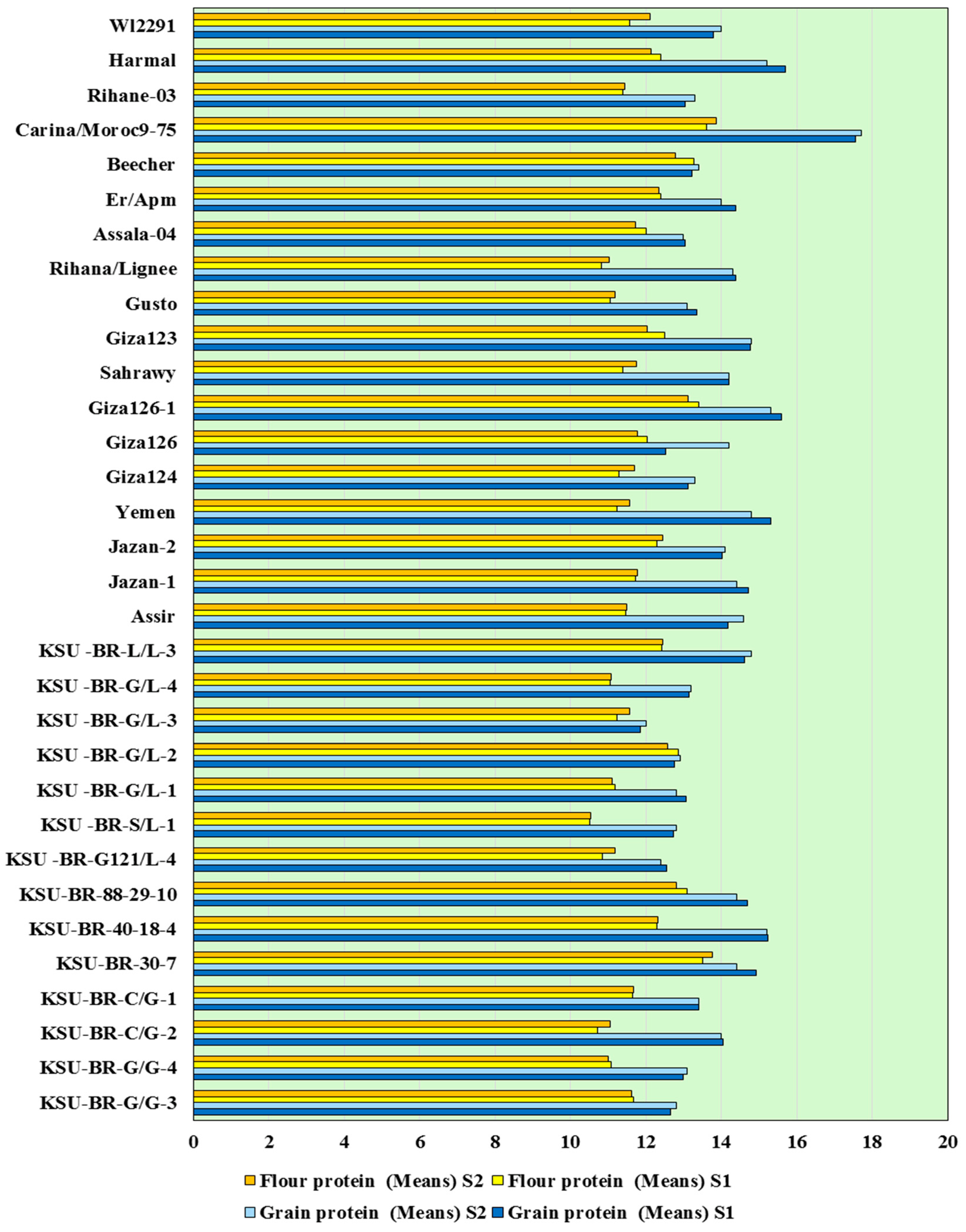
3.1. Agro-Morphological Diversity and Characterization
3.2. Correlation between Phenotypic Traits
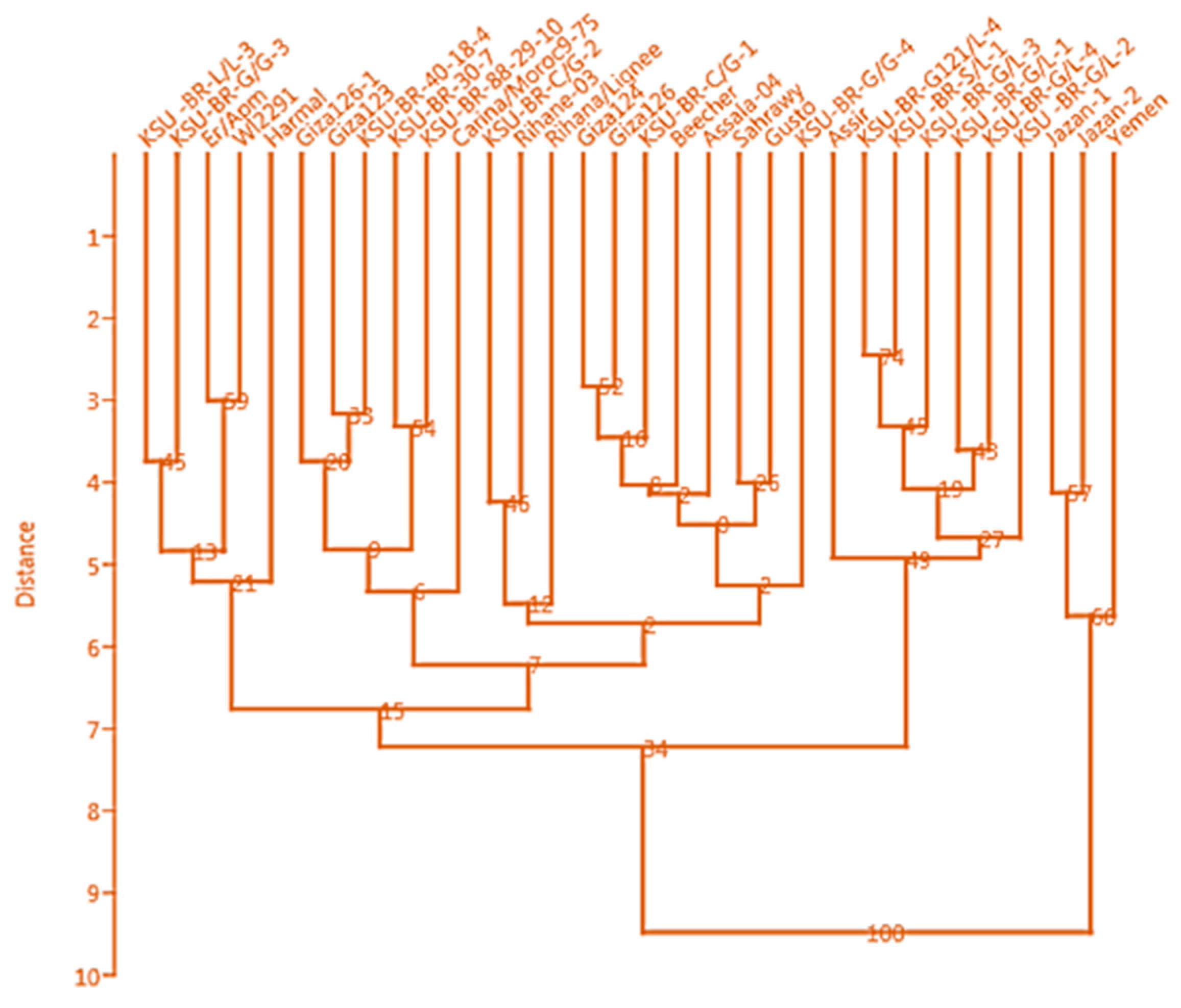
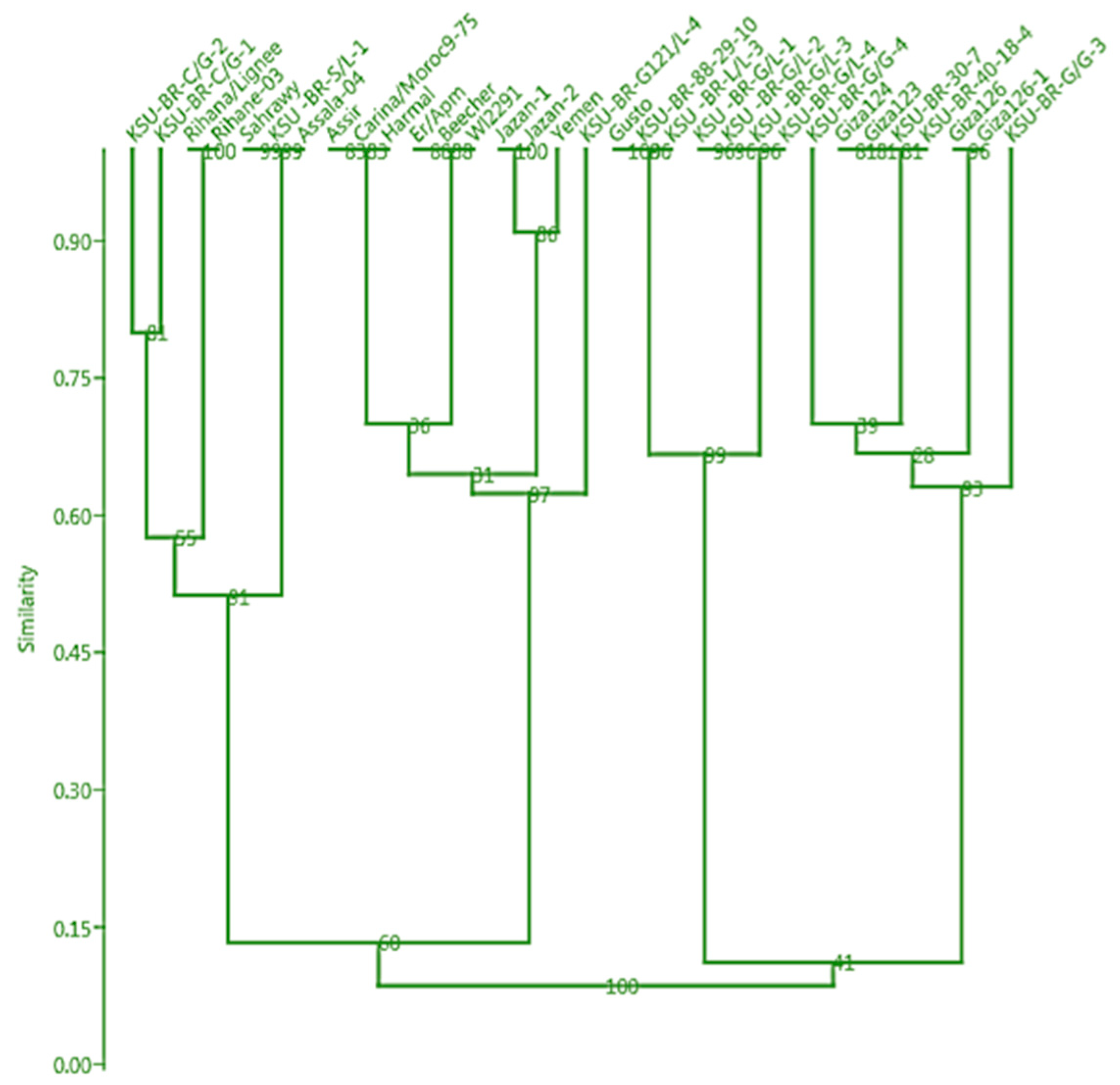
3.3. Clustering Based on Phenotypic Traits
3.4. Heat Map Analysis of the Whole Data
3.5. Molecular Characterization Using SSR Markers
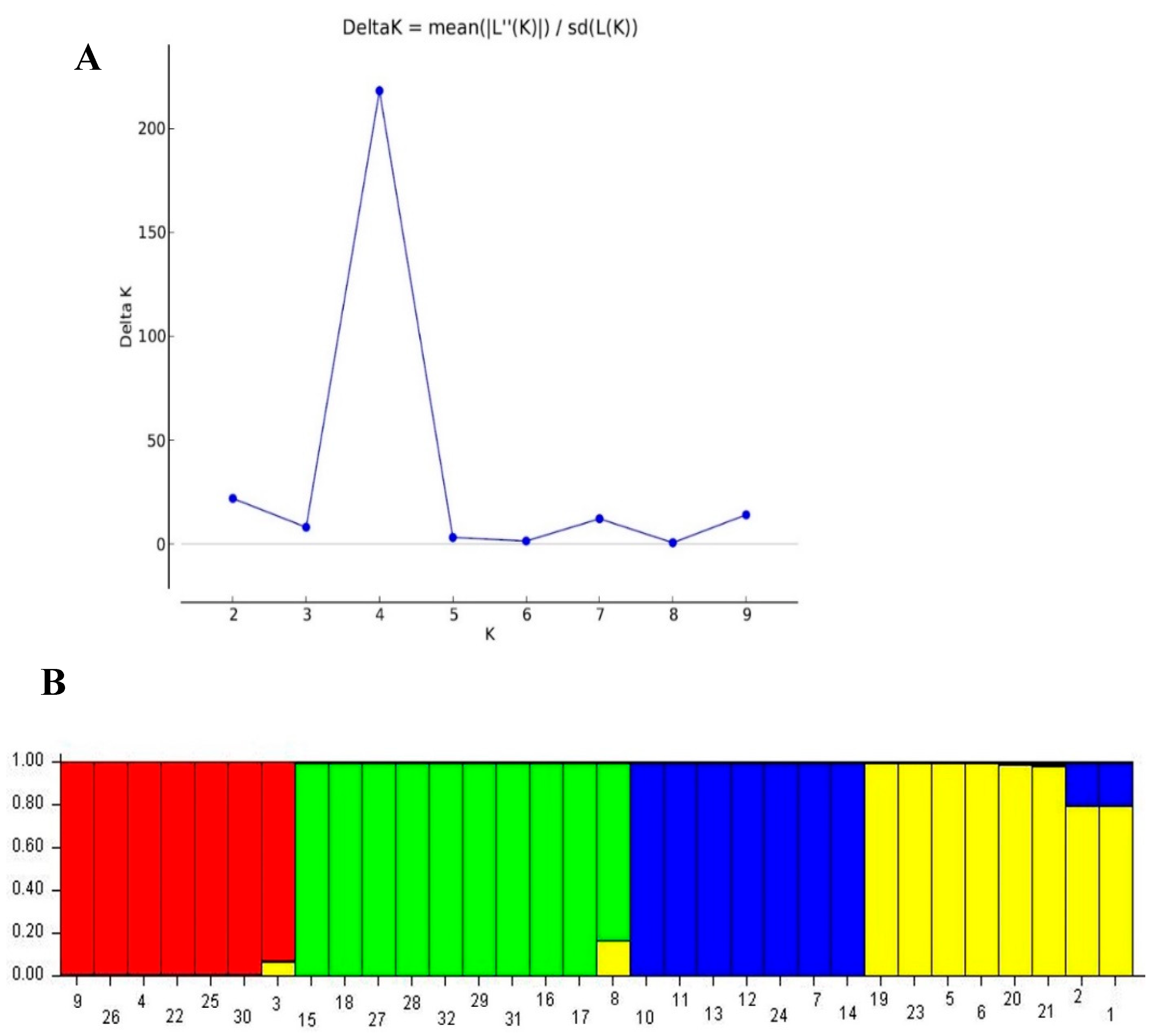
3.6. Population Structure Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Purugganan, M.D.; Fuller, D.Q. The nature of selection during plant domestication. Nature 2009, 457, 843–848. [Google Scholar] [CrossRef]
- Faostat, F. Food and Agriculture Organization of the United Nations-Statistic Division. 2019. Available online: https://www.fao.org/faostat/en/#data (accessed on 1 August 2024).
- Bhandari, H.; Bhanu, A.N.; Srivastava, K.; Singh, M.; Shreya, H.A. Assessment of genetic diversity in crop plants-an overview. Adv. Plants Agric. Res. 2017, 7, 279–286. [Google Scholar]
- Shakhatreh, Y.; Kafawin, O.; Ceccarelli, S.; Saoub, H.; Science, C. Selection of barley lines for drought tolerance in low-rainfall areas. J. Agron. 2001, 186, 119–127. [Google Scholar] [CrossRef]
- Renard, D.; Tilman, D. National food production stabilized by crop diversity. Nature 2019, 571, 257–260. [Google Scholar] [CrossRef] [PubMed]
- Sirami, C.; Gross, N.; Baillod, A.B.; Bertrand, C.; Carrié, R.; Hass, A.; Henckel, L.; Miguet, P.; Vuillot, C.; Alignier, A. Increasing crop heterogeneity enhances multitrophic diversity across agricultural regions. Proc. Natl. Acad. Sci. USA 2019, 116, 16442–16447. [Google Scholar] [CrossRef] [PubMed]
- Egli, L.; Schröter, M.; Scherber, C.; Tscharntke, T.; Seppelt, R. Crop asynchrony stabilizes food production. Nature 2020, 588, E7–E12. [Google Scholar] [CrossRef]
- Alghamdi, S.S.; Alfifi, S.A.; Migdadi, H.M.; Al-Rowaily, S.L.; El-Harty, E.H.; Muhammad Farooq, M.F. Morphological and Genetic Diversity of Cereal Genotypes in Kingdom of Saudi Arabia. Int. J. Agric. Biol. 2017, 19, 601–609. [Google Scholar] [CrossRef]
- Mariey, S.; Mohamed, E.N.; Ghareeb, Z.E.; Engy, S.; Abo Zaher, R. Genetic diversity of Egyptian barley using agro–physiological traits, grain quality and molecular markers. Curr. Sci. Int. 2021, 10, 58–71. [Google Scholar]
- Hussein, M.H.; Saker, M.; Moghaieb, R.; Hussein, H. Molecular characterization of salt tolerance in the genomes of some Egyptian and Saudi Arabian barely genotypes. Arab J. Biotechnol. 2005, 8, 241–252. [Google Scholar]
- El-Shazly, H.H.; El-Mutairi, Z. Genetic Relationships of Some Barley Cultivars, Based on Morphological Criteria and Rapd Fingerprinting. Int. J. Bot. 2006, 2, 252–260. [Google Scholar] [CrossRef]
- El-Awady, M.A.H.M.; El-Tarras, A.A.E.-S.; El-Assal, S.E.-D. Genetic diversity of some Saudi barley (Hordeum vulgare L.) landraces based on two types of molecular markers. Am. J. Appl. Sci. 2012, 9, 752. [Google Scholar]
- Peng, S.; Khush, G.S.; Virk, P.; Tang, Q.; Zou, Y. Progress in ideotype breeding to increase rice yield potential. Field Crops Res. 2008, 108, 32–38. [Google Scholar] [CrossRef]
- Donald, C.T. The breeding of crop ideotypes. Euphytica 1968, 17, 385–403. [Google Scholar] [CrossRef]
- Rasmusson, D. A plant breeder’s experience with ideotype breeding. Field Crops Res. 1991, 26, 191–200. [Google Scholar] [CrossRef]
- Carbajal-Friedrich, A.A.J.; Burgess, A.J. The role of the ideotype in future agricultural production. Front. Plant Physiol. 2024, 2, 1341617. [Google Scholar] [CrossRef]
- Roy, S.J.; Tucker, E.J.; Tester, M. Genetic analysis of abiotic stress tolerance in crops. Curr. Opin. Plant Biol. 2011, 14, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Bahieldin, A.; Ramadan, A.; Gadalla, N.; Alzohairy, A.; Edris, S.; Ahmed, I.; Shokry, A.; Hassan, S.; Saleh, O.; Baeshen, M.N. Molecular markers for salt tolerant wild barley Hordeum spontaneum. Life Sci. J. 2012, 9, 5838–5847. [Google Scholar]
- Mohammadi, S.A.; Abdollahi Sisi, N.; Sadeghzadeh, B. The influence of breeding history, origin and growth type on population structure of barley as revealed by SSR markers. Sci. Rep. 2020, 10, 19165. [Google Scholar] [CrossRef]
- Bouhlal, O.; Visioni, A.; Verma, R.P.S.; Kandil, M.; Gyawali, S.; Capettini, F.; Sanchez-Garcia, M. CGIAR Barley Breeding Toolbox: A diversity panel to facilitate breeding and genomic research in the developing world. Front. Plant Sci. 2022, 13. [Google Scholar] [CrossRef]
- Serrote, C.M.L.; Reiniger, L.R.S.; Silva, K.B.; dos Santos Rabaiolli, S.M.; Stefanel, C.M. Determining the polymorphism information content of a molecular marker. Gene 2020, 726, 144175. [Google Scholar] [CrossRef]
- Waits, L.P.; Luikart, G.; Taberlet, P. Estimating the probability of identity among genotypes in natural populations: Cautions and guidelines. Mol. Ecol. 2001, 10, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Zhivotovsky, L.A.; Feldman, M.W. Microsatellite variability and genetic distances. Proc. Natl. Acad. Sci. USA 1995, 92, 11549–11552. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef] [PubMed]
- Rafalski, A. Applications of single nucleotide polymorphisms in crop genetics. Curr. Opin. Plant Biol. 2002, 5, 94–100. [Google Scholar] [CrossRef]
- Elakhdar, A.; Abd EL-Sattar, M.; Amer, K.; Rady, A.; Kumamaru, T. Population structure and marker–trait association of salt tolerance in barley (Hordeum vulgare L.). Comptes Rendus Biol. 2016, 339, 454–461. [Google Scholar] [CrossRef]
- Elakhdar, A.; Kumamaru, T.; Qualset, C.O.; Brueggeman, R.S.; Amer, K.; Capo-chichi, L.; Evolution, C. Assessment of genetic diversity in Egyptian barley (Hordeum vulgare L.) genotypes using SSR and SNP markers. Genet. Resour. 2018, 65, 1937–1951. [Google Scholar] [CrossRef]
- Capo-Chichi, L.J.A.; Eldridge, S.; Elakhdar, A.; Kubo, T.; Brueggeman, R.; Anyia, A.O. QTL Mapping and Phenotypic Variation for Seedling Vigour Traits in Barley (Hordeum vulgare L.). Plants 2021, 10, 1149. [Google Scholar] [CrossRef] [PubMed]
- Capo-Chichi, L.J.A.; Elakhdar, A.; Kubo, T.; Nyachiro, J.; Juskiw, P.; Capettini, F.; Slaski, J.J.; Ramirez, G.H.; Beattie, A.D. Genetic diversity and population structure assessment of Western Canadian barley cooperative trials. Front. Plant Sci. 2022, 13, 1006719. [Google Scholar] [CrossRef]
- Yirgu, M.; Kebede, M.; Feyissa, T.; Lakew, B.; Woldeyohannes, A.B.; Fikere, M. Single nucleotide polymorphism (SNP) markers for genetic diversity and population structure study in Ethiopian barley (Hordeum vulgare L.) germplasm. BMC Genom. Data 2023, 24, 7. [Google Scholar] [CrossRef]
- Maanju, S.; Jasrotia, P.; Yadav, S.S.; Sharma, P.; Kashyap, P.L.; Kumar, S.; Jat, M.K.; Singh, G.P. Genetic diversity and population structure analyses in barley (Hordeum vulgare) against corn-leaf aphid, Rhopalosiphum maidis (Fitch). Front. Plant Sci. 2023, 14, 1188627. [Google Scholar] [CrossRef]
- American Association of Cereal Chemists; Approved Methods Committee. Approved Methods of the American Association of Cereal Chemists; American Association of Cereal Chemists: Eagan, MN, USA, 2000. [Google Scholar]
- Saghai-Maroof, M.A.; Soliman, K.M.; Jorgensen, R.A.; Allard, R.W. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA 1984, 81, 8014–8018. [Google Scholar] [CrossRef]
- Varshney, R.K.; Marcel, T.C.; Ramsay, L.; Russell, J.; Roder, M.S.; Stein, N.; Waugh, R.; Langridge, P.; Niks, R.E.; Graner, A. A high density barley microsatellite consensus map with 775 SSR loci. Theor. Appl. Genet. 2007, 114, 1091–1103. [Google Scholar] [CrossRef]
- StatSoft, I. STATISTICA (data analysis software system), version 6. Tulsa USA 2001, 150, 91–94. [Google Scholar]
- Hammer, O. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Rohlf, F.J. NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System; Exeter Publishing: Stony Brook, NY, USA, 1988. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Wen, X.; Falush, D. Documentation for Structure Software: Version 2.3; University of Chicago: Chicago, IL, USA, 2010; pp. 1–37. [Google Scholar]
- Earl, D.A.; VonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Dar, A.A.; Mahajan, R.; Lay, P.; Sharma, S. Genetic diversity and population structure of Cucumis sativus L. by using SSR markers. 3 Biotech 2017, 7, 307. [Google Scholar] [CrossRef] [PubMed]
- Vigouroux, Y.; Glaubitz, J.C.; Matsuoka, Y.; Goodman, M.M.; Sanchez, G.J.; Doebley, J. Population structure and genetic diversity of New World maize races assessed by DNA microsatellites. Am. J. Bot. 2008, 95, 1240–1253. [Google Scholar] [CrossRef]
- Agrama, H.; Eizenga, G.J.E. Molecular diversity and genome-wide linkage disequilibrium patterns in a worldwide collection of Oryza sativa and its wild relatives. Euphytica 2008, 160, 339–355. [Google Scholar] [CrossRef]
- Oliveira, M.; Sousa, L.; Reis, M.; Junior, E.S.; Cardoso, D.; Hamawaki, O.; Nogueira, A.; Research, M. Evaluation of genetic diversity among soybean (Glycine max) genotypes using univariate and multivariate analysis. Genetics 2017, 16. [Google Scholar] [CrossRef]
- Raja, W.H.; Yousuf, N.; Qureshi, I.; Sharma, O.C.; Singh, D.B.; Kumawat, K.L.; Nabi, S.U.; Mir, J.I.; Sheikh, M.A.; Kirmani, S.N. Morpho-molecular characterization and genetic diversity analysis across wild apple (Malus baccata) accessions using simple sequence repeat markers. S. Afr. J. Bot. 2022, 145, 378–385. [Google Scholar] [CrossRef]
- Keilwagen, J.; Kilian, B.; Özkan, H.; Babben, S.; Perovic, D.; Mayer, K.F.; Walther, A.; Poskar, C.H.; Ordon, F.; Eversole, K.J.S.R. Separating the wheat from the chaff–a strategy to utilize plant genetic resources from ex situ genebanks. Sci. Rep. 2014, 4, 5231. [Google Scholar] [CrossRef] [PubMed]
- Basnet, B.R.; Ali, M.B.; Ibrahim, A.M.; Payne, T.; Mosaad, M.G.; Science, C. Evaluation of genetic bases and diversity of Egyptian wheat cultivars released during the last 50 years using coefficient of parentage. Commun. Biometry Crop Sci. 2011, 6, 31–47. [Google Scholar]
- Farooqi, M.Q.U.; Moody, D.; Bai, G.; Bernardo, A.; St Amand, P.; Diggle, A.J.; Rengel, Z. Genetic characterization of root architectural traits in barley (Hordeum vulgare L.) using SNP markers. Front Plant Sci 2023, 14, 1265925. [Google Scholar] [CrossRef] [PubMed]
- Kebebew, F.; Tsehaye, Y.; McNeilly, T.; evolution, c. Morphological and farmers cognitive diversity of barley (Hordeum vulgare L. [Poaceae]) at Bale and North Shewa of Ethiopia. Genet. Resour. 2001, 48, 467–481. [Google Scholar]
- Marzougui, S.; Kharrat, M.; ben Younes, M. Assessment of genetic diversity and population structure of Tunisian barley accessions (Hordeum vulgare L.) using SSR markers. Acta Agrobot. 2020, 73, 1–9. [Google Scholar] [CrossRef]
- Mohamed, A.H.; Omar, A.A.; Attya, A.M.; Elashtokhy, M.M.; Zayed, E.M.; Rizk, R.M. Morphological and molecular characterization of some Egyptian six-rowed barley (Hordeum vulgare L.). Plants 2021, 10, 2527. [Google Scholar] [CrossRef] [PubMed]
- Güngör, H.; İlhan, E.; Kasapoğlu, A.G.; Filiz, E.; POUR, A.H.; Valchev, D.; Valcheva, D.; Haliloğlu, K.; Dumlupinar, Z. Genetic Diversity and Population Structure of Barley Cultivars Released in Turkey and Bulgaria using iPBS-retrotransposon and SCoT markers. J. Agric. Sci. 2022, 14, 1188627. [Google Scholar] [CrossRef]
- Nam, V.T.; Hang, P.L.B.; Linh, N.N.; Ly, L.H.; Hue, H.T.T.; Ha, N.H.; Hanh, H.H. Molecular markers for analysis of plant genetic diversity. Vietnam J. Biotechnol. 2020, 18, 589–608. [Google Scholar] [CrossRef]
- Krishnappa, G.; Savadi, S.; Tyagi, B.S.; Singh, S.K.; Mamrutha, H.M.; Kumar, S.; Mishra, C.N.; Khan, H.; Gangadhara, K.; Uday, G.; et al. Integrated genomic selection for rapid improvement of crops. Genomics 2021, 113, 1070–1086. [Google Scholar] [CrossRef] [PubMed]
- Brbaklić, L.; Trkulja, D.; Mikić, S.; Mirosavljević, M.; Momčilović, V.; Dudić, B.; Procházková, L.; Aćin, V.J.A. Genetic diversity and population structure of Serbian barley (Hordeum vulgare L.) collection during a 40-year long breeding period. Agronomy 2021, 11, 118. [Google Scholar] [CrossRef]
- Muller, R.; Hildebrand, T.; Ruegsegger, P. Non-invasive bone biopsy: A new method to analyse and display the three-dimensional structure of trabecular bone. Phys. Med. Biol. 1994, 39, 145–164. [Google Scholar] [CrossRef] [PubMed]
- Malysheva-Otto, L.V.; Ganal, M.W.; Roder, M.S. Analysis of molecular diversity, population structure and linkage disequilibrium in a worldwide survey of cultivated barley germplasm (Hordeum vulgare L.). BMC Genet. 2006, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Malysheva-Otto, L.; Ganal, M.W.; Law, J.R.; Reeves, J.C.; Röder, M.S. Temporal trends of genetic diversity in European barley cultivars (Hordeum vulgare L.). Mol. Breed. 2007, 20, 309–322. [Google Scholar] [CrossRef]
- Prasad, M.; Gupta, S.; Parida, S.K.; Kumari, K.; Muthamilarasan, M. Population structure and association mapping of yield contributing agronomic traits in foxtail millet. Plant Cell Rep. 2014, 33, 881–893. [Google Scholar]
- Westman, A.; Kresovich, S.L.; Genetics, A. The potential for cross-taxa simple-sequence repeat (SSR) amplification between Arabidopsis thaliana L. and crop brassicas. Theor. Appl. Genet. 1998, 96, 272–281. [Google Scholar] [CrossRef]



| Genotype | Pedigree | Row Type | Source | |
|---|---|---|---|---|
| 1 | KSU-BR-G/G-3 | Giza121/Gusto-SL3 | 6 | KSU Breeding Program |
| 2 | KSU-BR-G/G-4 | Giza123/Gusto-SL4 | 6 | KSU Breeding Program |
| 3 | KSU-BR-C/G-2 | KSU-BR-C/G-2 | 6 | KSU Breeding Program |
| 4 | KSU-BR-C/G-1 | KSU-BR-C/G-1 | 6 | KSU Breeding Program |
| 5 | KSU-BR-30-7 | Giza123/Local-SL 30-7 | 6 | KSU Breeding Program |
| 6 | KSU-BR-40-18-4 | Giza123/Local-SL 40-18-4 | 6 | KSU Breeding Program |
| 7 | KSU-BR-88-29-10 | Gusto/Local-SL 88-29-10 | 6 | KSU Breeding Program |
| 8 | KSU-BR-G121/L-4 | Giza121/Local-SL 4 | 6 | KSU Breeding Program |
| 9 | KSU-BR-S/L-1 | Sahrawy/Local-SL 1 | 6 | KSU Breeding Program |
| 10 | KSU-BR-G/L-1 | Gusto/Local-SL 1 | 6 | KSU Breeding Program |
| 11 | KSU-BR-G/L-2 | Gusto/Local-SL 2 | 6 | KSU Breeding Program |
| 12 | KSU-BR-G/L-3 | Gusto/Local-SL 3 | 6 | KSU Breeding Program |
| 13 | KSU-BR-G/L-4 | Gusto/Local-SL 4 | 6 | KSU Breeding Program |
| 14 | KSU-BR-L/L-3 | Lignee/Local-SL 3 | 6 | KSU Breeding Program |
| 15 | Assir | Local landraces | 2 | Saudi Arabia Landrace Assir |
| 16 | Jazan-1 | Local landraces | 2 | Saudi Arabia Landrace |
| 17 | Jazan-2 | Local landraces | 2 | Saudi Arabia Landrace |
| 18 | Yemen | Yemen landraces | 2 | Yemen Landrace |
| 19 | Giza124 | Giza 117/Bahteem 52// Giza 118/FAO 86 | 6 | Egyptian Cultivar (ARC-EGYPT) |
| 20 | Giza126 | BaladiBahteem/SD 729 Pour 12769-BC | 6 | Egyptian Cultivar (ARC-EGYPT) |
| 21 | Giza126-1 | Giza126 | 6 | Egyptian Cultivar (ARC-EGYPT) |
| 22 | Sahrawy | Baladi 16/Gem | 6 | Egyptian Cultivar (ARC-EGYPT) |
| 23 | Giza123 | Giza 117/FAO 86 | 6 | Egyptian Cultivar (ARC-EGYPT) |
| 24 | Gusto | Gusto | 6 | American Commercial Cultivar |
| 25 | Rihana/Lignee | Rihana/Lignee | 6 | ICARDA Selection |
| 26 | Assala-04 | Assala-04 | 6 | Old Landrace–ICARDA |
| 27 | Er/Apm | Er/Apm | 2 | ICARDA Selection |
| 28 | Beecher | Atlas/Vaughan | 6 | Old Landrace–ICARDA |
| 29 | Carina/Moroc9-75 | - | 2 | ICARDA Selection |
| 30 | Rihane-03 | As 46//Avt/Aths | 6 | Old Landrace–ICARDA |
| 31 | Harmal | Union/CI03576//Coho | 2 | Old Landrace–ICARDA |
| 32 | Wl2291 | CI3576/Union*2 | 2 | Waite Institute in South Australia |
| HD | PH | FLD | GFD | SPAD | SL | GN | TKW | BY | GY | HI | GP | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PH | 0.31 ** | |||||||||||
| FLD | 0.64 ** | 0.36 ** | ||||||||||
| GFD | 0.08 ** | 0.18 ** | 0.62 ** | |||||||||
| SPAD | 0.56 ** | 0.09 ** | 0.56 ** | 0.16 ** | ||||||||
| SL | 0.20 ** | 0.14 ** | 0.27 ** | 0.07 ns | 0.22 ** | |||||||
| GN | 0.46 ** | 0.28 ** | 0.55 ** | 0.27 ** | 0.53 ** | 0.25 ** | ||||||
| TKW | −0.35 ** | 0.03 ns | −0.40 ** | −0.17 ** | −0.46 ** | −0.08 ** | −0.45 ** | |||||
| BY | 0.34 ** | 0.22 ** | 0.37 ** | 0.08 ** | 0.23 ** | 0.23 ** | 0.25 ** | −0.10 ** | ||||
| GY | 0.31 ** | 0.23 ** | 0.18 ** | −0.07 ** | 0.17 ** | 0.12 ** | 0.19 ** | −0.15 ** | 0.51 ** | |||
| HI | −0.06 ns | 0.01 ns | −0.23 ** | −0.20 ** | −0.11 ** | −0.12 ** | −0.10 ** | 0.00 ns | −0.37 ** | 0.55 ** | ||
| GP | −0.03 ** | 0.18 ** | 0.12 ** | 0.15 ** | −0.15 ** | 0.32 ** | −0.20 ** | 0.37 ** | 0.13 ** | 0.01 | −0.08 ** | |
| FP | −0.01 ns | 0.12 ** | 0.17 ** | 0.11 ** | −0.07 ** | 0.21 ** | −0.04 ns | 0.10 ** | 0.28 ** | 0.11 ** | −0.14 ** | 0.42 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghazy, A.I.; Ali, M.A.; Ibrahim, E.I.; Sallam, M.; Al Ateeq, T.K.; Al-Ashkar, I.; Motawei, M.I.; Abdel-Haleem, H.; Al-Doss, A.A. Characterization of Improved Barley Germplasm under Desert Environments Using Agro-Morphological and SSR Markers. Agronomy 2024, 14, 1716. https://doi.org/10.3390/agronomy14081716
Ghazy AI, Ali MA, Ibrahim EI, Sallam M, Al Ateeq TK, Al-Ashkar I, Motawei MI, Abdel-Haleem H, Al-Doss AA. Characterization of Improved Barley Germplasm under Desert Environments Using Agro-Morphological and SSR Markers. Agronomy. 2024; 14(8):1716. https://doi.org/10.3390/agronomy14081716
Chicago/Turabian StyleGhazy, Abdelhalim I., Mohamed A. Ali, Eid I. Ibrahim, Mohammed Sallam, Talal K. Al Ateeq, Ibrahim Al-Ashkar, Mohamed I. Motawei, Hussein Abdel-Haleem, and Abdullah A. Al-Doss. 2024. "Characterization of Improved Barley Germplasm under Desert Environments Using Agro-Morphological and SSR Markers" Agronomy 14, no. 8: 1716. https://doi.org/10.3390/agronomy14081716
APA StyleGhazy, A. I., Ali, M. A., Ibrahim, E. I., Sallam, M., Al Ateeq, T. K., Al-Ashkar, I., Motawei, M. I., Abdel-Haleem, H., & Al-Doss, A. A. (2024). Characterization of Improved Barley Germplasm under Desert Environments Using Agro-Morphological and SSR Markers. Agronomy, 14(8), 1716. https://doi.org/10.3390/agronomy14081716









