Analysis of Total Flavonoid Variation and Other Functional Substances in RILs of Tartary Buckwheat, with Near-Infrared Model Construction for Rapid Non-Destructive Detection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Materials and Spectra Acquisition
2.1.1. Experimental Materials
2.1.2. Spectra Acquisition
2.2. Determination of the Contents of Total Flavonoid, VE, and GABA
2.2.1. Determination of Total Flavonoids of Tartary Buckwheat
2.2.2. Determination of VE in Tartary Buckwheat
2.2.3. Determination of GABA Content in Tartary Buckwheat
2.3. Data Processing and Model Evaluation
2.3.1. Data Processing
2.3.2. Model Evaluation
3. Results
3.1. Determination of Functional Components of Tartary Buckwheat and Analysis of Variation
3.2. Construction of the Near Infrared Model
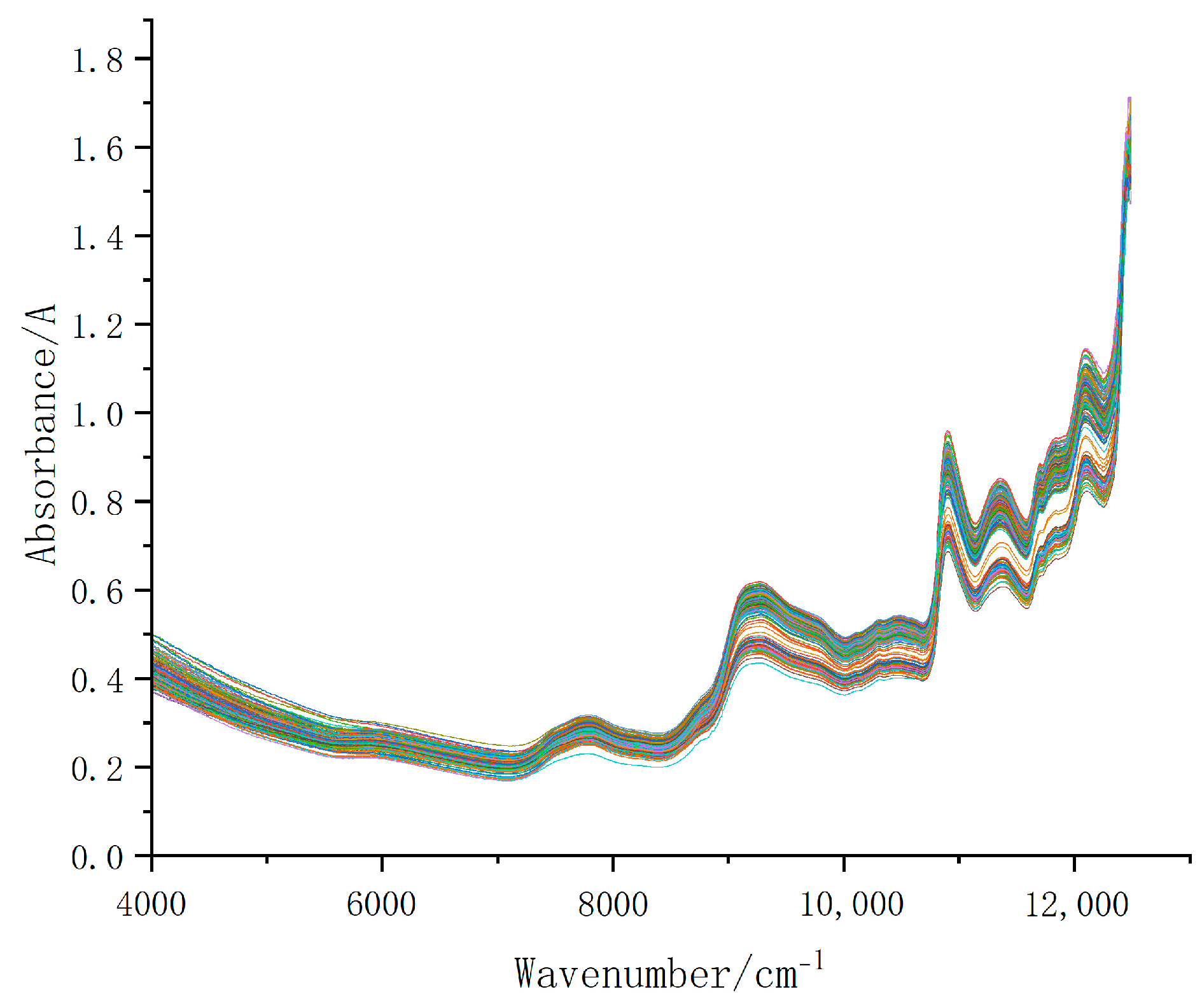
3.2.1. Near Infrared Spectrum of Tartary Buckwheat
3.2.2. Partitioning of the Sample Set
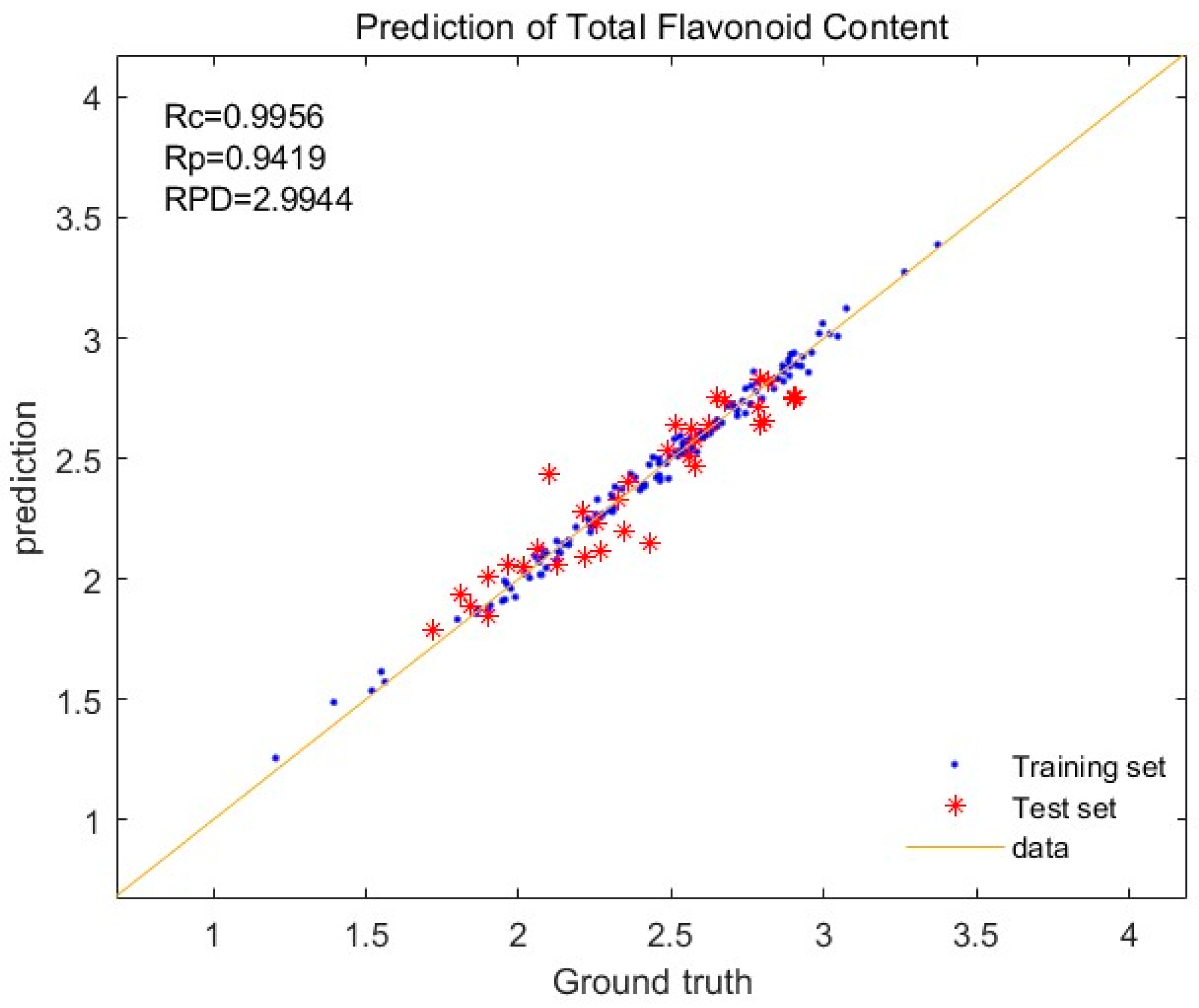
3.3. Creation of Total Flavonoid Prediction Models
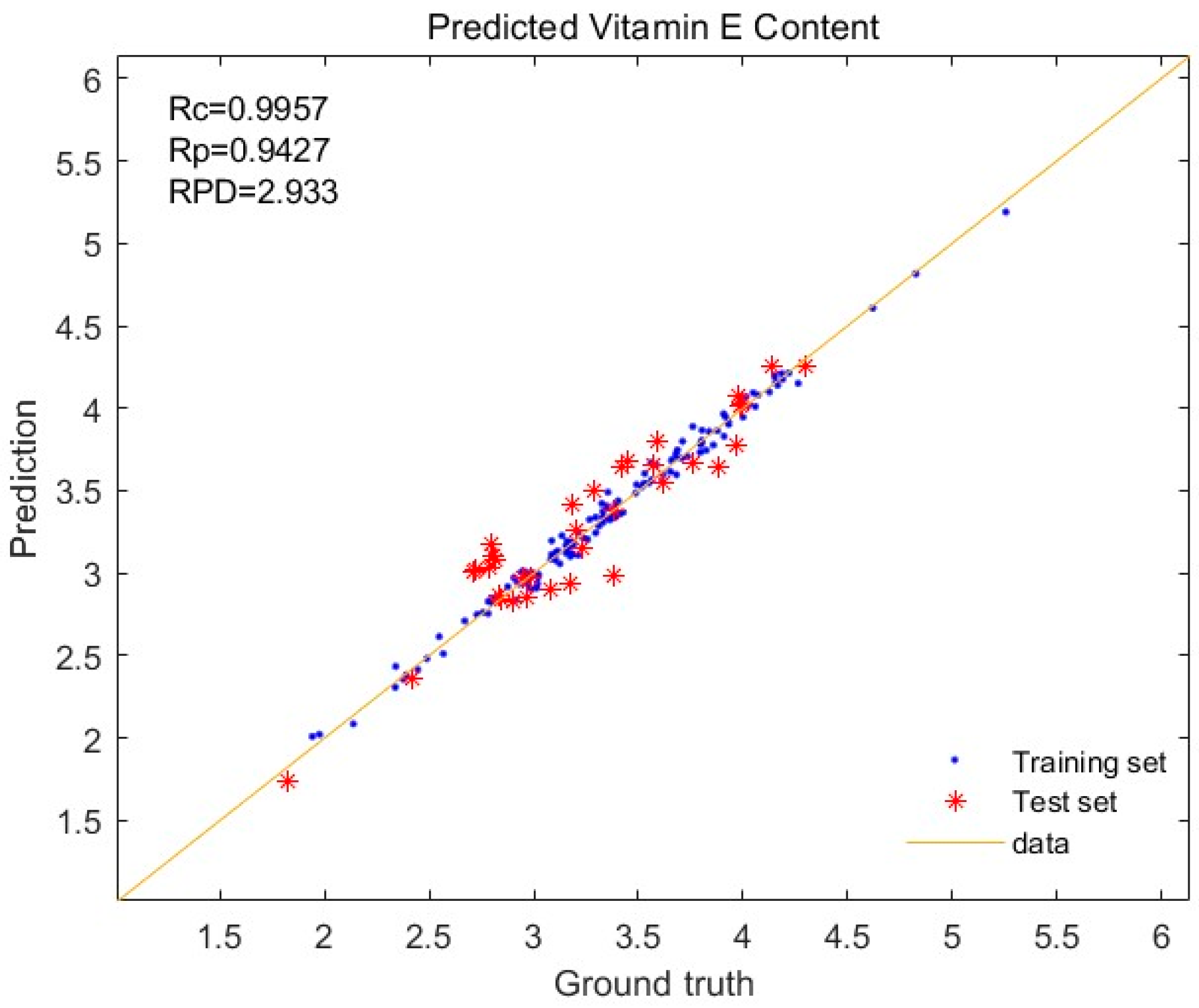
3.4. Effects of Different Pretreatment Methods on VE Modeling
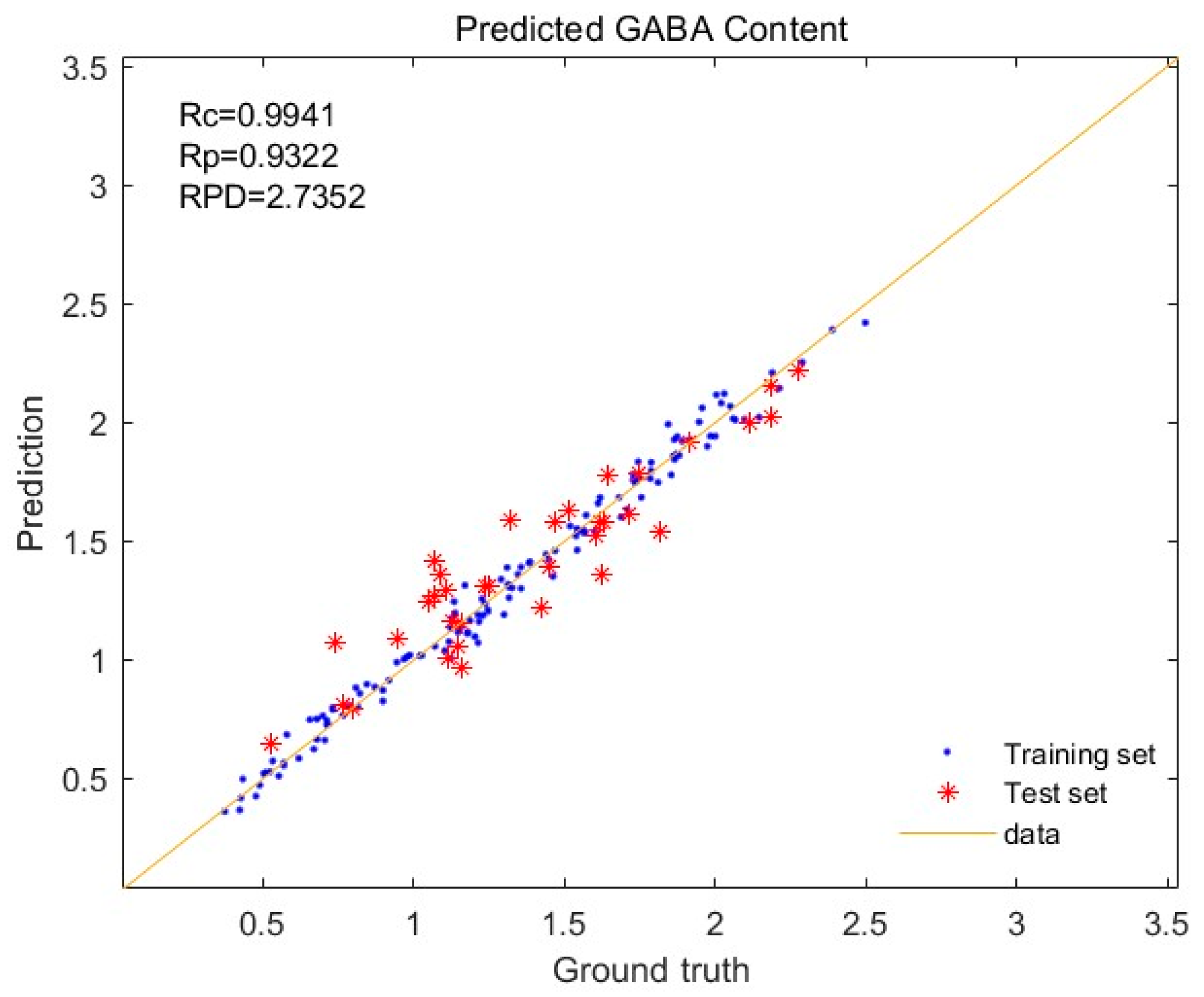
3.5. Effects of Different Pretreatment Methods on the Modeling of GABA
4. Discussion
4.1. Sample Diversity
4.2. Pre-Processing of the Spectrum
4.3. Sample Splitting
4.4. Extraction of the Characteristic Spectrum
4.5. Modeling of Whole Grains
4.6. Potential Limitations
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Luo, S.; Huang, J.M.; Yi, Y.; Zhang, Z.Y. Development status, problems, advantages and countermeasures of guizhou buckwheat industry. Tillage Cultiv. 2017, 6, 49–53+68. [Google Scholar]
- Qu, Z.H.; Bai, J.; Liu, R.M.; Wang, D.Q.; Zuo, W.B. Effects of variety and environment on flavonoid content in F. esculentum Moench. J. Shanxi Agric. Sci. 2023, 51, 1404–1409. [Google Scholar]
- Zhu, L.W.; Zhou, Y.; Cai, F.; Deng, J.; Huang, J.; Zhang, X.N.; Zhang, J.G.; Chen, Q.F. Quantitative analysis of perennial buckwheat leaves protein and GABA using near infrared spectroscopy. Spectrosc. Spectr. Anal. 2020, 40, 2421–2426. [Google Scholar]
- Zhang, Z.L.; Zhou, M.L.; Tang, Y.; Li, F.L.; Tang, Y.X.; Shao, J.R.; Xue, W.T.; Wu, Y.M. Bioactive compounds in functional buckwheat food. Food Res. Int. 2012, 49, 389–395. [Google Scholar] [CrossRef]
- Chang, Z.H.; NiMa, Y.Z.; Huang, H.J.; Laba, Z.X.; Gao, X.L.; Tian, P.J.; Yin, Z.J. Exploring the mechanism of Tartary Fagopyrum tataricum on pancreatic cancer based on data mining. Tibet J. Agric. Sci. 2023, 45, 29–34. [Google Scholar]
- Wang, Y.F.; Chen, X.Y.; Han, S.Y.; Zhu, L.S.; Liu, S.M.; Lv, H.; Chu, J.X. Experimental study on total flavones from buckwheat leaf in fighting pain and in flammation. Acad. J. Shanghai Univ. Tradit. Chin. Med. 2004, 11, 54–55. [Google Scholar]
- Zhu, L.S.; Ma, X.C.; Han, S.Y.; Liu, S.M.; Lv, H. Effects of total flavones of buckwheat leaf on blood lipid and lipid peroxides. Chin. J. Clin. Rehabil. 2004, 24, 5178–5179. [Google Scholar]
- Ke, J.; Ran, B.; Sun, P.; Cheng, Y.; Chen, Q.; Li, H. An evaluation of the absolute content of flavonoids and the identification of their relationship with the flavonoid biosynthesis genes in Tartary buckwheat seeds. Agronomy 2023, 13, 3006. [Google Scholar] [CrossRef]
- Tan, P.Y.; Guo, W.B. Research progress of Tartary buckwheat flavonoids on human body’s physiological function and mechanism. Med. Recapitul. 2018, 24, 1627–1632. [Google Scholar]
- Qiu, J.; Liu, Y.P.; Yue, Y.F.; Qin, Y.C.; Li, Z.G. Dietary Tartary buckwheat intake attenuates insulin resistance and improves lipid profiles in patients with type 2 diabetes: A randomized controlled trial. Nutr. Res. 2016, 36, 1392–1401. [Google Scholar] [CrossRef]
- Lei, L.T.; Zhou, Y.C.; Pu, X.Y. Preparation of flavonoid compounds from Tartary buckwheat and study on their antioxidative and hypoglycemic effects. Chin. Food Ind. 2023, 02, 102–104. [Google Scholar]
- Qin, P.Y.; Wang, Q.A.; Shan, F.; Hou, Z.H.; Ren, G.X. Nutritional composition and flavonoids content of flour from different buckwheat cultivars. Int. J. Food Sci. Tech. 2010, 45, 951–958. [Google Scholar] [CrossRef]
- Yao, P.; Huang, Y.; Dong, Q.; Wan, M.; Wang, A.; Chen, Y.; Li, C.; Wu, Q.; Chen, H.; Zhao, H. FtMYB6, a light-induced SG7 R2R3-MYB transcription factor, promotes flavonol biosynthesis in Tartary buckwheat (Fagopyrum tataricum). J. Agric. Food Chem. 2020, 68, 13685–13696. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.Y.; Xiong, X.; Jing, M.Y.; Gao, Y.; Hou, F.F.; Xing, G.M.; Li, S. Genetic analysis and QTL localization of vitamin E content in Hemerocallis citrina Baroni. J. Hebei Agric. Univ. 2023, 46, 38–45. [Google Scholar]
- Ma, J.T.; Qiu, D.Y.; Pang, Y.Z.; Gao, H.W.; Wang, X.M.; Qin, Y.C. Diverse roles of tocopherols in response to abiotic and biotic stresses and strategies for genetic biofortification in plants. Mol. Breed. 2020, 40, 18. [Google Scholar] [CrossRef]
- Sozen, E.; Demirel, T.; Ozer, N.K. Vitamin E: Regulatory role in the cardiovascular system. IUBMB Life 2019, 71, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Ekeuku, S.O.; Etim, E.P.; Pang, K.L.; Chin, K.Y.; Mai, C.W. Vitamin E in the management of pancreatic cancer: A scoping review. World J. Gastrointest. Oncol. 2023, 15, 943–958. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Wang, N.S.; Li, Y.H. Correlation of serum glutamate and gamma-aminobutyric acid levels with clinical symptoms in chronic schizophrenia patients and their diagnostic value for cognitive impairment. Int. J. Lab. Med. 2024, 45, 95–98+103. [Google Scholar]
- Yan, R. The levels and clinical significance of serum glutamate and T-aminobutyric acid in patients with depression. J. Int. Psychl. 2022, 49, 609–611. [Google Scholar]
- Tong, X.M.; Chai, C.X.; Wang, Y.Q. Effect of germination on grain quality of Tartary buckwheat and optimization of technology. Food Mach. 2021, 4, 176–183. [Google Scholar]
- Xi, Z.Y. Study on Non-Destructive Detection Method of Buckwheat Based on Near-Infrared Spectroscopy Technology. Master’s Thesis, Kunming University of Science and Technology, Kunming, China, 2013. [Google Scholar]
- Xu, B.C.; Ding, X.L. The quantitative methods of flavonoids in buckwheat (Fagopyrum tataricum). J. Food Sci. Biotechnol. 2003, 02, 98–101. [Google Scholar]
- Tang, C.Y.; Wang, T.; Tu, J.; Liu, G.H.; Li, P.; Zhao, J. Comparison of colorimetry and HPLC for determination of γ-aminobutyric acid in mulberry leaf tea. Food Sci. 2018, 39, 256–260. [Google Scholar]
- Zheng, Z.X. Application of near-infrared spectroscopy analysis technology in the feed processing industry. Fujian Agric. Mach. 2019, 1, 24–27. [Google Scholar]
- Yang, N.; Ren, G.X. Application of near-infrared reflectance spectroscopy to the evaluation of rutin and d-chiro-inositol contents in Tartary buckwheat. J. Agric. Food Chem. 2008, 761–764. [Google Scholar] [CrossRef] [PubMed]
- Li, W.L.; Han, H.F.; Cheng, Z.W.; Zhang, Y.; Liu, S.Y.; Qu, H.B. A feasibility research on the monitoring of traditional Chinese medicine production process using NIR-based multivariate process trajectories. Sens. Actuators B Chem. 2016, 231, 313–323. [Google Scholar] [CrossRef]
- Platov, Y.T.; Metlenkin, D.A.; Platova, R.A.; Rassulov, V.A.; Vereshchagin, A.I.; Marin, V.A. Buckwheat identification by combined UV-VIS-NIR spectroscopy and multivariate analysis. J. Appl. Spectrosc. 2021, 88, 723–730. [Google Scholar] [CrossRef]
- Zhu, L.W.; Yan, J.X.; Huang, J.; Shi, T.X.; Cai, F.; Li, H.Y.; Chen, Q.F.; Chen, Q.J. Rapid determination of amino acids in golden Tartary buckwheat based on near infrared spectroscopy and artificial neural network. Spectrosc. Spectr. Anal. 2022, 42, 49–55. [Google Scholar]
- Zhang, J.; Guo, J.; Zhang, M.L.; Zhang, X.; E, J.J. Establishment of rapid detection model of buckwheat nutritional components based on near infrared spectroscopy. J. Chin. Cereals Oils Assoc. 2020, 35, 151–158. [Google Scholar]
- Zhang, B.J.; Chen, S.S.; Li, K.Y.; Li, L.H.; Xu, R.H.; An, C.; Xiong, F.M.; Zhang, Y.; Dong, L.L.; Ren, M.J. Construction and application of detection model for amylose and amylopectin content in sorghum grains based on near infrared spectroscopy. Sci. Agric. Sin. 2022, 55, 26–35. [Google Scholar]
- Wang, Y.Y.; Dai, Y.J.; Wang, Y.Y.; Yang, J.L.; Xiang, D.H.; Yang, Y.Q.; Zeng, S.W. Research on non-destructive detection of protein and fat content in Torreya based on near-infrared spectroscopy technology. Sci. Technol. Food Ind. 2024, 45, 1–8. [Google Scholar]
- Wang, C.W. The Vegetable Quality Evaluation of Perennial Buckwheat. Master’s Thesis, Guizhou Normal University, Guiyang, China, 2015. [Google Scholar]
- Chen, M.M.; Qiu, Y.C.; Song, Y.; Yang, S.Q.; Zuo, F.; Qian, L.L. Study on origin tracing of mung bean based on near-infrared spectrum. J. Heilongjiang Bayi Agric. Univ. 2024, 36, 49–54. [Google Scholar]
- Yan, H.; Neves, M.D.G.; Wise, B.M.; Moraes, I.A.; Barbin, D.F.; Siesler, H.W. The application of handheld near-infrared spectroscopy and raman spectroscopic imaging for the identification and quality control of food products. Molecules 2023, 28, 7891. [Google Scholar] [CrossRef]
- Lv, D.; Li, R.Y.; Zheng, R.; Zheng, J.Q.; Meng, Z.Y.; Shi, T.X.; Chen, Q.F. Variation analysis of flavonoids content in seeds and seed traits of tartary buckwheat germplasn resources. Mol. Plant Breed. 2020, 18, 4762–4774. [Google Scholar]
- Ren, C.Z.; Shan, F.; Wang, M.; Li, Y.L. Review on nutrition and functionality and food product development of buckwheat. Chin. J. Grain Oil Sci. 2022, 37, 261–269. [Google Scholar]
- Galvao, R.K.H.; Araujo, M.C.U.; Jose, G.E.; Pontes, M.J.C.; Silva, E.C.; Saldanha, T.C.B. A method for calibration and validation subset partitioning. Talanta 2005, 67, 736–740. [Google Scholar] [CrossRef] [PubMed]
- Rabatel, G.; Marini, F.; Walczak, B.; Roger, J. VSN: Variable sorting for normalization. J. Chemom. 2019, 34, e3164. [Google Scholar] [CrossRef]
- Chu, X.L.; Li, Y.H. Practical Handbook of Near Infrared Spectroscopy, 1st ed.; Chemical Industry Press: Beijing, China, 2023; pp. 139–140. [Google Scholar]
- Barnes, R.J.; Dhanoa, M.S.; Lister, S.J. Correction to the description of standard normal variate (SNV) and detrend (DT) transformations in practical spectroscopy with applications in food and beverage analysis—2nd Edition. J. Near Infrared Spectrosc. 1993, 1, 185–186. [Google Scholar] [CrossRef]
- Sun, J.H.; Zhang, W.; Shi, J.Q.; Li, Y.K. Selection and application of spectral data preprocessing strategy. Acta Metrol. Sin. 2023, 44, 1284–1292. [Google Scholar]
- Wang, L.Q.; Yao, J.; Wang, R.Y.; Chen, Y.H.; Luo, S.N.; Wang, W.N.; Zhang, Y.R. Research on detection of soybean meal quality by nir based on PLS-GRNN. Spectrosc. Spect. Anal. 2022, 42, 1433–1438. [Google Scholar]
- Khamsopha, D.; Woranitta, S.; Teerachaichayut, S. Utilizing near infrared hyperspectral imaging for quantitatively predicting adulteration in tapioca starch. Food Control 2021, 123, 107781. [Google Scholar] [CrossRef]
- Li, S.P.; Wang, J.Y.; Wang, L. Construction and application of amylose content calibration model based on near-infrared spectroscopy. Grain Oil Sci. Technol. 2023, 36, 139–143. [Google Scholar]
- Chai, Y.H.; Yu, Y.; Zhu, H.; Li, Z.M.; Dong, H.; Yang, H.S. Identification of common buckwheat (Fagopyrum esculentum Moench) adulterated in Tartary buckwheat (Fagopyrum tataricum (L.) Gaertn) flour based on near-infrared spectroscopy and chemometrics. Curr. Res. Food Sci. 2023, 7, 100573. [Google Scholar] [CrossRef]
- Cui, C.; Liu, C.L.; Sun, X.R.; Wu, J.Z. Peanut frostbite detection method based on near infrared hyperspectral imaging technology. Food Ind. Technol. 2024, 45, 226–233. [Google Scholar]
- Centner, V.; Massart, D.L.; Denoord, O.E.; Dejong, S.; Vandeginste, B.M.; Sterna, C. Elimination of uninformative variables for multivariate calibration. Anal. Chem. 1996, 68, 3851–3858. [Google Scholar] [CrossRef]
- Li, B.; Su, C.T.; Yin, H.; Liu, Y.D. Hyperspectral imaging technology combined with machine learning for detection of moldy rice. Spectrosc. Spectr. Anal. 2023, 43, 2391–2396. [Google Scholar]
- Chen, P.; Dai, J.W.; Li, J.Y.; Xu, Y.P.; Liu, D.; Chu, X.L. Progress of chemometric methods in near- infrared spectroscopy. Chem. Reag. 2023, 45, 105–112. [Google Scholar]
- Li, Z.; Hong, M.Y.; Cui, S.L.; Chen, M.; Liu, X.K.; Chen, H.Y.; Liu, L.F. Rapid detection method of flavonoid content in peanut seed based on near infrared technology. Spectrosc. Spectr. Anal. 2024, 44, 1112–1116. [Google Scholar]
- Hashemi, S.M.; Perry, G.; Rajcan, I.; Eskandari, M. SoyMAGIC: An unprecedented platform for genetic studies and breeding activities in soybean. Front Plant Sci. 2022, 13, 945471. [Google Scholar] [CrossRef]
- Zhang, H.J.; Wu, J.H.; Luo, Y.; Li, L.J.; Yang, H.; Yu, X.Q.; Wang, X.S.; Chen, L.; Mei, H.W. Comparison of near infrared spectroscopy models for determining protein and amylose contents between calibration samples of recombinant inbred lines and conventional varieties of rice. Sci. Agric. Sin. 2007, 6, 941–948. [Google Scholar] [CrossRef]
- Jasinski, S.; Lécureuil, A.; Durandet, M.; Bernard-Moulin, P.; Guerche, P. Arabidopsis seed content QTL mapping using high-throughput phenotyping: The assets of near infrared spectroscopy. Front. Plant Sci. 2016, 7, 1682. [Google Scholar] [CrossRef]
- Illarionova, S.; Shadrin, D.; Trekin, A.; Ignatiev, V.; Oseledets, I. Generation of the nir spectral band for satellite images with convolutional neural networks. Sensors 2021, 21, 5646. [Google Scholar] [CrossRef] [PubMed]
- Moses, W.J.; Gitelson, A.A.; Berdnikov, S.; Povazhnyy, V. Satellite estimation of chlorophyll-α concentration using the red and NIR bands of MERIS—The Azovsea case study. IEEE Geosci. Remote Sens. Lett. 2009, 6, 845–849. [Google Scholar] [CrossRef]
- Holman, F.H.; Riche, A.B.; Michalski, A.; Castle, M.; Wooster, M.J.; Hawkesford, M.J. High throughput field phenotyping of wheat plant height and growth rate in field plot trials using UAV based remote sensing. Remote Sens. 2016, 8, 1031. [Google Scholar] [CrossRef]
| Functional Component | Number of Samples (NS) | Range | Mean Value (MV) | Skewness | Kurtosis | Standard Deviation (SD) | Coefficient of Variation (CV) |
|---|---|---|---|---|---|---|---|
| Total flavonoid | 175 | 1.20–3.37 | 2.42 | −0.462 | 0.276 | 0.36 | 15.06 |
| VE | 173 | 1.82–5.26 | 3.33 | 0.061 | 0.502 | 0.55 | 16.53 |
| GABA | 173 | 0.37–2.50 | 1.34 | 0.054 | −0.818 | 0.49 | 36.93 |
| Functional Component | NS | Sample Size | Range | MV | SD |
|---|---|---|---|---|---|
| Total flavonoid | training set | 140 | 1.20–3.37 | 2.44 | 0.37 |
| test set | 35 | 1.71–2.90 | 2.39 | 0.35 | |
| VE | training set | 138 | 1.82–5.26 | 3.37 | 0.54 |
| test set | 35 | 1.94–4.30 | 3.26 | 0.54 | |
| GABA | training set | 138 | 0.37–2.50 | 1.31 | 0.51 |
| test set | 35 | 0.53–2.27 | 1.39 | 0.43 |
| Pretreatment Method | Rc | Rp | RMSECV | RMSEP | RPD |
|---|---|---|---|---|---|
| Normalization | 0.9586 | 0.7725 | 0.1062 | 0.2245 | 1.5719 |
| Normalization + MSC | 0.9621 | 0.8324 | 0.1018 | 0.1959 | 1.8013 |
| Normalization + SNV | 0.9491 | 0.7303 | 0.1175 | 0.2509 | 1.4063 |
| Normalization + First derivative | 0.9956 | 0.9419 | 0.0350 | 0.1178 | 2.9944 |
| Normalization + Second derivative | 0.9954 | 0.9389 | 0.0356 | 0.1217 | 2.8988 |
| Normalization + SG | 0.9534 | 0.7483 | 0.1126 | 0.2401 | 1.4692 |
| Pretreatment Method | Rc | Rp | RMSECV | RMSEP | RPD |
|---|---|---|---|---|---|
| Normalization | 0.9856 | 0.7748 | 0.0915 | 0.3483 | 1.5562 |
| Normalization + MSC | 0.9861 | 0.8483 | 0.0900 | 1.5780 | 0.3435 |
| Normalization + SNV | 0.9870 | 0.8586 | 0.0869 | 0.2794 | 1.9399 |
| Normalization + First derivative | 0.9957 | 0.9427 | 0.0504 | 0.1848 | 2.9330 |
| Normalization + Second derivative | 0.9980 | 0.9329 | 0.0343 | 0.1974 | 2.7459 |
| Normalization + SG | 0.9807 | 0.8633 | 0.1057 | 0.3008 | 1.8021 |
| Pretreatment Method | Rc | Rp | RMSECV | RMSEP | RPD |
|---|---|---|---|---|---|
| Normalization | 0.9936 | 0.8903 | 0.0579 | 0.2007 | 2.1786 |
| Normalization + MSC | 0.9929 | 0.9161 | 0.0611 | 0.5031 | 0.8692 |
| Normalization + SNV | 0.9941 | 0.9322 | 0.0553 | 0.1599 | 2.7352 |
| Normalization + First derivative | 0.9973 | 0.9032 | 0.0378 | 0.2028 | 2.1560 |
| Normalization + Second derivative | 0.0.9989 | 0.8950 | 0.0245 | 0.1944 | 2.2494 |
| Normalization + SG | 0.9943 | 0.9067 | 0.0547 | 0.1793 | 2.4385 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, L.; Du, Q.; Shi, T.; Huang, J.; Deng, J.; Li, H.; Cai, F.; Chen, Q. Analysis of Total Flavonoid Variation and Other Functional Substances in RILs of Tartary Buckwheat, with Near-Infrared Model Construction for Rapid Non-Destructive Detection. Agronomy 2024, 14, 1826. https://doi.org/10.3390/agronomy14081826
Zhu L, Du Q, Shi T, Huang J, Deng J, Li H, Cai F, Chen Q. Analysis of Total Flavonoid Variation and Other Functional Substances in RILs of Tartary Buckwheat, with Near-Infrared Model Construction for Rapid Non-Destructive Detection. Agronomy. 2024; 14(8):1826. https://doi.org/10.3390/agronomy14081826
Chicago/Turabian StyleZhu, Liwei, Qianxi Du, Taoxiong Shi, Juan Huang, Jiao Deng, Hongyou Li, Fang Cai, and Qingfu Chen. 2024. "Analysis of Total Flavonoid Variation and Other Functional Substances in RILs of Tartary Buckwheat, with Near-Infrared Model Construction for Rapid Non-Destructive Detection" Agronomy 14, no. 8: 1826. https://doi.org/10.3390/agronomy14081826






