Detection of Clubroot Disease Resistance in Brassica juncea Germplasm at the Seedling Stage
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Identification of the Physiological Race of Plasmodiophora brassicae
2.2.2. Clubroot Pathogen Inoculation
2.2.3. Identification of Clubroot Disease Resistance in Mustard Germplasm
2.2.4. Molecular Marker Detection of Loci Associated to Mustard Clubroot Resistance
2.3. Data Analyses
3. Results
3.1. Molecular Identification of the Physiological Race of Plasmodiophora brassicae
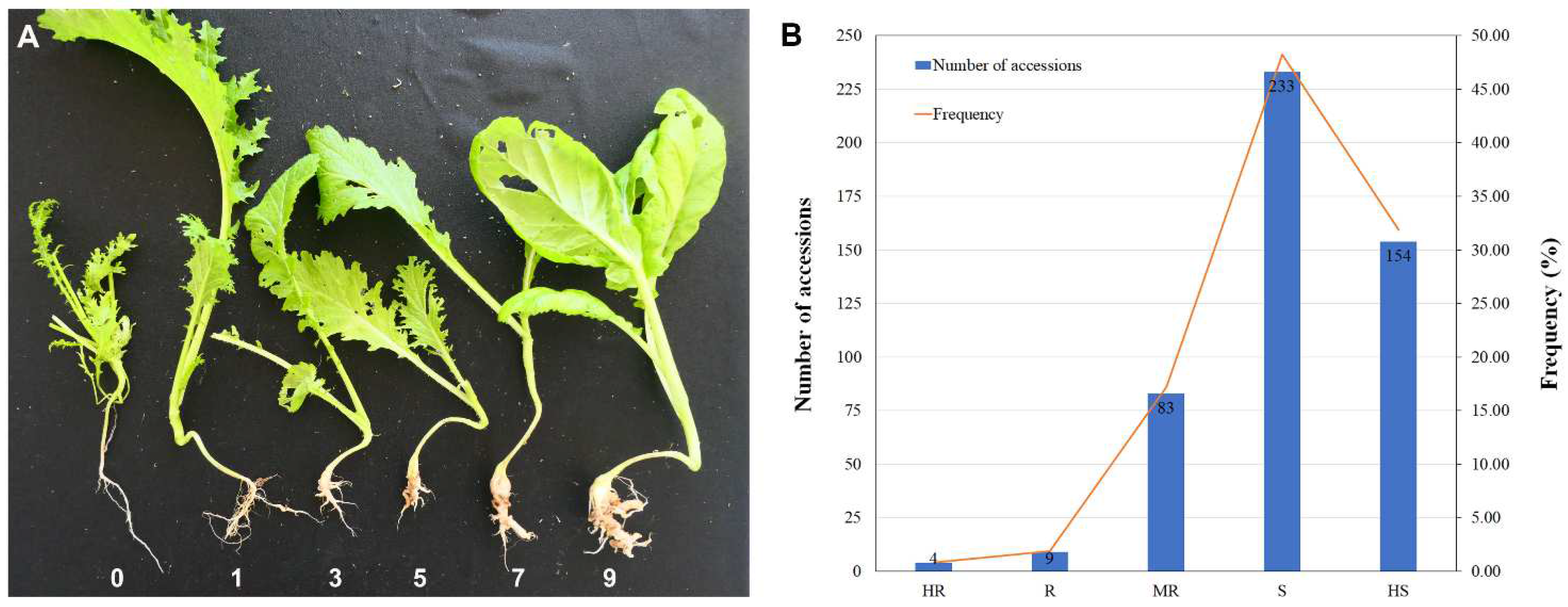
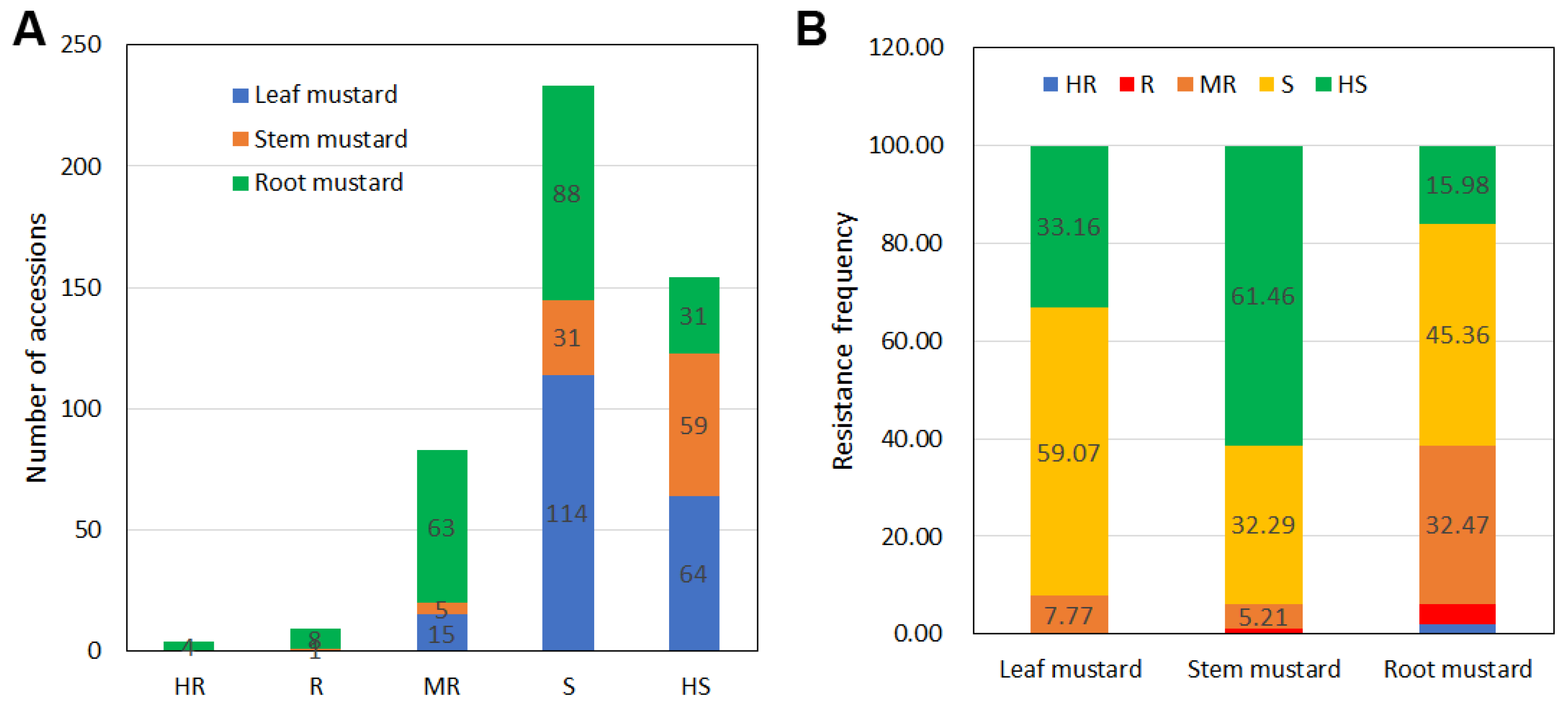
3.2. Detection of Seedling Resistance to Clubroot Disease in Mustard Resources
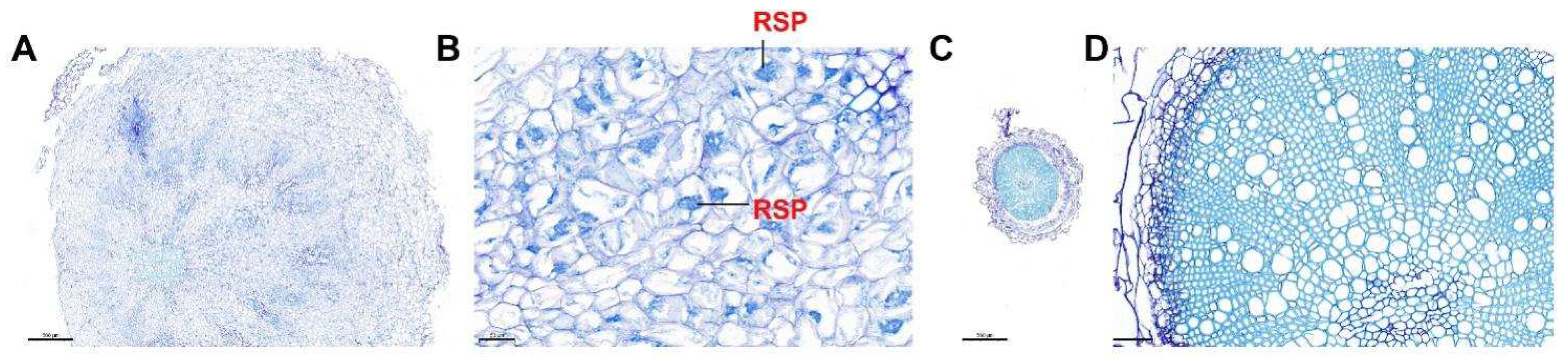
3.3. Histological Observation of Resistant and Susceptible Mustard Root Cross Sections
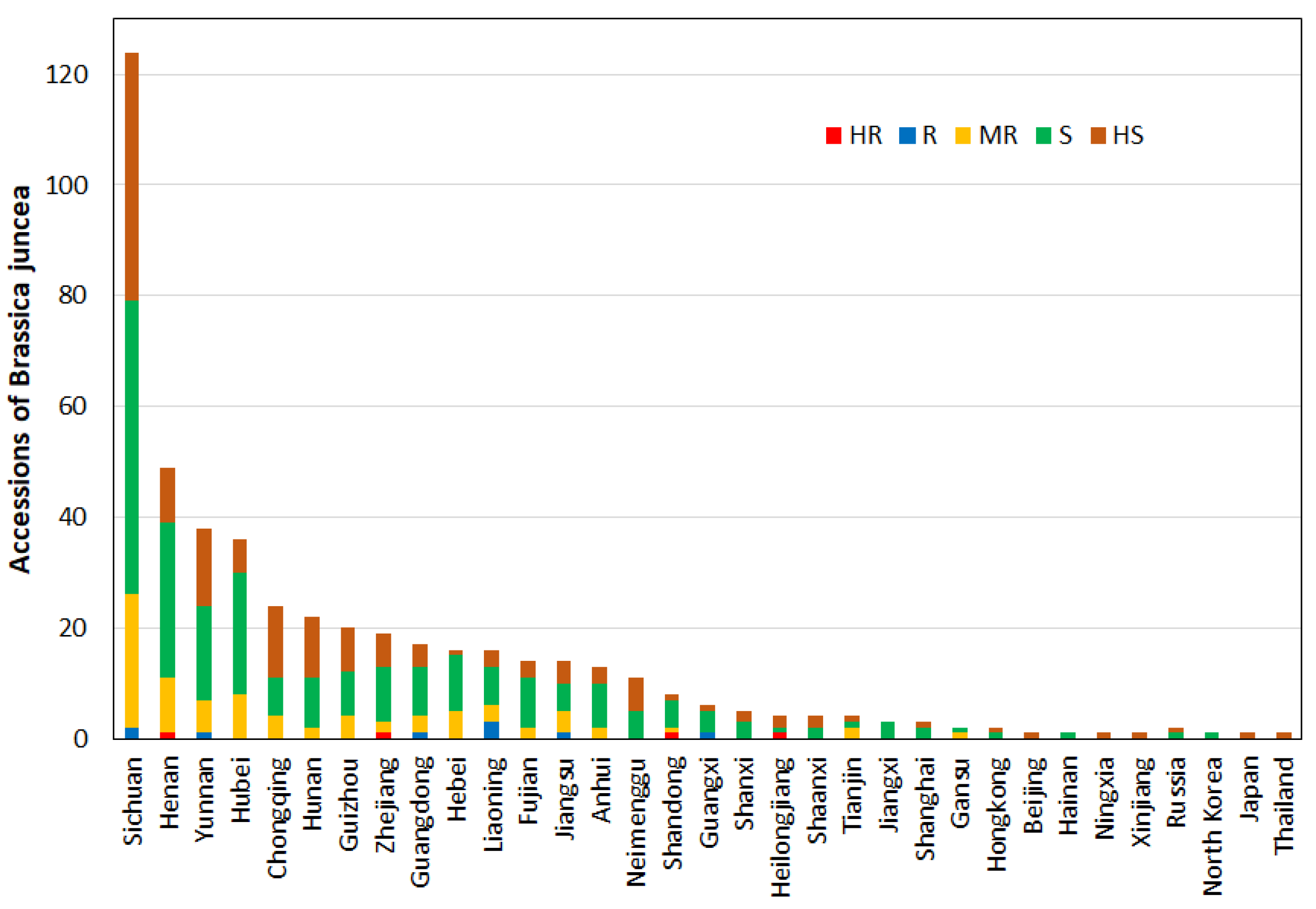
3.4. Geographical Distribution of Mustard Resources with Clubroot Disease Resistance
3.5. Comparison of Clubroot Resistance among Types of Mustard Resources at the Seedling Stage
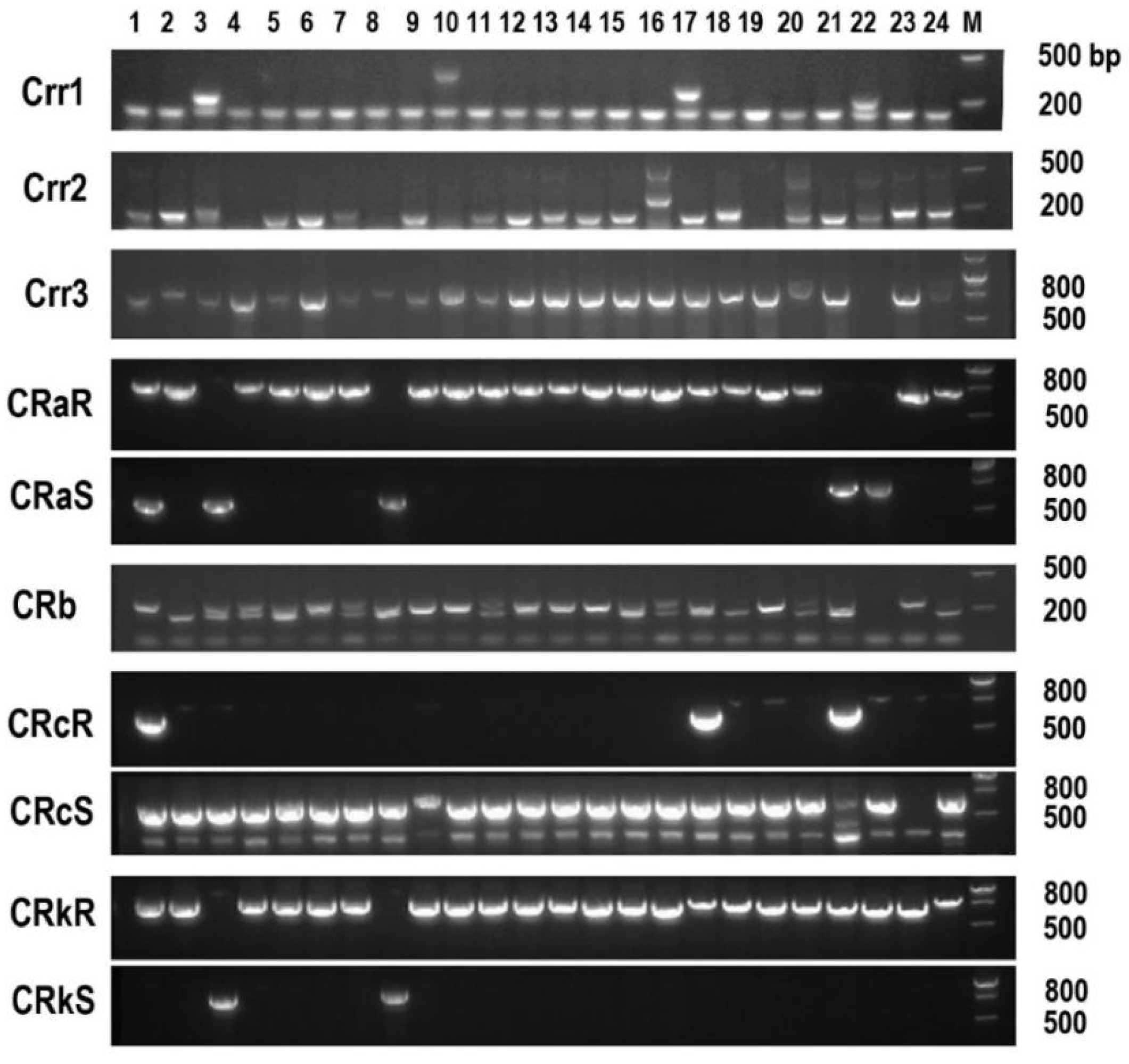
3.6. Molecular Detection of Clubroot Resistance in Mustard Resources
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liu, P. Chinese Mustard; China Agricultural Publishing House: Beijing, China, 1996; pp. 24–26. [Google Scholar]
- Fan, Y.; Shen, J.; Dong, D. Development status and research prospect of mustard vegetables industry. J. Agric. 2016, 6, 65–71. [Google Scholar]
- Wan, Z.; Li, H.; Yao, P.; Liu, X.; Xu, Y.; Fu, T.; Zou, R.; Xu, Y. Progress and prospect of heterosis utilization in mustard (Brassica juncea L.) vegetables. J. Huazhong Agric. Univ. 2018, 37, 115–120. [Google Scholar]
- Guo, Y. Pathogenesis characteristics and prevention of Cruciferae clubroot disease. Fujian Agric. 2003, 11, 22. [Google Scholar]
- Huang, Y.; Xu, L.; Xiao, C.; Liu, C.; Chen, G. Effects of Plasmodiophora brassicae infection on the contents of zeatin and indoleacetic acid in the roots of tumour stem mustard. China Veg. 2014, 2, 41–44. [Google Scholar]
- Diederichsen, E.; Frauen, M.; Linders, E.G.A.; Hatakeyama, K.; Hirai, M. Status and perspectives of clubroot resistance breeding in crucifer crops. J. Plant Growth Regul. 2009, 28, 265–281. [Google Scholar] [CrossRef]
- Hwang, S.F.; Strelkov, S.E.; Feng, J.; Gossen, B.D.; Howard, R.J. Plasmodiophora brassicae: A review of an emerging pathogen of the Canadian canola (Brassica napus) crop. Mol. Plant Pathol. 2012, 13, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Howard, R.J.; Strelkov, S.E.; Harding, M.W. Clubroot of cruciferous crops–new perspectives on an old disease. Can. J. Plant Pathol. 2010, 32, 43–57. [Google Scholar] [CrossRef]
- Sun, B.; Shen, X.; Guo, H.; Zhou, Y. Research progress on clubroot disease and disease resistance breeding of cruciferous plants. China Veg. 2005, 4, 34–37. [Google Scholar]
- Zhao, L.; Hui, M.; Dong, J.; Jiang, J.; Cheng, Y.; Zhang, E.; Wang, J. Green integrated control technology of clubroot in cruciferous vegetables. China Veg. 2022, 9, 111–114. [Google Scholar]
- Zhang, Y.; Ma, X.; Yu, H.; Dong, Z.; Lei, N.; Yu, X. Research progress on integrated control of clubroot in cruciferous crops. China Veg. 2022, 10, 27–37. [Google Scholar]
- He, H.; Si, J. Clubroot disease of cruciferae vegetables and its comprehensive prevention and control technology. J. Chang. Veg. 2017, 14, 1–4. [Google Scholar]
- Zeng, L.; Ren, L.; Liu, F.; Chen, W.; Chen, K.; Xu, L.; Fang, X. Reaction and resistant gene loci detection of 28 Chinese cabbage cultivars to different Plasmodiophora brassicae strains. Chin. J. Oil Crop Sci. 2017, 39, 532–539. [Google Scholar]
- Wang, L.; Wang, X.; Wu, H.; He, M.; Li, Y. Resistance identification and evaluation of clubroot resistant varieties of Chinese cabbage to Plasmodiophora brassicae. China Veg. 2017, 8, 46–50. [Google Scholar]
- Zhu, M.; Zhang, S.; Zhang, H.; Zhang, S.; Li, F.; Wang, X.; Wu, J.; Li, G.; Wei, Y.; Yue, L.; et al. Anti-sources screening and molecular marker identification of Chinese cabbage against clubroot. China Veg. 2018, 3, 40–45. [Google Scholar]
- Zhang, S.; Zhang, Q.; Si, C.; Wang, Y.; Yang, Z.; Yang, X. Identification of resistance of Chinese cabbage varieties to Plasmodiophora brassicae. Shandong Agric. Sci. 2018, 50, 98–102+108. [Google Scholar]
- Wen, J.; Wang, C.; Hua, D.; Zhang, H.; Xu, Y.; Zhang, B. Reaction and resistant gene loci detection of different Chinese cabbage varieties to different Plasmodiophora brassicae strains. Acta Agric. Boreali-Sin. 2019, 34, 103–109. [Google Scholar]
- Li, H.; Dong, J.; Wang, Y.; Xiao, C.; Zhao, L. Resistance identification to Plasmodiophora brassicae and character evaluation of clubroot resistant varieties of Chinese cabbage. Chin. Melon Veg. 2020, 33, 39–43. [Google Scholar]
- Ma, X.; Zhang, H.; Zhang, S.; Li, F.; Zhang, S.; Li, G.; Liu, X.; Sun, R. Evaluation on resistance of different Chinese cabbage varieties to clubroot pathogens occurred in different regions. China Veg. 2021, 9, 41–47. [Google Scholar]
- Wei, X.; Yuan, Y.; Zhao, Y.; Yang, S.; Wang, Z.; Su, H.; Li, J.; Li, L.; Niu, L.; Zhang, X. Identification and detection of clubroot resistance loci of 96 Chinese cabbage cultivars resistance to Plasmodiophora brassicae. Chin. Melon Veg. 2022, 35, 27–34. [Google Scholar]
- Chen, X. Identification of Pathogen of Cabbage Clubroot Disease and Screening of Resistant Materials; Northeast Agricultural University: Harbin, China, 2015. [Google Scholar]
- Sun, C.; Ma, J.; Lei, L.; Jie, J.; Kang, J. Identification of cabbage germplasm resources with resistance to clubroot and sequence analysis of CRa and Crr1a homologous genes from cabbage. J. Plant Genet. Resour. 2016, 17, 1058–1064. [Google Scholar]
- Wang, S.; Wu, Q.; Wang, H.; Yu, L.; Li, J.; Ding, W. Identification of Plasmodiophora brassicae Wor. and resistance assessment of Cabbage (Brassica oleracea var. capitata L.) germplasm. J. Plant Genet. Resour. 2016, 17, 1123–1132. [Google Scholar]
- Chen, J.; Ren, X.; Song, H.; Li, C.; Yuan, T.; Si, J. Identification of races of Plasmodiophora brassicae from four different regions and resistance identification of new cabbage cross combinations. J. Southwest Univ. Nat. Sci. Ed. 2016, 38, 68–72. [Google Scholar]
- Ma, D.; Pang, S.; Ding, Y.; Jian, Y.; Geng, L. Study on clubroot resistance of different Brassica crops. J. Shihezi Univ. Nat. Sci. Ed. 2016, 34, 164–169. [Google Scholar]
- Peng, L.; Zhou, L.; Ren, X.; Song, H.; Li, C.; Si, J. Creating new materials with resistance to clubroot disease by Embryo Rescuing cabbage × Chinese cabbage. J. Plant Prot. 2016, 43, 419–426. [Google Scholar]
- Zheng, J.; Wang, X.; Li, Q.; Yuan, S.; Wei, S.; Tian, X.; Huang, Y.; Wang, W.; Yang, H. Characterization of five molecular markers for pathotype identification of the clubroot pathogen Plasmodiophora brassicae. Phytopathology 2018, 108, 1486–1492. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, H.; Song, J.; Yang, W.; Jia, H.; Agerbirk, N.; Chen, Y.; Li, C.; Piao, Y.; Li, S.; et al. Identification of Clubroot-Resistant Germplasm in a Radish (Raphanus sativus L.) Core Collection. Agronomy 2024, 14, 157. [Google Scholar] [CrossRef]
- Si, J.; Li, C.; Song, H.; Ren, X.; Song, M.; Wang, X. Identification and evaluation of resistance of cabbage to clubroot. J. Southwest Univ. Nat. Sci. Ed. 2009, 31, 26–30. [Google Scholar]
- Yuan, T. Identification of Resistance to Cabbage Clubroot Disease and Screening of Molecular Markers; Southwest University: Chongqing, China, 2014. [Google Scholar]
- Piao, Z.; Ramchiary, N.; Lim, Y.P. Genetics of clubroot resistance in Brassica species. J. Plant Growth Regul. 2009, 28, 252–264. [Google Scholar] [CrossRef]
- Liu, S.; Liu, Y.; Yang, X.; Tong, C.; Edwards, D.; Parkin, I.A.; Zhao, M.; Ma, J.; Yu, J.; Huang, S.; et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014, 5, 3930. [Google Scholar] [CrossRef]
- Chen, Q. Occurrence and control of clubroot disease of winter sown Chaozhou mustard. Jiangxi Plant Prot. 2004, 3, 122–123. [Google Scholar]
- Jiang, H.; Peng, Y.; Dong, D.; Yan, Y.; Wu, C.; Wang, X. Relationship between soil pH and clubroot of Brassica juncea var. tumida Tsen & Lee. J. South. Agric. 2017, 48, 1617–1623. [Google Scholar]
- Jiang, H.; Peng, Y.; Yan, Y.; Zhang, Y.; Wu, C.; He, B.; Wang, X. Screening, identification and evaluation of antagonistic bacteria against clubroot of mustard. Plant Prot. 2018, 44, 104–110. [Google Scholar]
- Leng, R.; Shen, J.; Dong, Z.; Tang, Y.; Zhao, S.; Yao, Q.; Ren, X. Effects of different chemicals and fertilizers on the control of Fuling stem mustard clubroot disease. China Veg. 2012, 19, 31–32. [Google Scholar]
- Liu, D.; Ran, M.; Wang, X.; Zhang, Q. Integrated control of clubroot of leaf mustard (Brassica juncea L.) at seedling stage. J. Anhui Agric. Sci. 2015, 43, 77–79. [Google Scholar]
- Liu, L.; Ding, W. Comprehensive prevention and treatment of clubroot in Brassica juncea var. tumida Tsen et Lee. Plant Dr. 2018, 31, 38–39. [Google Scholar]
- Liu, L.; Dong, P.; Kuang, M.; Li, S.; Yuan, G.; Ding, W. A preliminary analysis of the functional diversity of the microbial communities in healthy and clubroot infected soil rhizosphere planted with Brassica juncea var. tumida Tsen et Lee. Plant Dr. 2020, 33, 33–38. [Google Scholar]
- Tang, K.; He, Y.; Shen, M.; Huang, Q.; Wei, Y.; Huang, R.; Zhao, K. Isolation and screening of antagonistic strains of tuber mustard rhizoma and analysis of their growth promoting characteristics. Sichuan Agric. Sci. Technol. 2022, 8, 53–56. [Google Scholar]
- Wang, D.; Tan, Y.; Tian, X.; Zhang, C.; Qin, M.; Pan, L. High throughput analysis on change characteristics of rhizosphere soil fungal community during different occurance times of tuber mustard clubroot disease. China Veg. 2016, 5, 33–37. [Google Scholar]
- Wang, X.; Gao, M.; Peng, H.; Zhang, J. Study on controlling of clubroot of tumour mustard (Brassica juncea var. tumida Tsen et Lee). J. Chang. Veg. 2003, 5, 38–39. [Google Scholar]
- Wang, X.; Peng, H.; Gao, M.; Han, H.; Zhang, J.; Xiao, C. Preliminary identification of the pathogen of root tumour of stem mustard and factors influencing the incidence of the disease. J. Southwest Agric. 2002, 4, 75–78. [Google Scholar]
- Xiao, C.; Guo, X.; Han, H.; Peng, H. Identification of the pathogen for clubroot on diseased stem mustard in Fuling, Chongqing and its major characteristics. J. Southwest Agric. Univ. 2002, 6, 539–541. [Google Scholar]
- Hasan, M.J.; Strelkov, S.E.; Howard, R.J.; Rahman, H. Screening of Brassica germplasm for resistance to Plasmodiophora brassicae pathotypes prevalent in Canada for broadening diversity in clubroot resistance. Can. J. Plant Sci. 2012, 92, 501–515. [Google Scholar] [CrossRef]
- Sharma, P.; Siddiqui, S.A.; Rai, P.K.; Meena, P.D.; Kumar, J.; Chauhan, J.S. Evaluation of Brassica germplasm for field resistance against clubroot (Plasmodiophora brassicae Woron). Arch. Phytopathol. Plant Prot. 2012, 45, 356–359. [Google Scholar] [CrossRef]
- Wei, X.; Li, J.; Zhang, X.; Zhao, Y.; Nath, U.K.; Mao, L.; Xie, Z.; Yang, S.; Shi, G.; Wang, Z.; et al. Fine mapping and functional analysis of major QTL, CRq for clubroot resistance in Chinese cabbage (Brassica rapa ssp. pekinensis). Agronomy 2022, 12, 1172. [Google Scholar] [CrossRef]
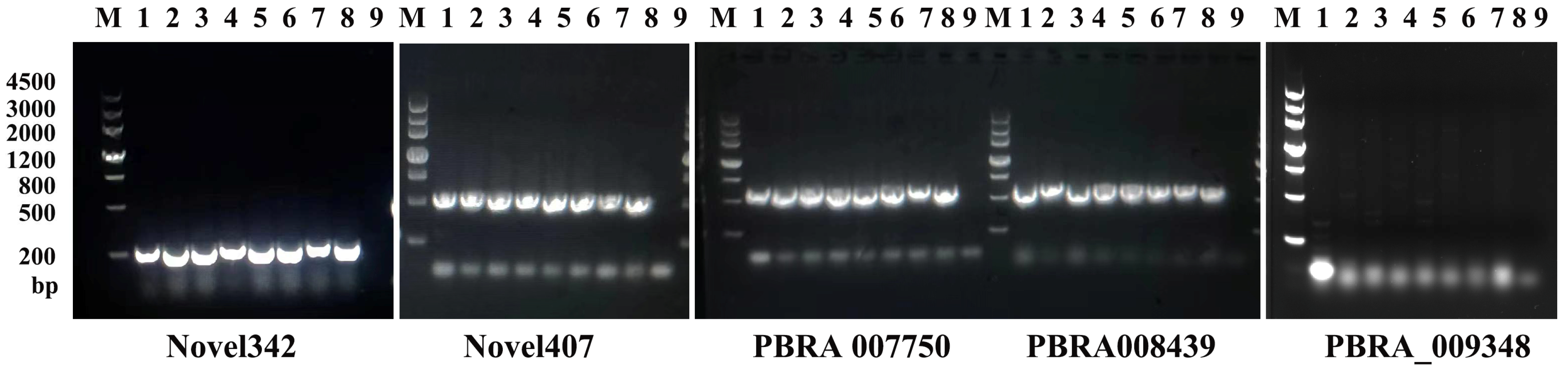

| Primer Name | Primer Sequences (5′ → 3′) | Product Size (bp) |
|---|---|---|
| Novel342-F | TCCTCTTGAACCGACACTGC | 249 |
| Novel342-R | CTTCTCTCGCACTAGCCAGG | |
| Novel407-F | ATTGCGTTGCTGAACTGCTG | 549 |
| Novel407-R | GTGCCCAATAGCAATCGCAG | |
| PBRA_007750-F | FCTTCGTGCTGACCGATTCCT | 638 |
| PBRA_007750-R | ATAATGCTCTGCGTCAGCCA | |
| PBRA_008439-F | TCGGCGACCTGAGCGAGAA | 651 |
| PBRA_008439-R | TCAACATGCGCATAGTAC | |
| PBRA_009348-F | CACTGCTATCGTCTCCCTGG | 509 |
| PBRA_009348-R | RCCTGCAATGTTTCGCTGCAA |
| Loci | Primer Name | Primer Sequences (5′ → 3′) | Product Size (bp) | Reference |
|---|---|---|---|---|
| Crr1 | BRMS088F | TATCGGTACTGATTCGCTCTTCAAC | R263/S233 | [29] |
| BRMS088R | ATCGGTTGTTATTTGAGAGCAGATT | |||
| Crr2 | BRMS096F | AGTCGAGATCTCGTTCGTGTCTCCC | R220/S189 | [29] |
| BRMS096R | TGAAGAAGGATTGAAGCTGTTGTTG | |||
| Crr3 | OPC11-2F | GTAACTTGGTACAGAACAGCATAG | R1300/S1000 | [30] |
| OPC11-2R | ACTTGTCTAATGAATGATGATGG | |||
| CRa | SC2930TF | TAGACCTTTTTTTTGTCTTTTTTTTTAC | R800 | [31] |
| SC2930QF | CAGACTAGACTTTTTGTCATTTAGA | S800 | ||
| SC2930R | CTAAGGCCATAGAAATCAGGTC | |||
| CRb | KB-F | AGAGCAGAGTGAAACCAGAACT | R254/S194 | [32] |
| KB-R | GTTTCAGTTCAGTCAGGTTTTTGCAG | |||
| CRc | B50-C9F | GATTCAATGCATTTCTCTCGAT | R800 | [31] |
| B50-6RF | AATGCATTTTCGCTCAACC | S800 | ||
| B50R | CGTATTATATCTCTTTCTCCATCCC | |||
| CRk | HC688-4F | TCTCTGTATTGCGTTGACTG | ||
| HC688-6R | ATATGTTGAAGCCTATGTCT | R1000 | [31] | |
| HC688-7R | AAATATATGTGAAGTCTTATGATC | S1000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, W.; Song, J.; Zhang, X.; Xu, C.; Han, J.; Li, Z.; Wang, Y.; Jia, H.; Wang, H. Detection of Clubroot Disease Resistance in Brassica juncea Germplasm at the Seedling Stage. Agronomy 2024, 14, 2042. https://doi.org/10.3390/agronomy14092042
Yang W, Song J, Zhang X, Xu C, Han J, Li Z, Wang Y, Jia H, Wang H. Detection of Clubroot Disease Resistance in Brassica juncea Germplasm at the Seedling Stage. Agronomy. 2024; 14(9):2042. https://doi.org/10.3390/agronomy14092042
Chicago/Turabian StyleYang, Wenlong, Jiangping Song, Xiaohui Zhang, Chu Xu, Jiaqi Han, Zhijie Li, Yang Wang, Huixia Jia, and Haiping Wang. 2024. "Detection of Clubroot Disease Resistance in Brassica juncea Germplasm at the Seedling Stage" Agronomy 14, no. 9: 2042. https://doi.org/10.3390/agronomy14092042
APA StyleYang, W., Song, J., Zhang, X., Xu, C., Han, J., Li, Z., Wang, Y., Jia, H., & Wang, H. (2024). Detection of Clubroot Disease Resistance in Brassica juncea Germplasm at the Seedling Stage. Agronomy, 14(9), 2042. https://doi.org/10.3390/agronomy14092042








