Overexpression of the GmPM35 Gene Significantly Enhances Drought Tolerance in Transgenic Arabidopsis and Soybean
Abstract
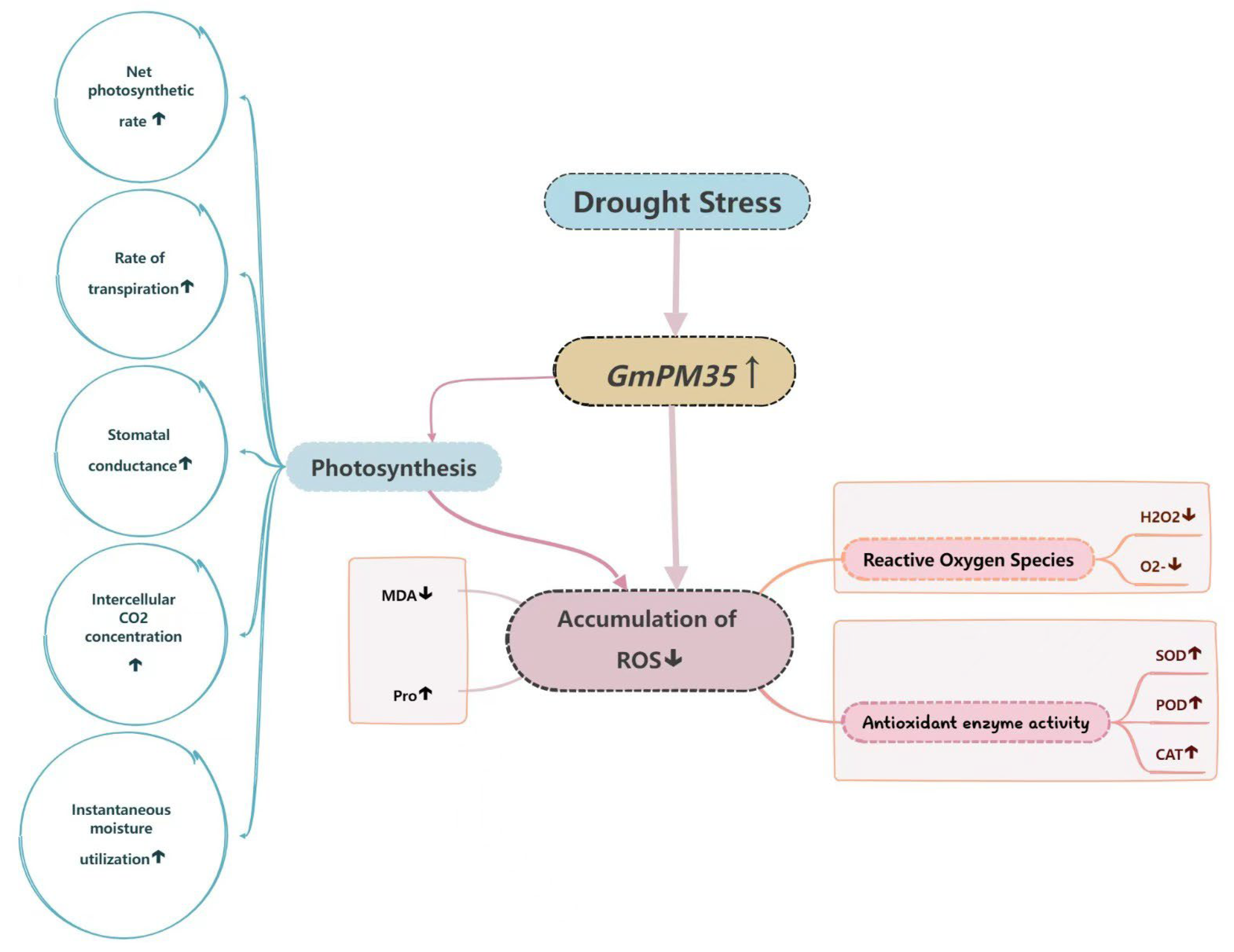
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growing Conditions
2.2. Preparation of Experimental Materials
2.2.1. Construction of Overexpression and Gene-Edited Vectors
2.2.2. Transformation and Screening of Transgenic Arabidopsis
2.2.3. Induction and Detection of Soybean Hairy Roots
2.2.4. Transformation and Screening of Transgenic Soybean
2.3. Experimental Procedures
2.3.1. Subcellular Localization
2.3.2. RNA Extraction and qRT-PCR Analysis
2.3.3. Analysis of Tissue-Specific Expression and Abiotic Stress Expression Patterns
2.3.4. Detection of Seedling Germination, Green Seedling Rate, Root Length, and Fresh Weight of Arabidopsis thaliana Seedlings
2.3.5. Rehydration Test and Physiological and Biochemical Detection Under Drought Stress in Transgenic Arabidopsis thaliana
2.3.6. Root System Changes and Determination of Physiological and Biochemical Indices of Soybean Hairy Roots Under Drought Stress
2.3.7. Determination of Germination Rate and Shoot Length of Transgenic Soybean Plants of T3 Generation
2.3.8. Rehydration Experiment of T3-Generation Transgenic Soybean Plants
2.3.9. Determination of Root Length in T3-Generation Transgenic Soybean Plants
2.3.10. Determination of Photosynthetic Physiological Indexes in T3-Generation Positive Plants
2.3.11. Determination of Biochemical Parameters in T3-Generation Positive Plants
2.3.12. T3-Generation Transgenic Plants Reactive Oxygen Species Assay
2.3.13. Investigation of Agronomic Traits in T2-Generation Transgenic Soybean Plants
2.4. Statistical Analysis
3. Results
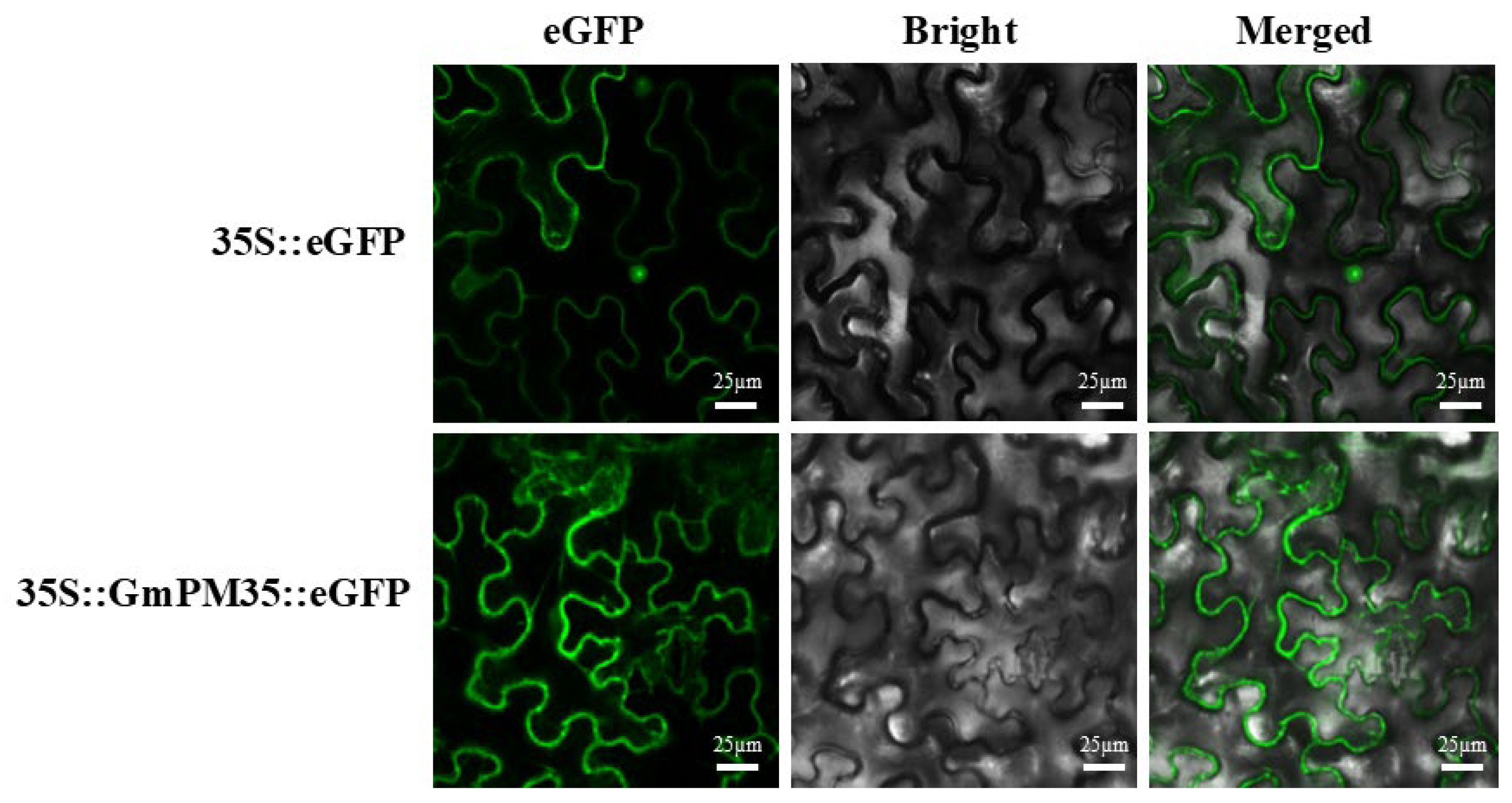
3.1. Subcellular Localization
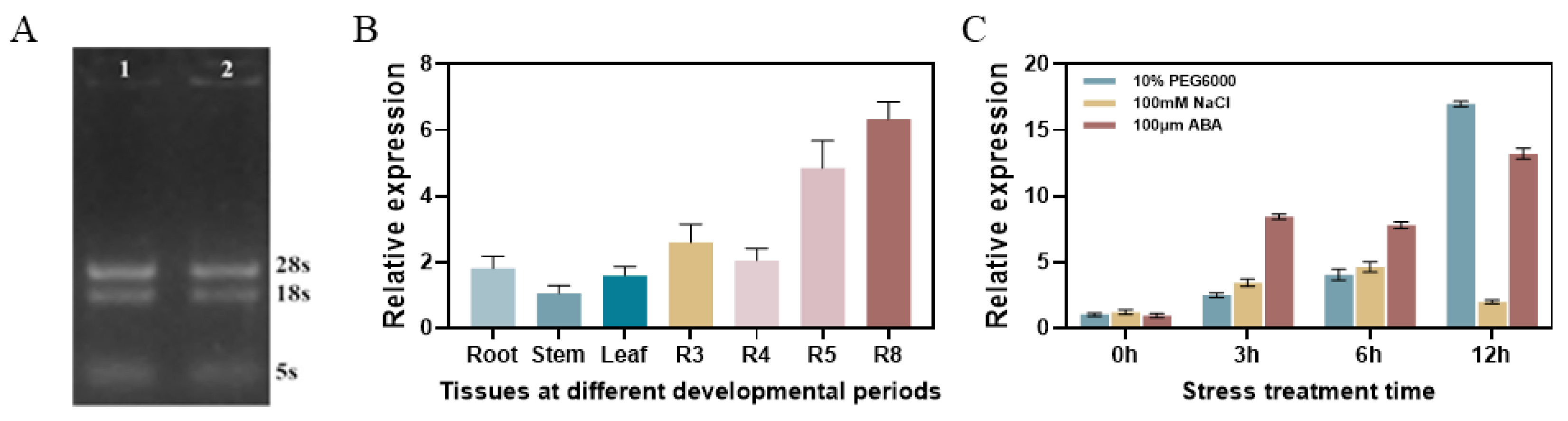
3.2. Analysis of Tissue-Specific Expression of GmPM35 Gene in Soybean and Its Expression Pattern Under Abiotic Stresses
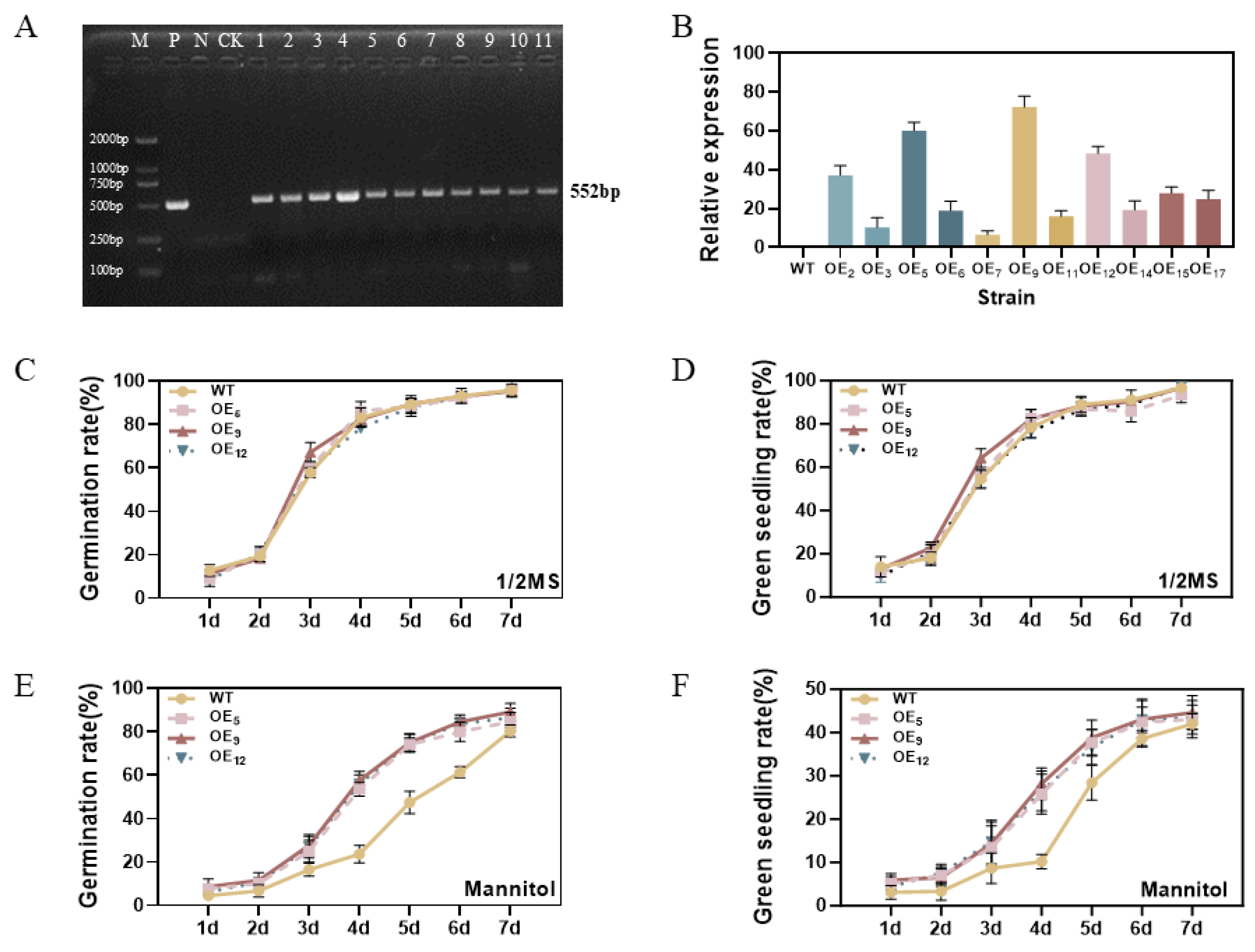
3.3. Overexpression of GmPM35 Gene Enhances Germination and Green Shooting in Transgenic Arabidopsis thaliana
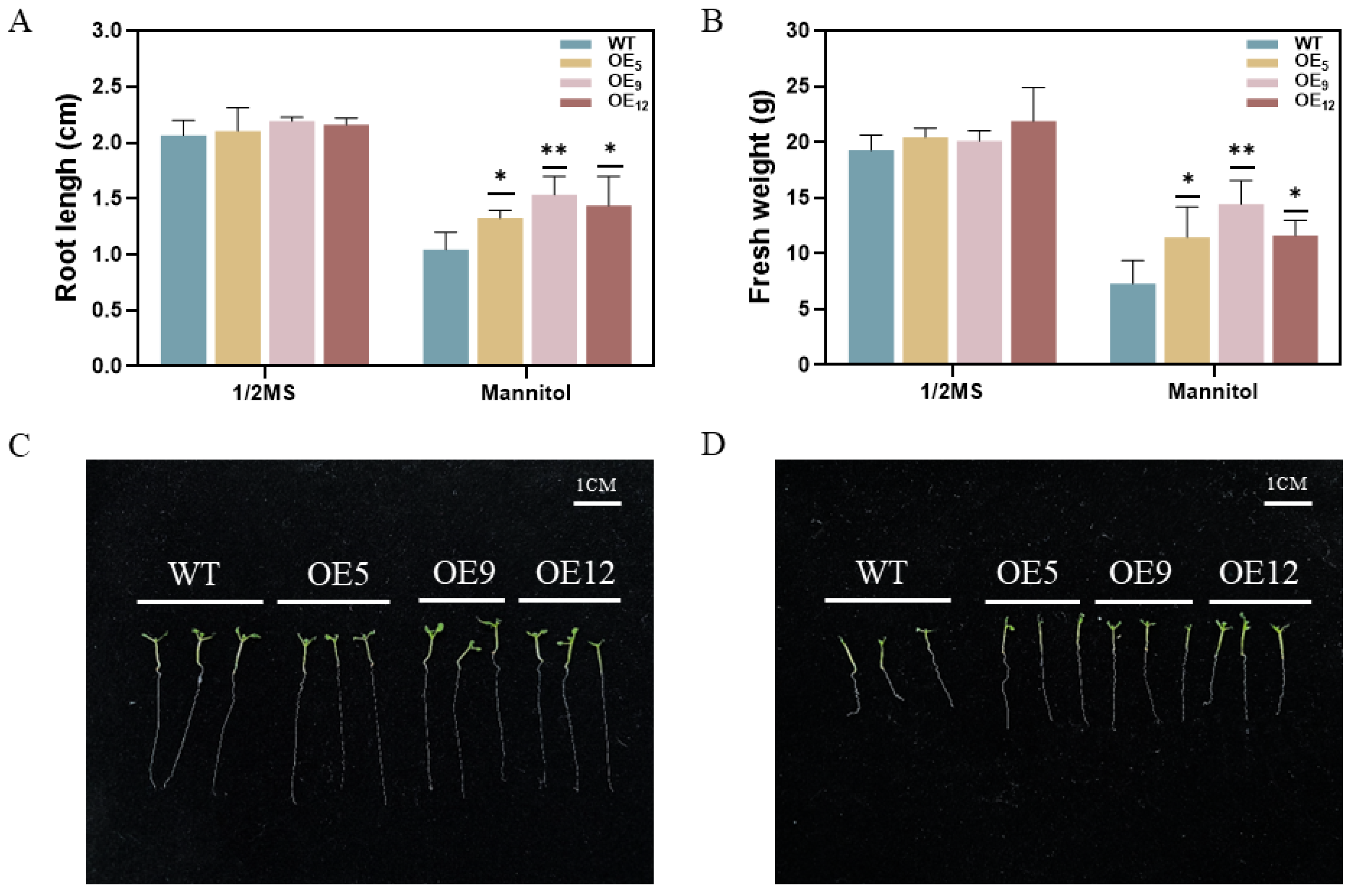
3.4. Overexpression of the GmPM35 Gene Promotes Water Retention and Root Elongation in Transgenic Arabidopsis thaliana Under Drought Stress
3.5. T2-Generation Transgenic Arabidopsis Rehydration Test and Physiological and Biochemical Detection of Transgenic Arabidopsis Under Drought Stress
3.6. PCR Detection of Soybean Hairy Roots Overexpressing GmPM35 Gene and Expression Analysis of Overexpressed Soybean Hairy Roots Under Drought Stress
3.7. Root Changes in Overexpressed Soybean Hairy Roots Under Drought Stress
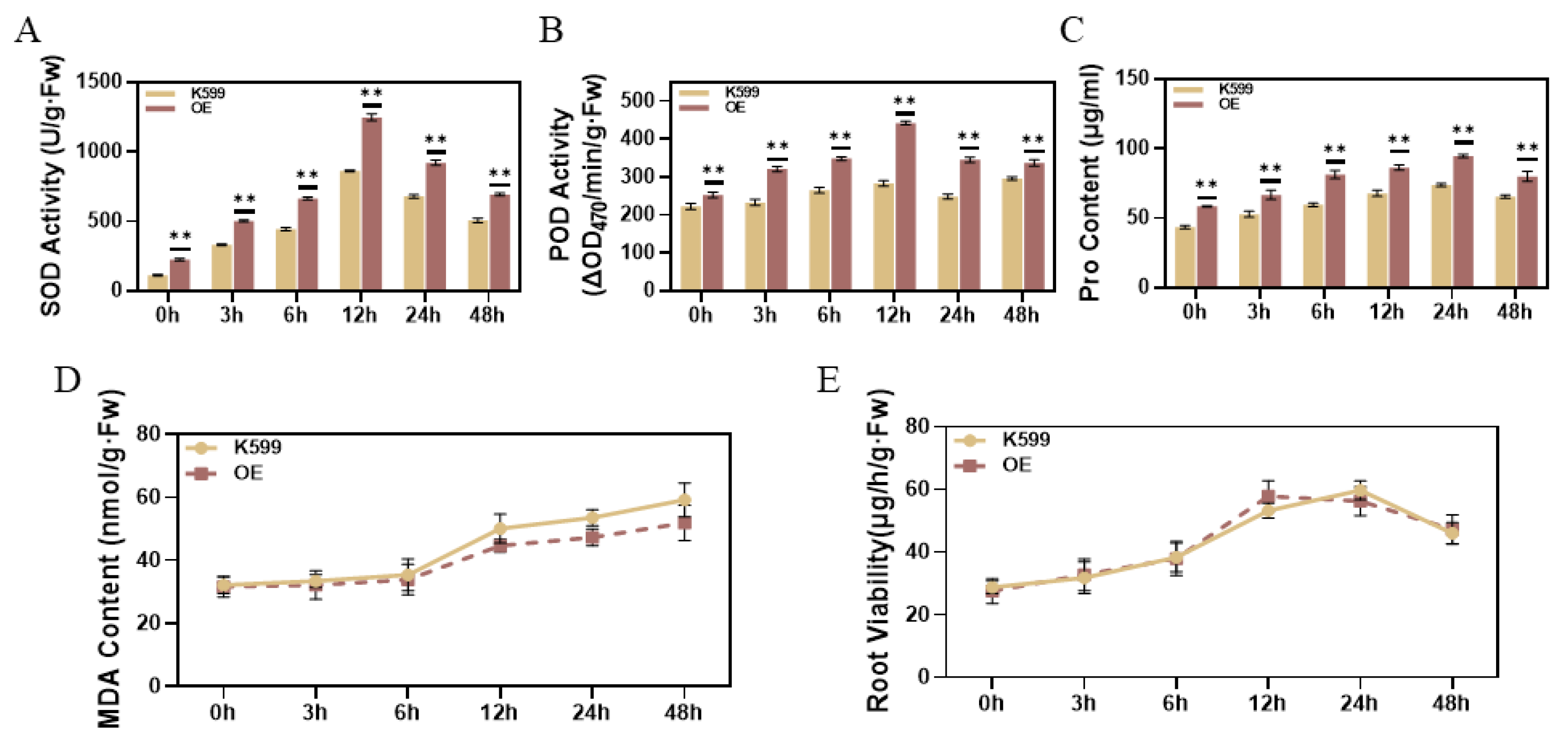
3.8. Physiological and Biochemical Assay of Overexpressed Soybean Hairy Roots Under Drought Stress
3.9. Mutation Efficiency Statistics for Editing Vectors
3.10. Gene Expression and Editing in Different Strains of Soybean
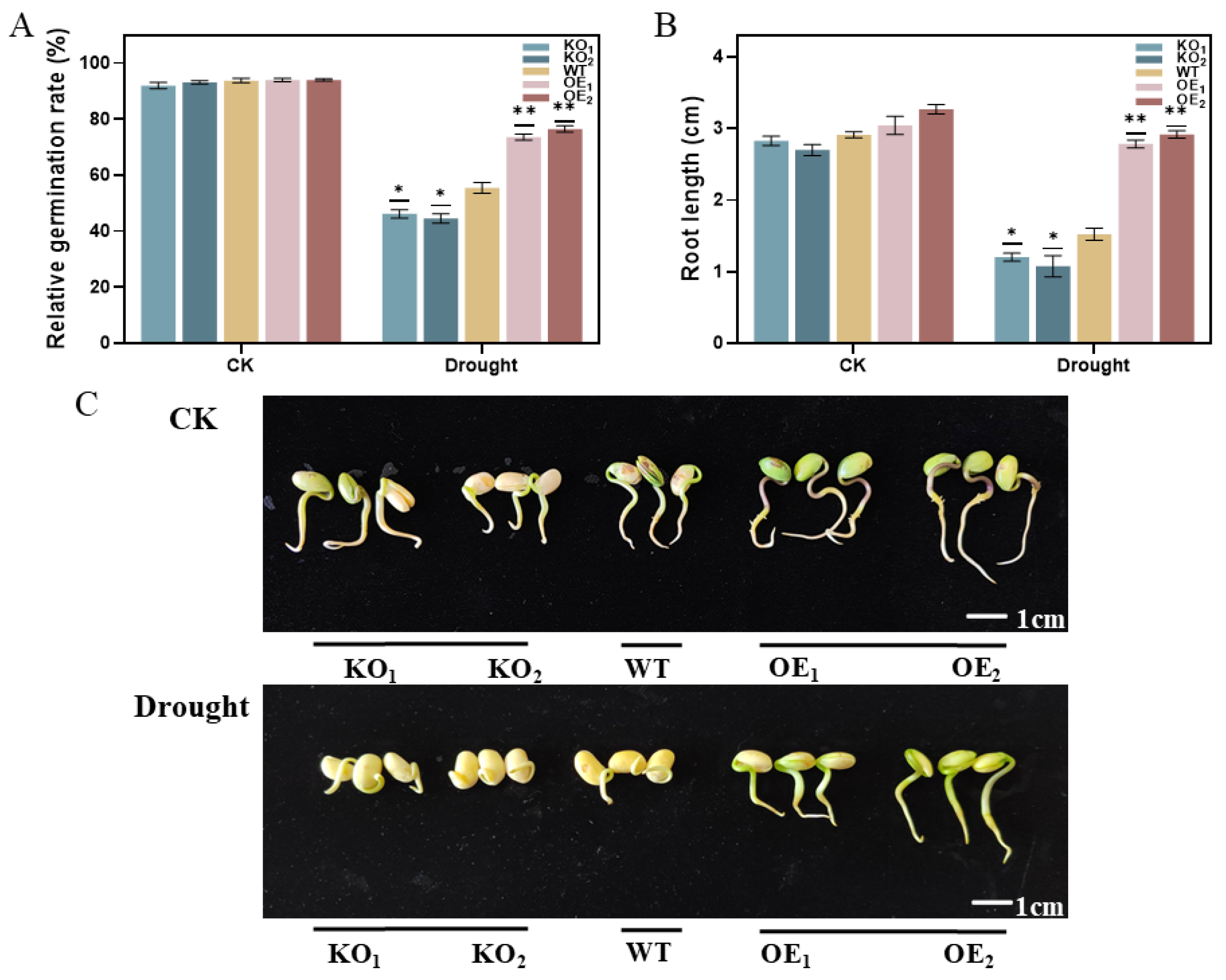
3.11. Determination of Germination Rate and Shoot Length of T3-Generation Positive Soybean Plants Under Drought Stress
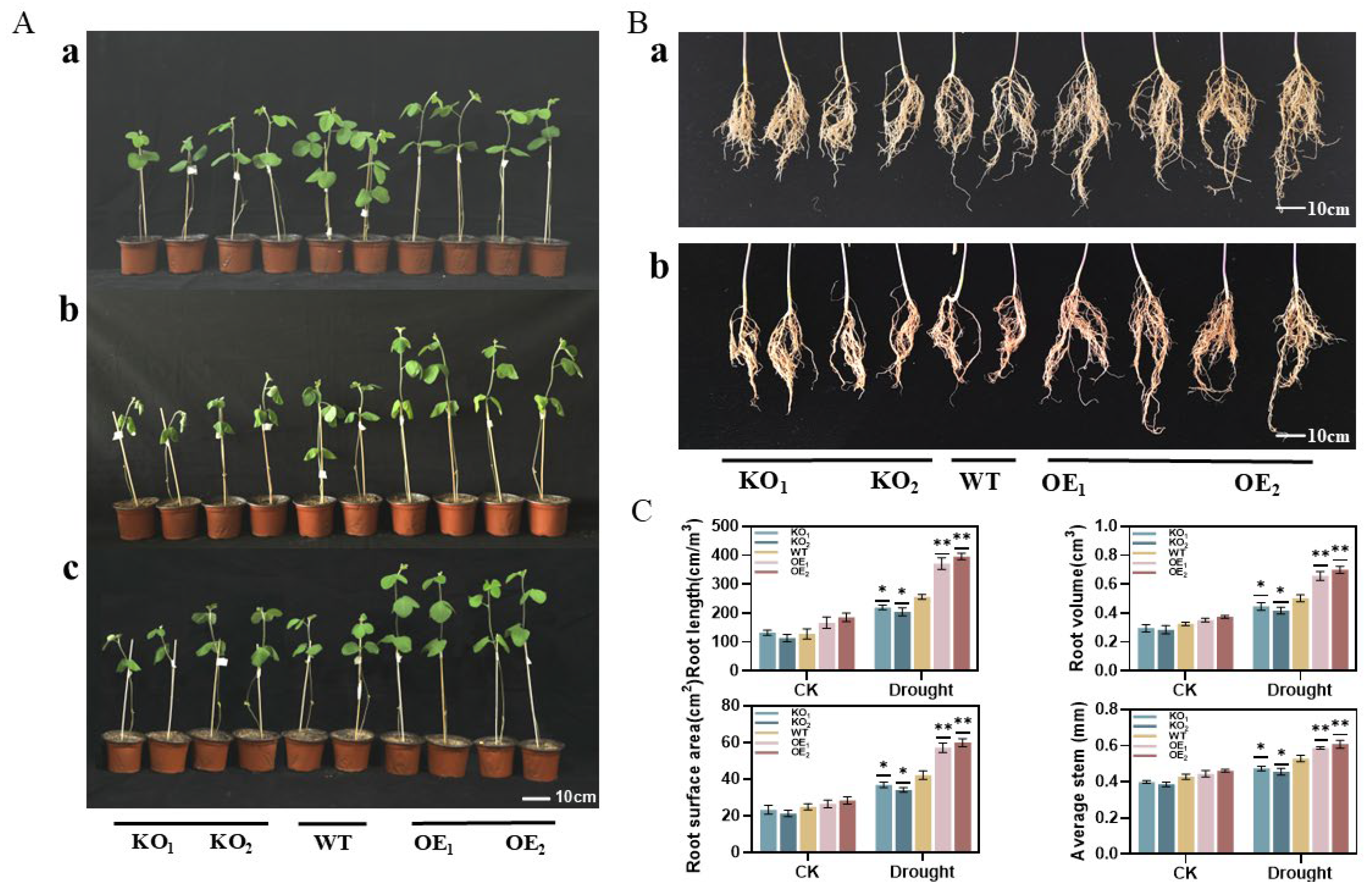
3.12. Rehydration Test and Root Phenotyping of T3-Generation Positive Soybean Plants Under Drought Stress
3.13. Determination of Photosynthetic Physiological Indexes in T3-Generation Transgenic Soybean Plants Under Drought Stress
3.14. Determination of Biochemical Indexes in T3-Generation Transgenic Soybean Plants Under Drought Stress
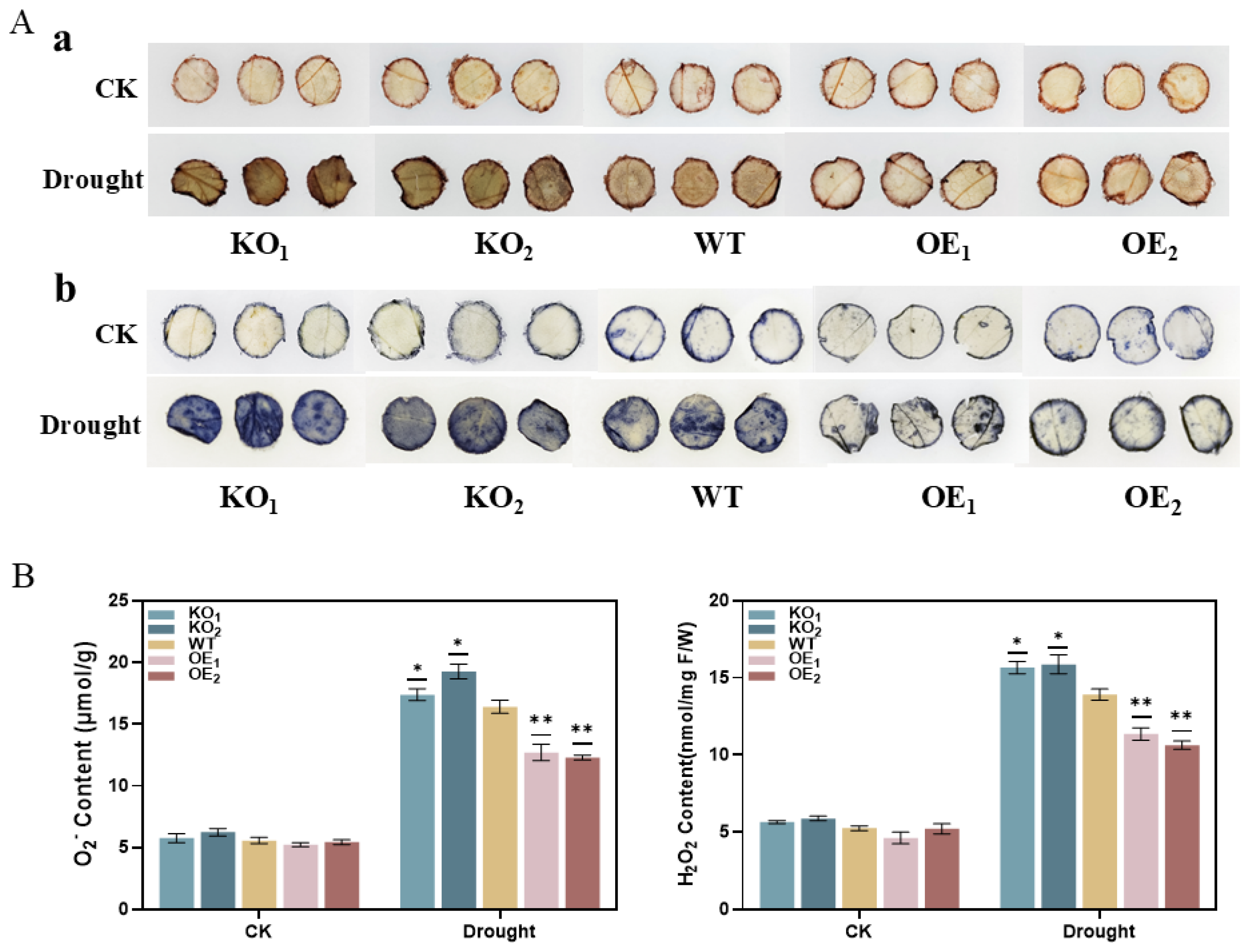
3.15. Determination of Reactive Oxygen Species in T3-Generation Transgenic Soybean Plants Under Drought Stress
3.16. Investigation of Agronomic Traits in T2-Generation Positive Soybean Plants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nogrady, B. How to address agriculture’s water woes. Nature 2024, 630, S26–S27. [Google Scholar] [CrossRef]
- Tariq, A.; Sardans, J.; Zeng, F.; Graciano, C.; Hughes, A.C.; Farré-Armengol, G.; Peñuelas, J. Impact of aridity rise and arid lands expansion on carbon-storing capacity, biodiversity loss, and ecosystem services. Glob. Chang. Biol. 2024, 30, e17292. [Google Scholar] [CrossRef] [PubMed]
- Yimer, E.A.; De Trift, L.; Dondeyne, S.; Speijer, L.; Huysmans, M.; Cools, J.; Nossent, J.; van Griensven, A. Framework for mapping large-scale nature-based solutions for drought mitigation: Regional application in Flanders. Water Res. 2024, 261, 122003. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Lam, H.-M.; Nguyen, H.T.; Siddique, K.H.M.; Varshney, R.K.; Colmer, T.D.; Cowling, W.; Bramley, H.; Mori, T.A.; Hodgson, J.M.; et al. Neglecting legumes has compromised human health and sustainable food production. Nat. Plants 2016, 2, 16112. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhao, W.; Wei, X.; Sun, Y.; Dong, S. Molecular mechanism of drought resistance in soybean roots revealed using physiological and multi-omics analyses. Plant Physiol. Biochem. 2024, 208, 108451. [Google Scholar] [CrossRef]
- Xu, M.; Tong, Q.; Wang, Y.; Wang, Z.; Xu, G.; Elias, G.K.; Li, S.; Liang, Z. Transcriptomic Analysis of the Grapevine LEA Gene Family in Response to Osmotic and Cold Stress Reveals a Key Role for VamDHN3. Plant Cell Physiol. 2020, 61, 775–786. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Zhang, X.; Yao, W.; Zhao, K.; Liu, L.; Fan, G.; Zhou, B.; Jiang, T. Genome-wide search and structural and functional analyses for late embryogenesis-abundant (LEA) gene family in poplar. BMC Plant Biol. 2021, 21, 110. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Cao, D.; Wang, Z.; Ma, L.; Tian, K.; Liu, Y.; Gong, Z.; Zhu, X.; Jiang, C.; Li, Y. Genome-wide identification and expression analyses of the LEA protein gene family in tea plant reveal their involvement in seed development and abiotic stress responses. Sci. Rep. 2019, 9, 14123. [Google Scholar] [CrossRef]
- Borovskii, G.B.; Stupnikova, I.V.; Antipina, A.I.; Vladimirova, S.V.; Voinikov, V.K. Accumulation of dehydrin-like proteins in the mitochondria of cereals in response to cold, freezing, drought and ABA treatment. BMC Plant Biol. 2002, 2, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Zhu, T.; Wang, H.; Zhou, H.; Shao, L.; Ding, Q.; Zhang, D.; Ma, L. Genome-wide identification, phylogenetic analysis and expression profiling of the late embryogene-sis-abundant (LEA) gene family in Brachypodium distachyon. Funct. Plant Biol. 2021, 48, 386–401. [Google Scholar] [CrossRef] [PubMed]
- Hundertmark, M.; Hincha, D.K. LEA (Late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BioMed Central 2008, 9, 118. [Google Scholar] [CrossRef]
- Mowla, S.B.; Cuypers, A.; Driscoll, S.P.; Kiddle, G.; Thomson, J.; Foyer, C.H.; Theodoulou, F.L. Yeast complementation reveals a role for an Arabidopsis thaliana late embryogenesis abundant (LEA)-like protein in oxidative stress tolerance. Plant J. 2006, 48, 743–756. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.; Zhu, L.; Wang, L.; Zhang, H.; Zhang, X.; Xu, S.; Xue, J. Identification of the Maize LEA Gene Family and Its Relationship with Kernel Dehydration. Plants 2023, 12, 3674. [Google Scholar] [CrossRef]
- Bies-Ethève, N.; Gaubier-Comella, P.; Debures, A.; Lasserre, E.; Jobet, E.; Raynal, M.; Cooke, R.; Delseny, M. Inventory, evolution and expression profiling diversity of the LEA (late embryogenesis abundant) protein gene family in Arabidopsis thaliana. Plant Mol. Biol. 2008, 67, 107–124. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, Y.; Kang, Y.; Liu, W.; Li, S.; Wang, Z.; He, X. Genome-wide characterization of LEA gene family reveals a positive role of BnaA.LEA6.a in freezing tolerance in rapeseed (Brassica napus L.). BMC Plant Biol. 2024, 24, 433. [Google Scholar] [CrossRef]
- Huang, R.; Xiao, D.; Wang, X.; Zhan, J.; Wang, A.; He, L. Genome-wide identification, evolutionary and expression analyses of LEA gene family in peanut (Arachis hypogaea L.). BMC Plant Biol. 2022, 22, 155. [Google Scholar] [CrossRef]
- Muvunyi, B.P.; Yan, Q.; Wu, F.; Min, X.; Yan, Z.Z.; Kanzana, G.; Zhang, J. Mining Late Embryogenesis Abundant (LEA) Family Genes in Cleistogenes songorica, a Xerophyte Perennial Desert Plant. Int. J. Mol. Sci. 2018, 19, 3430. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Hao, Y.; Dong, Z.; Tang, W.; Wang, X.; Li, J.; Zhang, Z. Identification and expression analysis of LEA gene family members in pepper (Capsicum annuum L.). FEBS Open Bio 2023, 13, 2246–2262. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.-S.; Ge, N.; Wang, Q.-Y.; Zhao, L.-T.; Chen, C.; Chen, J.-W. Genome-wide identification and characterization of members of the LEA gene family in Panax notoginseng and their transcriptional responses to dehydration of recalcitrant seeds. BMC Genom. 2023, 24, 126. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, N.; Wang, X.; Meng, X.; Cui, X.; Chen, Z.; Liu, H. Late embryogenesis abundant (LEA) gene family in Salvia miltiorrhiza: Identification, expression analysis, and response to drought stress. Plant Signal Behav. 2021, 16, 1891769. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Lei, X.; Wang, Y.; Di, P.; Meng, X.; Peng, W.; Rong, J.; Wang, Y. Genome-wide identification of the LEA gene family in Panax ginseng: Evidence for the role of PgLEA2-50 in plant abiotic stress response. Plant Physiol. Biochem. 2024, 212, 108742. [Google Scholar] [CrossRef] [PubMed]
- Cuevas-Velazquez, C.L.; Saab-Rincón, G.; Reyes, J.L.; Covarrubias, A.A. The unstructured N-terminal region of Arabidops is group 4 late embryogenesis abundant proteins (LEA) is required for folding and for chaperone-like activity under water deficit. J. Biol. Chem. 2016, 291, 10893–10903. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, Q.; Qin, J.; Xiao, G.; Zhu, S.; Hu, T. OsLEA1a overexpression enhances tolerance to diverse abiotic stresses by inhibiting cell membrane damage and enhancing ROS scavenging capacity in transgenic rice. Funct Plant Biol. 2021, 48, 860–870. [Google Scholar] [CrossRef]
- Luo, D.; Hou, X.; Zhang, Y.; Meng, Y.; Zhang, H.; Liu, S.; Wang, X.; Chen, R. CaDHN5, a Dehydrin Gene from Pepper, Plays an Important Role in Salt and Osmotic Stress Responses. Int. J. Mol. Sci. 2019, 20, 1989. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liang, J.; Sun, L.; Yang, X.; Li, D. Group 3 LEA Protein, ZmLEA3, Is Involved in Protection from Low Temperature Stress. Front. Plant Sci. 2016, 7, 1011. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fan, J.; Zhang, Y.; Sun, H.; Duan, R.; Jiang, Y.; Wang, X.; Sun, Y.; Luo, Z.; Wang, P.; Guan, S.; et al. Overexpression of soybean GmDHN9 gene enhances drought resistance of transgenic Arabidopsis. GM Crops Food 2024, 15, 118–129. [Google Scholar] [CrossRef]
- Guo, B.; Zhang, J.; Yang, C.; Dong, L.; Ye, H.; Valliyodan, B.; Nguyen, H.T.; Song, L. The Late Embryogenesis Abundant Proteins in Soybean: Identification, Expression Analysis, and the Roles of GmLEA4_19 in Drought Stress. Int. J. Mol. Sci. 2023, 24, 14834. [Google Scholar] [CrossRef]
- Zhang, X.; Henriques, R.; Lin, S.-S.; Niu, Q.-W.; Chua, N.-H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef]
- Kereszt, A.; Li, D.; Indrasumunar, A.; Nguyen, C.D.; Nontachaiyapoom, S.; Kinkema, M.; Gresshoff, P.M. Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2007, 2, 948–952. [Google Scholar] [CrossRef]
- Freitas-Alves, N.S.; Moreira-Pinto, C.E.; Távora, F.T.; Paes-de-Melo, B.; Arraes, F.B.; Lourenço-Tessutti, I.T.; Grossi-de-Sa, M.F. CRISPR/Cas genome editing in soybean: Challenges and new insights to overcome existing bottle-necks. J. Adv. Res. 2024. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Fang, X.; Yuan, X.; Zhang, Y.; Li, H.; Zhou, Y.; Cui, X. Overexpression of Transcription Factor GmTGA15 Enhances Drought Tolerance in Transgenic Soybean Hairy Roots and Arabidopsis Plants. Agronomy 2021, 11, 170. [Google Scholar] [CrossRef]
- Kumar, M.S.; Mawlong, I.; Ali, K.; Tyagi, A. Regulation of phytosterol biosynthetic pathway during drought stress in rice. Plant Physiol. Biochem. 2018, 129, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Tanveer, M.; Shahzad, B.; Sharma, A.; Khan, E.A. 24-Epibrassinolide application in plants: An implication for improving drought stress tolerance in plants. Plant Physiol. Biochem. 2018, 135, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Nadarajah, K.K. ROS Homeostasis in Abiotic Stress Tolerance in Plants. Int. J. Mol. Sci. 2020, 21, 5208. [Google Scholar] [CrossRef] [PubMed]
- Mishra, N.; Jiang, C.; Chen, L.; Paul, A.; Chatterjee, A.; Shen, G. Achieving abiotic stress tolerance in plants through antioxidative defense mechanisms. Front. Plant Sci. 2023, 14, 1110622. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Reichheld, J.P.; Foyer, C.H. Accepted manuscript title: ROS-related redox regulation and signaling in plants. Semin. Cell Dev. Biol. 2019, 15, 4–18. [Google Scholar]
- Zandalinas, S.I.; Mittler, R. ROS-induced ROS release in plant and animal cells. Free. Radic. Biol. Med. 2018, 122, 21–27. [Google Scholar] [CrossRef]
- Huang, H.; Ullah, F.; Zhou, D.-X.; Yi, M.; Zhao, Y. Mechanisms of ROS Regulation of Plant Development and Stress Responses. Front. Plant Sci. 2019, 10, 800. [Google Scholar] [CrossRef]
- Ogura, T.; Goeschl, C.; Filiault, D.; Mirea, M.; Slovak, R.; Wolhrab, B.; Satbhai, S.B.; Busch, W. Root System Depth in Arabidopsis Is Shaped by EXOCYST70A3 via the Dynamic Modulation of Auxin Transport. Cell 2019, 178, 400–412.e16. [Google Scholar] [CrossRef]
- Fàbregas, N.; Lozano-Elena, F.; Blasco-Escámez, D.; Tohge, T.; Martínez-Andújar, C.; Albacete, A.; Osorio, S.; Bustamante, M.; Riechmann, J.L.; Nomura, T.; et al. Overexpression of the vascular brassinosteroid receptor BRL3 confers drought resistance without penalizing plant growth. Nat. Commun. 2018, 9, 4680. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.J.; Andrews, J.; Mulholland, B.J.; McKee, J.M.; Hilton, H.W.; Horridge, J.S.; Taylor, I.B. Overproduction of abscisic acid in tomato increases transpiration efficiency and root hydraulic con-ductivity and influences leaf expansion. Plant Physiol. 2007, 143, 1905–1917. [Google Scholar] [CrossRef] [PubMed]
- Meena, M.; Divyanshu, K.; Kumar, S.; Swapnil, P.; Zehra, A.; Shukla, V.; Yadav, M.; Upadhyay, R.S. Regulation of L-proline biosynthesis, signal transduction, transport, accumulation and its vital role in plants during variable environmental conditions. Heliyon 2019, 5, e02952. [Google Scholar] [CrossRef] [PubMed]
- Samtani, H.; Sharma, A.; Khurana, P. Overexpression of HVA1 Enhances Drought and Heat Stress Tolerance in Triticum aestivum Doubled Haploid Plants. Cells 2022, 11, 912. [Google Scholar] [CrossRef] [PubMed]
- Molinari, M.D.C.; Fuganti-Pagliarini, R.; Marin, S.R.R.; Ferreira, L.C.; Barbosa, D.D.A.; Marcolino-Gomes, J.; Nepomuceno, A.L. Overexpression of AtNCED3 gene improved drought tolerance in soybean in greenhouse and field conditions. Genet Mol. Biol. 2020, 43, e20190292. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Zhang, W.; Qin, M.; Li, S.; Qiao, M.; Liu, Z.; Xiang, F. Drought Tolerance Conferred in Soybean (Glycine max. L) by GmMYB84, a Novel R2R3-MYB Transcription Factor. Plant Cell Physiol. 2017, 58, 1764–1776. [Google Scholar] [CrossRef]
- Qiao, M.; Hong, C.; Jiao, Y.; Hou, S.; Gao, H. Impacts of Drought on Photosynthesis in Major Food Crops and the Related Mechanisms of Plant Responses to Drought. Plants 2024, 13, 1808. [Google Scholar] [CrossRef]
- Wahab, A.; Abdi, G.; Saleem, M.H.; Ali, B.; Ullah, S.; Shah, W.; Mumtaz, S.; Yasin, G.; Muresan, C.C.; Marc, R.A. Plants’ Physio-Biochemical and Phyto-Hormonal Responses to Alleviate the Adverse Effects of Drought Stress: A Comprehensive Review. Plants 2022, 11, 1620. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; He, X.; Zhao, Y.; Lu, S.; Guo, Z. Constitutive expression of a group 3 LEA protein from Medicago falcata (MfLEA3) increases cold and drought tolerance in transgenic tobacco. Plant Cell Rep. 2020, 39, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Fiorillo, A.; Mattei, M.; Aducci, P.; Visconti, S.; Camoni, L. The Salt Tolerance Related Protein (STRP) Mediates Cold Stress Responses and Abscisic Acid Signalling in Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 1251. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Huang, G.; Zhang, Q.; Wang, X.; Li, C.; Tao, Y.; Zhang, S.; Lai, J.; Yang, C.; Wang, Y. The LEA protein, ABR, is regulated by ABI5 and involved in dark-induced leaf senescence in Arabidopsis thaliana. Plant Sci. 2016, 247, 93–103. [Google Scholar] [CrossRef]
- He, L.; Bian, J.; Xu, J.; Yang, K. Novel Maize NAC Transcriptional Repressor ZmNAC071 Confers Enhanced Sensitivity to ABA and Osmotic Stress by Downregulating Stress-Responsive Genes in Transgenic Arabidopsis. J. Agric. Food Chem. 2019, 67, 8905–8918. [Google Scholar] [CrossRef]
- Xiaoqin, L.; Yue, L.; Shangwei, Z. Interplay between Light and Plant Hormones in the Control of Arabidopsis Seedling Chlo-rophyll Biosynthesis. Front. Plant Sci. 2017, 8, 1433–1454. [Google Scholar]
- Li, W.; Zheng, X.; Cheng, R.; Zhong, C.; Zhao, J.; Liu, T.H.; Liu, S. Soybean ZINC FINGER PROTEIN03 targets two SUPEROXIDE DISMUTASE1s and confers resistance to Phytophthora sojae. Plant Physiol. 2023, 192, 633–647. [Google Scholar] [CrossRef]
- Sharma, A.; Wang, J.; Xu, D.; Tao, S.; Chong, S.; Yan, D.; Li, Z.; Yuan, H.; Zheng, B. Melatonin regulates the functional components of photosynthesis, antioxidant system, gene expression, and metabolic pathways to induce drought resistance in grafted Carya cathayensis plants. Sci. Total Environ. 2020, 713, 136675. [Google Scholar] [CrossRef]
- Yao, T.; Ding, C.; Che, Y.; Zhang, Z.; Cui, C.; Ji, G.; Song, J.; Zhang, H.; Ao, H.; Zhang, H. Heterologous expression of Zygophyllum xanthoxylon zinc finger protein gene (ZxZF) enhances the tolerance of poplar photosynthetic function to drought stress. Plant Physiol. Biochem. 2023, 199, 107748. [Google Scholar] [CrossRef]







| Photosynthetic Indexes | Line | Normal | 7 Days of Drought |
|---|---|---|---|
| Net photosynthetic rate (Pn) μmol/(m2·s) | KO1 | 6.564 ± 0.48 b | 3.51 ± 0.51 c |
| KO2 | 7.481 ± 0.42 b | 3.847 ± 0.43 b | |
| WT | 6.860 ± 0.38 b | 3.715 ± 0.47 b | |
| OE1 | 8.981 ± 0.44 a | 6.494 ± 0.50 a | |
| OE2 | 8.893 ± 0.41 a | 6.679 ± 0.44 a | |
| Rate of transpiration (Tr) mol/(m2·s) | KO1 | 2.458 ± 0.01 c | 1.886 ± 0.64 b |
| KO2 | 2.679 ± 0.05 b | 1.652 ± 0.66 c | |
| WT | 2.697 ± 0.07 b | 1.942 ± 0.08 b | |
| OE1 | 2.971 ± 0.11 a | 2.680 ± 0.11 a | |
| OE2 | 3.028 ± 0.08 a | 2.745 ± 0.15 a | |
| Stomatal conductance (Gs) mmol/(m2·s) | KO1 | 1.208 ± 0.06 c | 0.203 ± 0.05 c |
| KO2 | 0.954 ± 0.03 d | 0.198 ± 0.08 c | |
| WT | 1.348 ± 0.07 c | 0.234 ± 0.05 c | |
| OE1 | 1.697 ± 0.04 a | 0.491 ± 0.06 a | |
| OE2 | 1.801 ± 0.04 a | 0.572 ± 0.07 a | |
| Intercellular CO2 concentration (Ci) μmol/mol | KO1 | 370.294 ± 16.28 b | 272.588 ± 15.85 c |
| KO2 | 367.912 ± 15.46 b | 264.568 ± 16.11 bc | |
| WT | 378.128 ± 13.85 b | 248.716 ± 14.67 bc | |
| OE1 | 394.295 ± 18.78 a | 358.191 ± 12.49 a | |
| OE2 | 400.344 ± 13.66 a | 376.564 ± 15.72 a | |
| Instantaneous water use efficiency (WUEi) Pn/Tr | KO1 | 2.668 ± 0.03 c | 1.861 ± 0.02 c |
| KO2 | 2.791 ± 0.05 b | 2.328 ± 0.03 b | |
| WT | 2.543 ± 0.04 c | 1.913 ± 0.04 c | |
| OE1 | 3.023 ± 0.05 a | 2.423 ± 0.04 a | |
| OE2 | 2.937 ± 0.02 a | 2.433 ± 0.05 a |
| Line | Plant Height (cm) | Number of Main Stem Segments | Effective Branch Number | Number of Pods per Plant | Number of Main Stem Pods | Yield per Plant (g) | Hundred-Grain Weight (g) |
|---|---|---|---|---|---|---|---|
| KO1 | 99.38 ± 6.22 a | 20.11 ± 1.9 b | 2.11 ± 1.93 a | 107.11 ± 34.84 bc | 78.33 ± 15.07 b | 34.76 ± 2.38 b | 17.57 ± 1.59 a |
| KO2 | 97.36 ± 5.25 a | 19.81 ± 1.16 ab | 3.25 ± 1.53 a | 115.42 ± 41.81 c | 81.21 ± 10.36 b | 36.56 ± 2.95 b | 17.79 ± 0.78 a |
| WT | 100.15 ± 3.87 a | 19.48 ± 1.03 ab | 2.14 ± 0.51 a | 81.67 ± 13.53 d | 54.16 ± 7.96 c | 28.19 ± 1.76 c | 17.76 ± 1.51 a |
| OE1 | 102.36 ± 5.52 a | 20.86 ± 1.48 a | 5.11 ± 1.07 a | 132.44 ± 23.98 a | 72.5 ± 7.07 a | 42.58 ± 3.17 a | 17.79 ± 1.85 a |
| OE2 | 106.19 ± 5.05 a | 21.22 ± 1.28 a | 5.54 ± 1.72 a | 135.5 ± 47.92 a | 82.11 ± 8.48 a | 38.78 ± 2.85 a | 18.66 ± 1.84 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Sun, Y.; Wang, R.; Li, X.; Li, Y.; Wang, T.; Guo, Z.; Li, Y.; Qiu, W.; Guan, S.; et al. Overexpression of the GmPM35 Gene Significantly Enhances Drought Tolerance in Transgenic Arabidopsis and Soybean. Agronomy 2025, 15, 192. https://doi.org/10.3390/agronomy15010192
Wang X, Sun Y, Wang R, Li X, Li Y, Wang T, Guo Z, Li Y, Qiu W, Guan S, et al. Overexpression of the GmPM35 Gene Significantly Enhances Drought Tolerance in Transgenic Arabidopsis and Soybean. Agronomy. 2025; 15(1):192. https://doi.org/10.3390/agronomy15010192
Chicago/Turabian StyleWang, Xinyu, Yao Sun, Rui Wang, Xinyang Li, Yongyi Li, Tianyu Wang, Zhaohao Guo, Yan Li, Wenxi Qiu, Shuyan Guan, and et al. 2025. "Overexpression of the GmPM35 Gene Significantly Enhances Drought Tolerance in Transgenic Arabidopsis and Soybean" Agronomy 15, no. 1: 192. https://doi.org/10.3390/agronomy15010192
APA StyleWang, X., Sun, Y., Wang, R., Li, X., Li, Y., Wang, T., Guo, Z., Li, Y., Qiu, W., Guan, S., Zhang, Q., Wang, P., Li, M., Liu, S., & Fan, X. (2025). Overexpression of the GmPM35 Gene Significantly Enhances Drought Tolerance in Transgenic Arabidopsis and Soybean. Agronomy, 15(1), 192. https://doi.org/10.3390/agronomy15010192






