Genome-Wide Identification of the Kinesin Gene Family in Soybean and Its Response to Salt Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of GmKIN Family Members
2.2. Motif, Gene Structure, and Phylogenetic Analysis
2.3. Promoter Cis-Element Analysis
2.4. Chromosomal Localization
2.5. Gene Synteny Analysis
2.6. Transcriptomic Gene Expression Analysis
2.7. RNA Extraction and Gene Expression Analysis
2.8. Yeast Salinity Tolerance Assay Using INVSc1 Strain
2.9. Genetic Transformation and Salinity Tolerance Evaluation of Soybean Hairy Roots
2.10. Data Analysis
3. Results
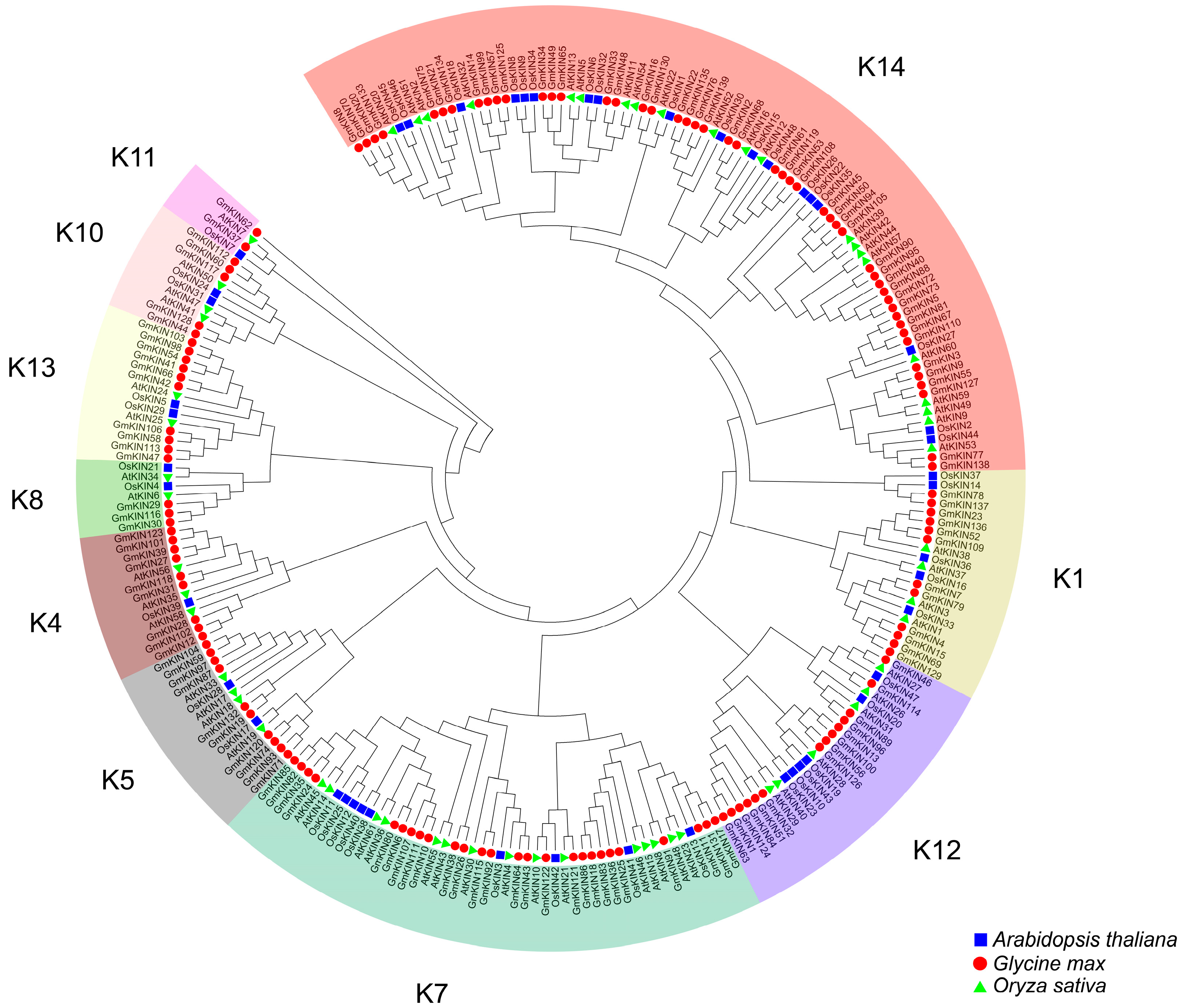
3.1. Identification of GmKIN Family Members and Phylogenetic Tree Analysis
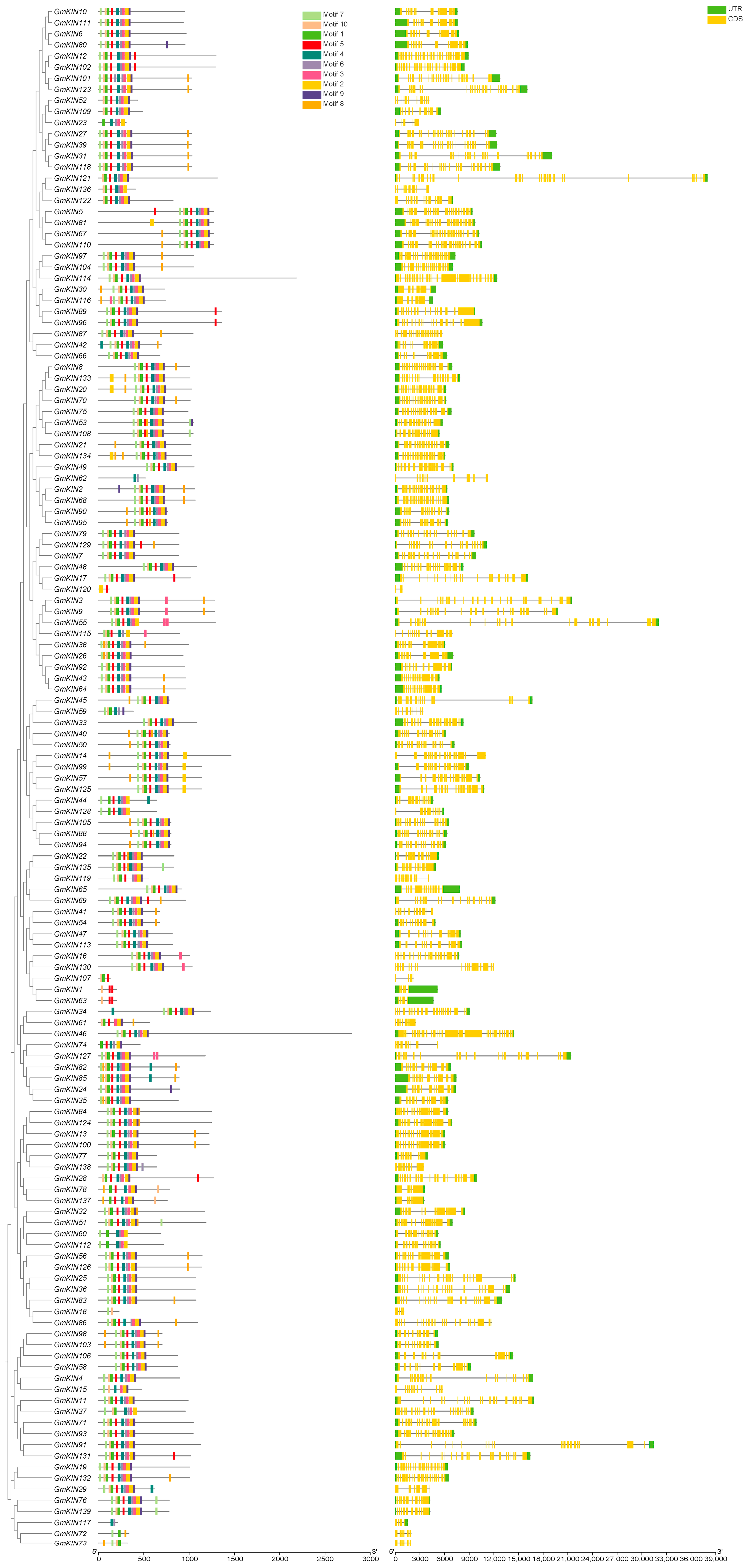
3.2. Analysis of Conserved Motifs and Gene Structure of GmKIN
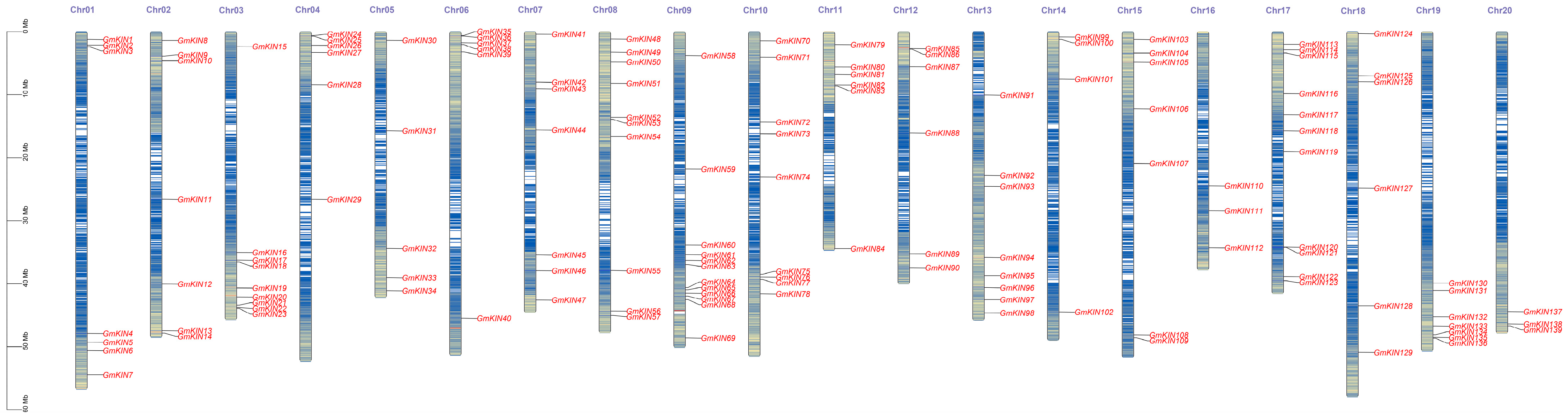
3.3. Chromosomal Localization of GmKINs
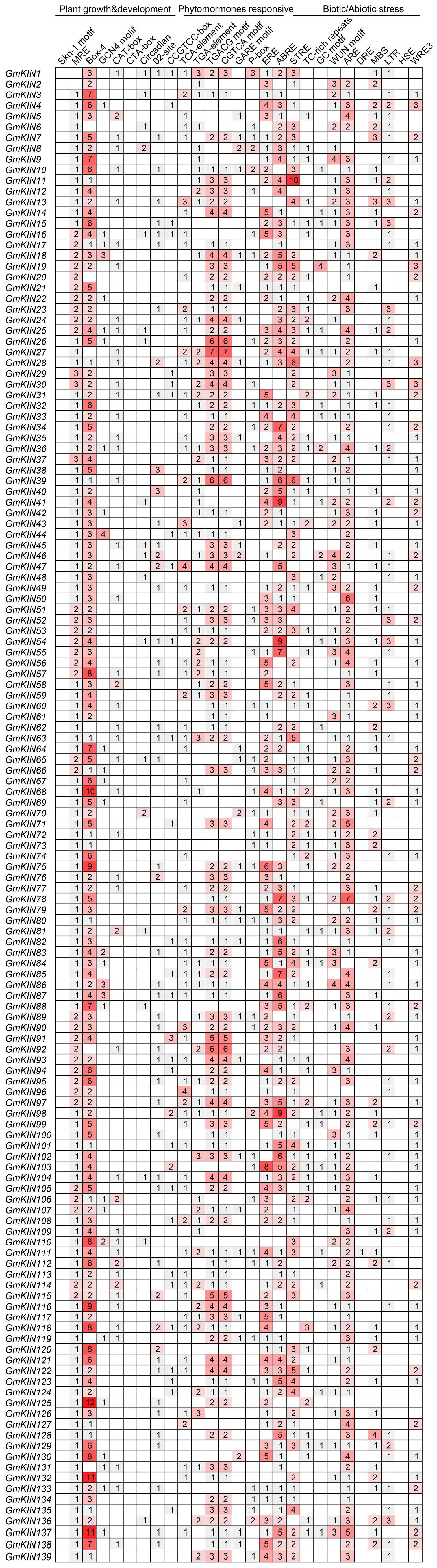
3.4. Cis-Acting Element Analysis of Promoters
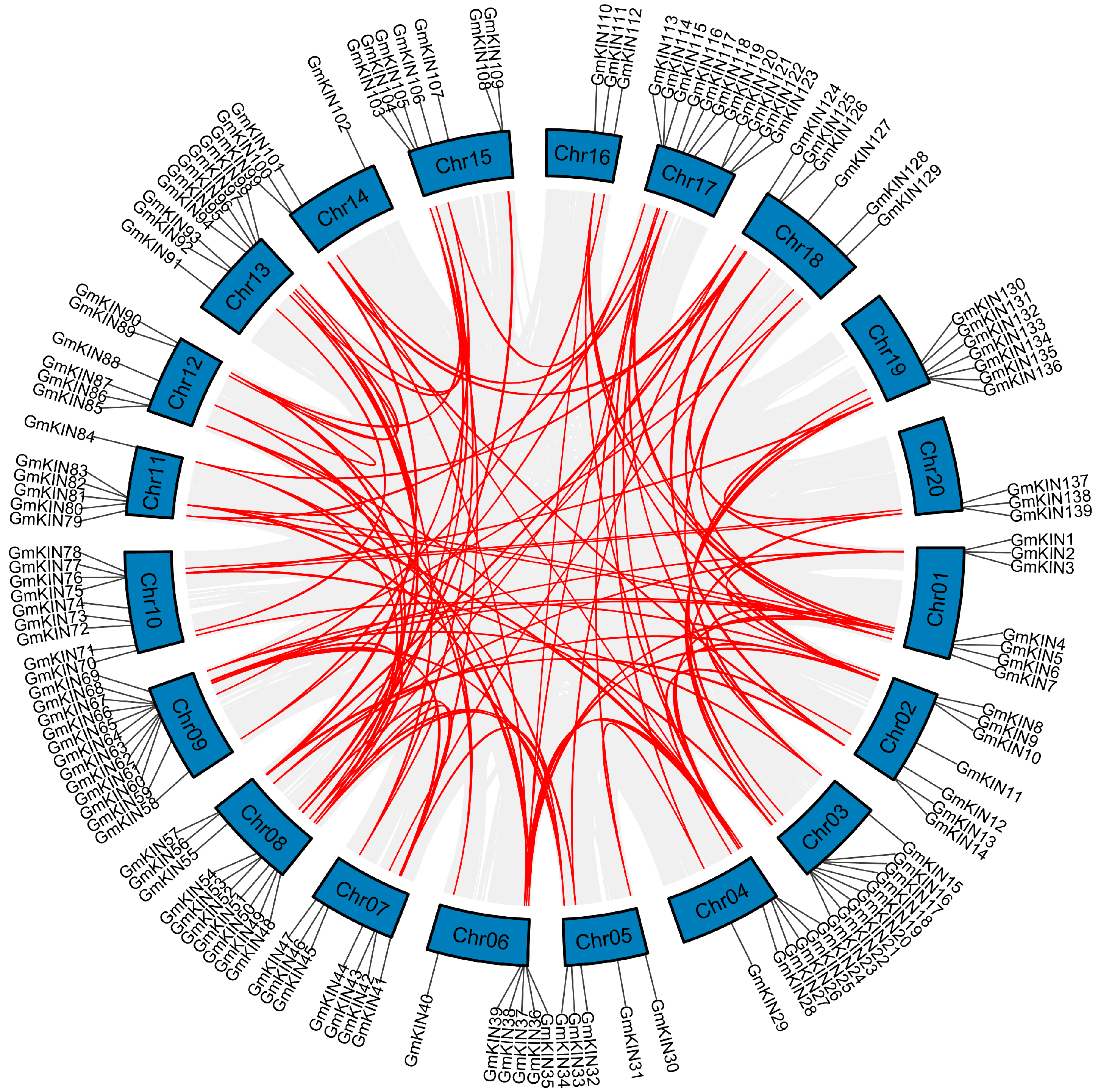
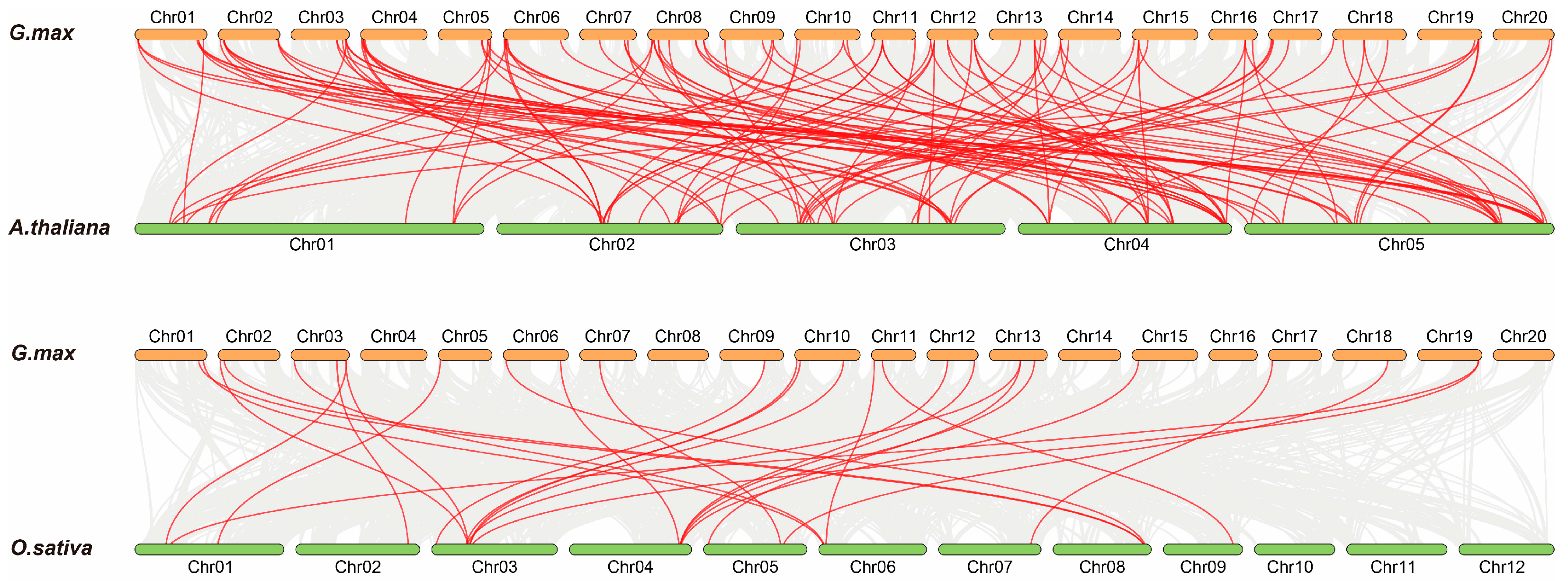
3.5. Gene Duplication and Synteny Analysis
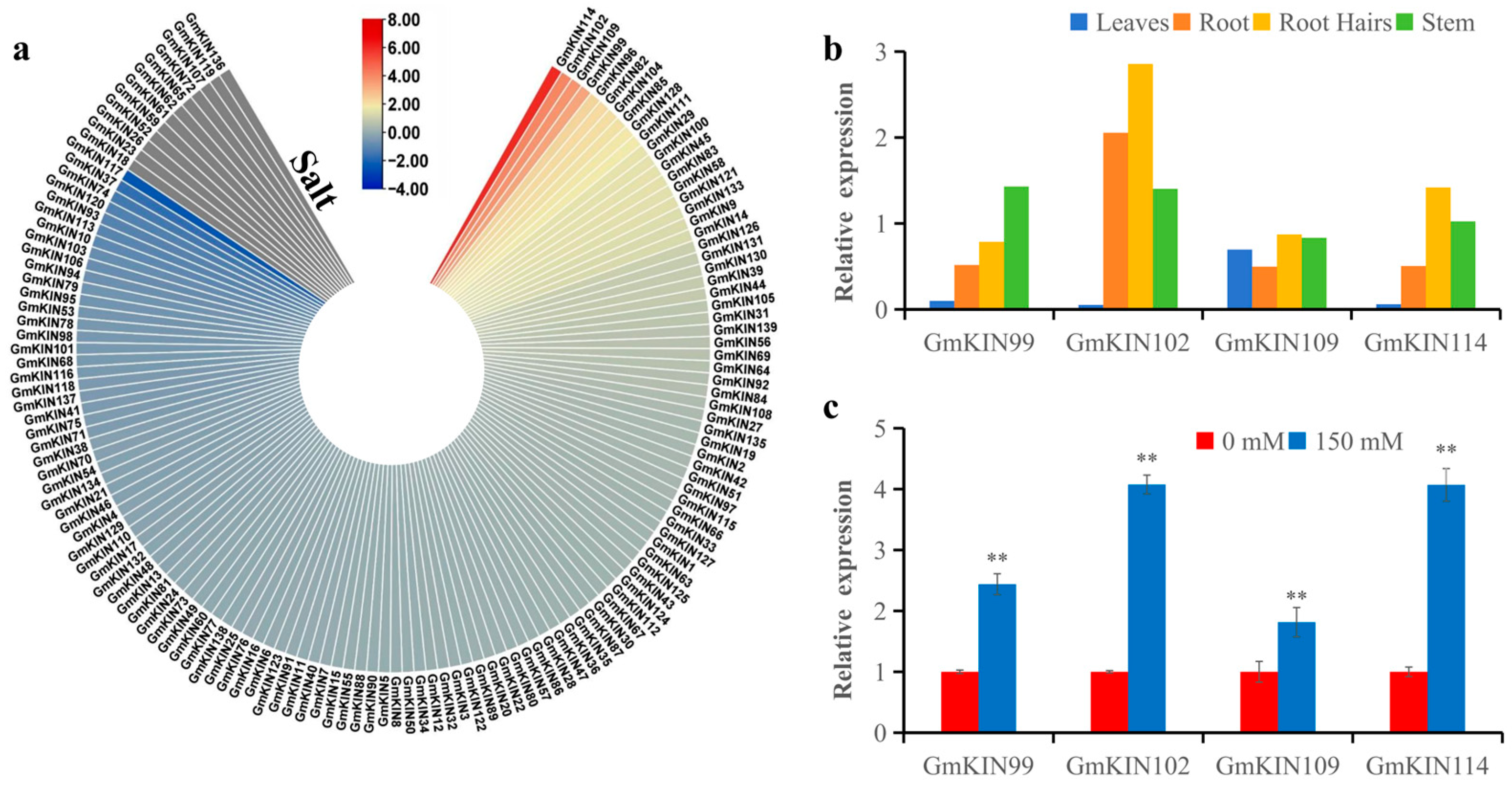
3.6. Expression Profiles of GmKIN Members Under NaCl Stress and in Different Plant Tissues
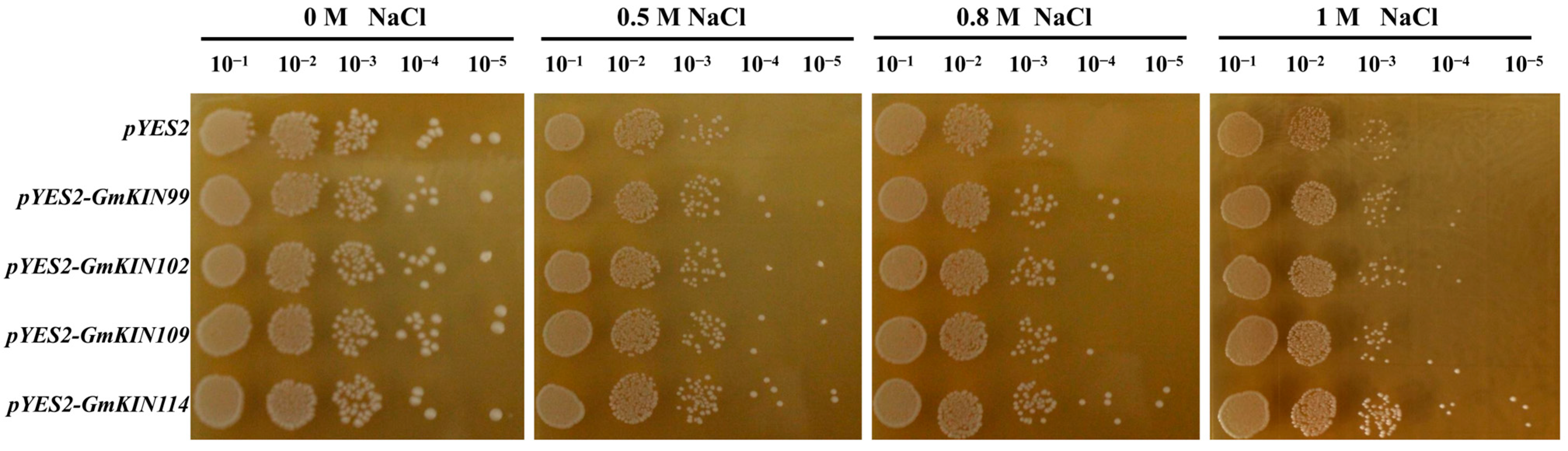
3.7. Effect of Overexpressing GmKINs on Yeast Salinity Tolerance
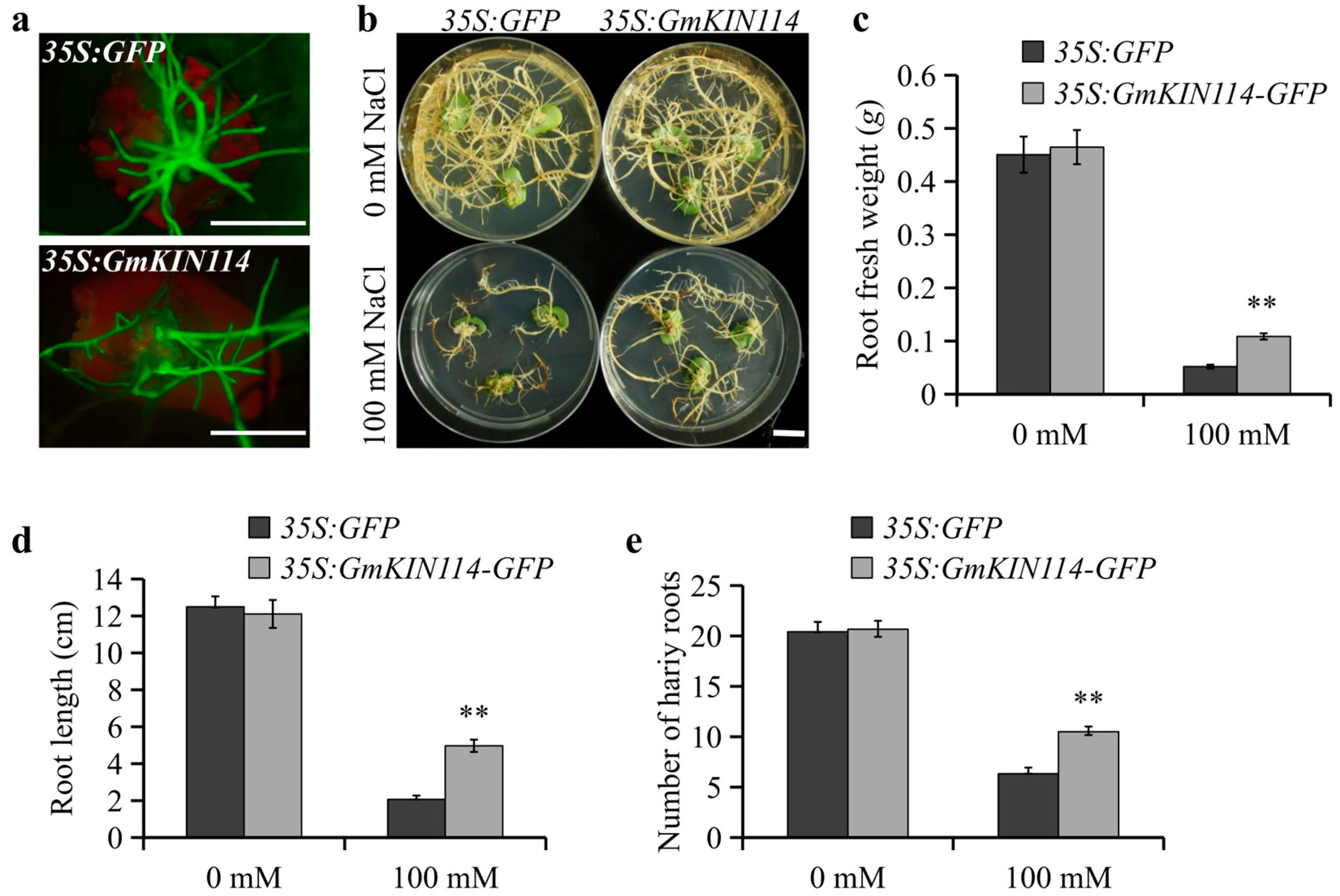
3.8. Effect of Overexpressing GmKIN114 on Salt Tolerance in Soybean Hairy Roots
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| At | Arabidopsis thaliana |
| Gm | Glycine max |
| GUS | Glucuronidase |
| KHC | Kinesin heavy chain |
| KIN | Kinesin |
| KLC | Kinesin light chain |
| KP1 | Kinesin-like protein 1 |
| MEME | Multiple em for motif elicitation |
| MS | Murashige and skoog |
| MW | Molecular weight |
| Os | Oryza sativa |
| pI | Isoelectric point |
| qRT-PCR | Quantitative real-time PCR |
| UTRs | Untranslated regions |
| VDAC3 | Voltage-dependent anion channel 3 |
References
- Sharma, M.; Tisarum, R.; Kohli, R.K.; Batish, D.R.; Cha-Um, S.; Singh, H.P. Inroads into saline-alkaline stress response in plants: Unravelling morphological, physiological, biochemical, and molecular mechanisms. Planta 2024, 259, 130. [Google Scholar] [CrossRef] [PubMed]
- van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Abdalla, M.; Ahmed, M.A. Coupled effects of soil drying and salinity on soil-plant hydraulics. Plant Physiol. 2022, 190, 1228–1241. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yu, F. A Gγ protein regulates alkaline sensitivity in crops. Science 2023, 379, eade8416. [Google Scholar] [CrossRef] [PubMed]
- Molla, K.A. Molecular switch to regulate salt tolerance in rice. Plant Cell 2023, 35, 3396–3397. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Soni, K.; Frew, R.; Kebede, B. A review of conventional and rapid analytical techniques coupled with multivariate analysis for origin traceability of soybean. Crit. Rev. Food Sci. Nutr. 2024, 64, 6616–6635. [Google Scholar] [CrossRef]
- Vianna, G.R.; Cunha, N.B.; Rech, E.L. Soybean seed protein storage vacuoles for expression of recombinant molecules. Curr. Opin. Plant Biol. 2023, 71, 102331. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, M.; Feng, F.; Tian, Z. Toward a “Green Revolution” for soybean. Mol. Plant 2020, 13, 688–697. [Google Scholar] [CrossRef] [PubMed]
- Phang, T.H.; Shao, G.; Lam, H.M. Salt tolerance in soybean. J. Integr. Plant Biol. 2008, 50, 1196–1212. [Google Scholar] [CrossRef] [PubMed]
- Jin, T.; Sun, Y.; Shan, Z.; He, J.; Wang, N.; Gai, J.; Li, Y. Natural variation in the promoter of GsERD15B affects salt tolerance in soybean. Plant Biotechnol. J. 2021, 19, 1155–1169. [Google Scholar] [CrossRef]
- Ali, A.; Petrov, V.; Yun, D.J.; Gechev, T. Revisiting plant salt tolerance: Novel components of the SOS pathway. Trends Plant Sci. 2023, 28, 1060–1069. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Zhu, C. Genome-wide association studies identify OsWRKY53 as a key regulator of salt tolerance in rice. Nat. Commun. 2023, 14, 3550. [Google Scholar] [CrossRef] [PubMed]
- Sukumaran, S.; Lethin, J.; Liu, X.; Pelc, J.; Zeng, P.; Hassan, S.; Aronsson, H. Genome-wide analysis of MYB transcription factors in the wheat genome and their roles in salt stress response. Cells 2023, 12, 1431. [Google Scholar] [CrossRef]
- Zheng, L.; Hu, Y.; Yang, T.; Wang, Z.; Wang, D.; Jia, L.; Xie, Y. A root cap-localized NAC transcription factor controls root halotropic response to salt stress in Arabidopsis. Nat. Commun. 2024, 15, 2061. [Google Scholar] [CrossRef] [PubMed]
- Bi, C.; Yu, Y.; Dong, C.; Yang, Y.; Zhai, Y.; Du, F.; Xia, C.; Ni, Z.; Kong, X. The bZIP transcription factor TabZIP15 improves salt stress tolerance in wheat. Plant Biotechnol. J. 2021, 19, 209–211. [Google Scholar] [CrossRef] [PubMed]
- Ali, I.; Yang, W.C. The functions of kinesin and kinesin-related proteins in eukaryotes. Cell Adhes. Migr. 2020, 14, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Nejabat, M.; Hadizadeh, F.; Sahebkar, A. The application of kinesin inhibitors in medical issues. Curr. Rev. Clin. Exp. Pharmacol. 2024, 19, 370–378. [Google Scholar] [CrossRef]
- Angerani, S.; Lindberg, E.; Klena, N. Kinesin-1 activity recorded in living cells with a precipitating dye. Nat. Commun. 2021, 12, 1463. [Google Scholar] [CrossRef]
- Yoshida, M.W.; Hakozaki, M.; Goshima, G. Armadillo repeat-containing kinesin represents the versatile plus-end-directed transporter in Physcomitrella. Nat. Plants 2023, 9, 733–748. [Google Scholar] [CrossRef]
- Sonar, P.; Youyen, W.; Cleetus, A.; Wisanpitayakorn, P.; Mousavi, S.I.; Stepp, W.L.; Hancock, W.O.; Tüzel, E.; Ökten, Z. Kinesin-2 from C. reinhardtii is an atypically fast and auto-inhibited motor that is activated by heterotrimerization for intraflagellar transport. Curr. Biol. 2020, 30, 1160–1166.e1165. [Google Scholar] [CrossRef] [PubMed]
- Gicking, A.M.; Swentowsky, K.W.; Dawe, R.K. Functional diversification of the kinesin-14 family in land plants. FEBS Lett. 2018, 592, 1918–1928. [Google Scholar] [CrossRef] [PubMed]
- Ogren, A.; Parmar, S.; Mukherjee, S.; Gonzalez, S.J. Kinesin-14 motors participate in a force balance at microtubule plus-ends to regulate dynamic instability. Proc. Natl. Acad. Sci. USA 2022, 119, e2108046119. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Ren, J. The architecture of kinesin-3 KLP-6 reveals a multilevel-lockdown mechanism for autoinhibition. Nat. Commun. 2022, 13, 4281. [Google Scholar] [CrossRef] [PubMed]
- Nithianantham, S.; Iwanski, M.K.; Gaska, I.; Pandey, H.; Bodrug, T.; Inagaki, S.; Major, J.; Brouhard, G.J.; Gheber, L.; Rosenfeld, S.S.; et al. The kinesin-5 tail and bipolar minifilament domains are the origin of its microtubule crosslinking and sliding activity. Mol. Biol. Cell 2023, 34, ar111. [Google Scholar] [CrossRef]
- Cross, J.A.; Woolfson, D.N.; Dodding, M.P. Kinesin-1 captures RNA cargo in its adaptable coils. Genes Dev. 2021, 35, 937–939. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Dong, G.; Chen, F.; Xiong, E.; Liu, H.; Jiang, Y.; Xiong, G.; Ruan, B.; Qian, Q.; Zeng, D.; et al. The kinesin-13 protein BR HYPERSENSITIVE 1 is a negative brassinosteroid signaling component regulating rice growth and development. Theor. Appl. Genet. 2022, 135, 1751–1766. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Yuan, Z.; Xu, H.; Pan, L.; Chen, J.; Gatera, A.; Uzair, M. Genome-wide identification and expression analysis of kinesin family in barley (Hordeum vulgare). Genes 2022, 13, 2376. [Google Scholar] [CrossRef]
- Li, J.; Yu, D.; Qanmber, G.; Lu, L.; Wang, L.; Zheng, L.; Liu, Z.; Wu, H.; Liu, X.; Chen, Q.; et al. GhKLCR1, a kinesin light chain-related gene, induces drought-stress sensitivity in Arabidopsis. Sci. China Life Sci. 2019, 62, 63–75. [Google Scholar] [CrossRef]
- Xing, X.; Liu, M.; Jiang, F.; Zhou, R.; Bai, Y.; Wei, H.; Zhang, D.; Wei, J.; Wu, Z. Abscisic acid induces the expression of AsKIN during the recovery period of garlic cryopreservation. Plant Cell Rep. 2022, 41, 1955–1973. [Google Scholar] [CrossRef]
- Chen, G.; Xuan, W. OsTUB1 confers salt insensitivity by interacting with Kinesin13A to stabilize microtubules and ion transporters in rice. New Phytol. 2022, 235, 1836–1852. [Google Scholar] [CrossRef]
- Chen, N.; Pan, L.; Yang, Z.; Su, M.; Xu, J.; Jiang, X.; Yin, X.; Wang, T.; Wan, F.; Chi, X. A MYB-related transcription factor from peanut, AhMYB30, improves freezing and salt stress tolerance in transgenic Arabidopsis through both DREB/CBF and ABA-signaling pathways. Front. Plant Sci. 2023, 14, 1136626. [Google Scholar] [CrossRef] [PubMed]
- Nadeem, M.; Chen, A.; Hong, H.; Li, D.; Li, J.; Zhao, D.; Wang, W.; Wang, X.; Qiu, L. GmMs1 encodes a kinesin-like protein essential for male fertility in soybean (Glycine max L.). J. Integr. Plant Biol. 2021, 63, 1054–1064. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Sun, X.; Yang, X.; Li, Q.; Lin, C.; Xu, J.; Gong, W.; Wang, Y.; Liu, L.; Zhao, L.; et al. MS1 is essential for male fertility by regulating the microsporocyte cell plate expansion in soybean. Sci. China Life Sci. 2021, 64, 1533–1545. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Xu, J.; Yu, K.; Wu, J.; Xu, H.; Wang, S.; Wen, T. Genome-wide identification of the key Kinesin genes during fiber and boll development in upland cotton (Gossypium hirsutum L.). Mol. Genet. Genom. 2024, 299, 2. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Li, S.; Chen, B.; Jian, C.; Mei, F.; Zhang, Y.; Li, F.; Chen, N.; Li, T.; Du, L.; et al. Variation in cis-regulation of a NAC transcription factor contributes to drought tolerance in wheat. Mol. Plant 2022, 15, 276–292. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Jia, T.; Hou, J.; Iqbal, M.Z.; Zhang, Y.; Cheng, B.; Feng, H.; Li, Z.; Liu, L.; Zhou, J.; Feng, G.; et al. Overexpression of the white clover TrSAMDC1 gene enhanced salt and drought resistance in Arabidopsis thaliana. Plant Physiol. Biochem. 2021, 165, 147–160. [Google Scholar] [CrossRef]
- Gietz, R.D.; Woods, R.A. Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods Mol. Biol. 2006, 313, 107–120. [Google Scholar] [PubMed]
- Jin, T.; Sun, Y.; Zhao, R.; Shan, Z.; Gai, J.; Li, Y. Overexpression of peroxidase gene GsPRX9 confers salt tolerance in soybean. Int. J. Mol. Sci. 2019, 20, 3745. [Google Scholar] [CrossRef] [PubMed]
- Kereszt, A.; Li, D.; Indrasumunar, A.; Nguyen, C.D.; Nontachaiyapoom, S.; Kinkema, M.; Gresshoff, P.M. Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2007, 2, 948–952. [Google Scholar] [CrossRef]
- Qi, X.; Li, M.W.; Xie, M.; Liu, X.; Ni, M.; Shao, G.; Song, C.; Kay-Yuen Yim, A.; Tao, Y.; Wong, F.L.; et al. Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat. Commun. 2014, 5, 4340. [Google Scholar] [CrossRef]
- Zhu, C.; Dixit, R. Functions of the Arabidopsis kinesin superfamily of microtubule-based motor proteins. Protoplasma 2012, 249, 887–899. [Google Scholar] [CrossRef] [PubMed]
- Vale, R.D.; Fletterick, R.J. The design plan of kinesin motors. Annu. Rev. Cell Dev. Biol. 1997, 13, 745–777. [Google Scholar] [CrossRef]
- Cross, R.A. Review: Mechanochemistry of the kinesin-1 ATPase. Biopolymers 2016, 105, 476–482. [Google Scholar] [CrossRef] [PubMed]
- Fajardo, D.; Saint Jean, R.; Lyons, P.J. Acquisition of new function through gene duplication in the metallocarboxypeptidase family. Sci. Rep. 2023, 13, 2512. [Google Scholar] [CrossRef]
- Wang, D.R.; Yang, K.; Wang, X.; You, C.X. A C2H2-type zinc finger transcription factor, MdZAT17, acts as a positive regulator in response to salt stress. J. Plant Physiol. 2022, 275, 153737. [Google Scholar] [CrossRef]
- Kavas, M.; Gökdemir, G.; Seçgin, Z.; Bakhsh, A. Ectopic expression of common bean ERF transcription factor PvERF35 promotes salt stress tolerance in tobacco. Plant Biol. 2020, 22, 1102–1112. [Google Scholar] [CrossRef]
- Shiraku, M.L.; Magwanga, R.O.; Cai, X.; Kirungu, J.N.; Xu, Y.; Mehari, T.G.; Hou, Y.; Wang, Y.; Agong, S.G.; Peng, R.; et al. Functional characterization of GhACX3 gene reveals its significant role in enhancing drought and salt stress tolerance in cotton. Front. Plant Sci. 2021, 12, 658755. [Google Scholar] [CrossRef]
- Jin, D.; Li, S.; Li, Z.; Yang, L.; Han, X.; Hu, Y.; Jiang, Y. Arabidopsis ABRE-binding factors modulate salinity-induced inhibition of root hair growth by interacting with and suppressing RHD6. Plant Sci. 2023, 332, 111728. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Jiang, J.; Qian, Q.; Xu, Y.; Zhang, C.; Xiao, J.; Du, C.; Luo, W.; Zou, G.; Chen, M.; et al. Mutation of rice BC12/GDD1, which encodes a kinesin-like protein that binds to a GA biosynthesis gene promoter, leads to dwarfism with impaired cell elongation. Plant Cell 2011, 23, 628–640. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.Y.; Chen, Z.W.; Xu, T.; Qu, Z.; Pan, X.D.; Qin, X.H.; Ren, D.T.; Liu, G.Q. Arabidopsis kinesin KP1 specifically interacts with VDAC3, a mitochondrial protein, and regulates respiration during seed germination at low temperature. Plant Cell 2011, 23, 1093–1106. [Google Scholar] [CrossRef]
- Hwarari, D.; Guan, Y.; Ahmad, B.; Movahedi, A. ICE-CBF-COR signaling cascade and its regulation in plants responding to cold stress. Int. J. Mol. Sci. 2022, 23, 1549. [Google Scholar] [CrossRef]
- Fang, C.; Kong, F. Soybean. Curr. Biol. 2022, 32, R902–R904. [Google Scholar] [CrossRef]
- Antos-Krzeminska, N.; Kicinska, A. Acanthamoeba castellanii Uncoupling Protein: A Complete Sequence, Activity, and Role in Response to Oxidative Stress. Int. J. Mol. Sci. 2023, 24, 12501. [Google Scholar] [CrossRef]
- Song, J.; Tóth, K.; Montes-Luz, B.; Stacey, G. Soybean hairy root transformation: A rapid and highly efficient method. Curr. Protoc. 2021, 1, e195. [Google Scholar] [CrossRef] [PubMed]
- Tóth, K.; Batek, J.; Stacey, G. Generation of soybean (Glycine max) transient transgenic roots. Curr. Protoc. Plant Biol. 2016, 1, 1–13. [Google Scholar] [CrossRef]
- Noman, M.; Jameel, A.; Qiang, W.D.; Ahmad, N.; Liu, W.C.; Wang, F.W.; Li, H.Y. Overexpression of GmCAMTA12 enhanced drought tolerance in Arabidopsis and soybean. Int. J. Mol. Sci. 2019, 20, 4849. [Google Scholar] [CrossRef]
- Xu, C.; Shan, J. CONSTANS-LIKE 1a positively regulates salt and drought tolerance in soybean. Plant Physiol. 2023, 191, 2427–2446. [Google Scholar] [CrossRef] [PubMed]
- Quintero, F.J.; Blatt, M.R.; Pardo, J.M. Functional conservation between yeast and plant endosomal Na+/H+ antiporters. FEBS Lett. 2000, 471, 224–228. [Google Scholar] [CrossRef]
- Karlova, R.; Boer, D. Root plasticity under abiotic stress. Plant Physiol. 2021, 187, 1057–1070. [Google Scholar] [CrossRef]
- Müller, S.; Livanos, P. Plant kinesin-12: Localization heterogeneity and functional implications. Int. J. Mol. Sci. 2019, 20, 4213. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.J.; Shan, M.M.; Wan, X.; Liu, J.C.; Zhang, K.H.; Ju, J.Q.; Xing, C.H.; Sun, S.C. Kinesin KIF15 regulates tubulin acetylation and spindle assembly checkpoint in mouse oocyte meiosis. Cell Mol. Life Sci. 2022, 79, 422. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.X.; Ding, M.Q.; Yi, Y.; Zou, Y.J.; Liao, B.Y.; Sun, S.C. Insufficient KIF15 during porcine oocyte ageing induces HDAC6-based microtubule instability. Theriogenology 2024, 226, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Salazar, B.M.; Ohi, R. Antiparallel microtubule bundling supports KIF15-driven mitotic spindle assembly. Mol. Biol. Cell 2024, 35, ar84. [Google Scholar] [CrossRef]
- Wang, J.; Tu, Q.; Zhang, S.; He, X.; Ma, C.; Qian, X.; Wu, R.; Shi, X.; Yang, Z.; Liu, Y.; et al. Kif15 deficiency contributes to depression-like behavior in mice. Metab. Brain Dis. 2023, 38, 2369–2381. [Google Scholar] [CrossRef] [PubMed]
- Sturgill, E.G.; Norris, S.R.; Guo, Y.; Ohi, R. Kinesin-5 inhibitor resistance is driven by kinesin-12. J. Cell Biol. 2016, 213, 213–227. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, T.; Zhang, K.; Zhang, X.; Wu, C.; Long, W. Genome-Wide Identification of the Kinesin Gene Family in Soybean and Its Response to Salt Stress. Agronomy 2025, 15, 275. https://doi.org/10.3390/agronomy15020275
Jin T, Zhang K, Zhang X, Wu C, Long W. Genome-Wide Identification of the Kinesin Gene Family in Soybean and Its Response to Salt Stress. Agronomy. 2025; 15(2):275. https://doi.org/10.3390/agronomy15020275
Chicago/Turabian StyleJin, Ting, Kai Zhang, Xiujie Zhang, Chunhua Wu, and Weihua Long. 2025. "Genome-Wide Identification of the Kinesin Gene Family in Soybean and Its Response to Salt Stress" Agronomy 15, no. 2: 275. https://doi.org/10.3390/agronomy15020275
APA StyleJin, T., Zhang, K., Zhang, X., Wu, C., & Long, W. (2025). Genome-Wide Identification of the Kinesin Gene Family in Soybean and Its Response to Salt Stress. Agronomy, 15(2), 275. https://doi.org/10.3390/agronomy15020275




