Abstract
Managing foliar corn diseases like northern leaf blight (NLB) and gray leaf spot (GLS), which can occur rapidly and impact yield, requires proactive measures including early scouting and fungicides to mitigate these effects. Decision support tools, which use data from in-field monitors and predicted leaf wetness duration (LWD) intervals based on meteorological conditions, can help growers to anticipate and manage crop diseases effectively. Effective crop disease management programs integrate crop rotation, tillage practices, hybrid selection, and fungicides. However, growers often struggle with correctly timing fungicide applications, achieving only a 30–55% positive return on investment (ROI). This paper describes the development of a disease-warning and fungicide timing system, equally effective at predicting NLB and GLS with ~70% accuracy, that utilizes historical and forecast hourly weather data. This scalable recommendation system represents a valuable tool for proactive, practicable crop disease management, leveraging in-season weather data and advanced modeling techniques to guide fungicide applications, thereby improving profitability and reducing environmental impact. Extensive on-farm trials (>150) conducted between 2020 and 2023 have shown that the predicted fungicide timing out-yielded conventional grower timing by 5 bushels per acre (336 kg/ha) and the untreated check by 9 bushels per acre (605 kg/ha), providing a significantly improved ROI.
1. Introduction
Crop diseases can severely impact plant survival and yield. Proactive measures, such as preventive fungicides, are essential if there is sufficient warning about conditions conducive to disease development [1]. Rowlandson et al. showed that decision support tools, using data from in-field sensors and predicted leaf wetness duration (LWD) intervals based on meteorological conditions, can help growers to anticipate and manage crop diseases effectively. Access to accurate and timely weather data is crucial for effective crop disease management, and the internet provides meteorological data for thousands of locations across the United States. Effective crop disease management programs combine crop rotation, tillage practices, hybrid selection, and fungicide use. Third-party market research conducted by Kynetec—https://www.kynetec.com/agriculture (accessed on 24 January 2020), for Corteva, showed that Midwest growers’ top two challenges are application timing and return on investment (ROI). Growers can increase their ROI and reduce pathogen resistance, and manufacturers can enhance customer loyalty and retention, by applying fungicides only when conditions are favorable for disease outbreaks.
Leaf wetness is a critical parameter in plant disease development [1]. Studies have shown that leaf wetness duration (LWD) is influenced by factors such as dew and rainfall. Models that incorporate daily meteorological conditions and surface energy balances, such as the Penman–Monteith model, can estimate dew formation and duration [2]. These models are essential for predicting disease conditions and guiding fungicide applications. The number of leaf wetness hours required for disease development varies according to the pathogen, ranging from 30 min for Phytophthora in strawberries to more than 100 h for Diaporthe in soybeans [3,4]. LWD can be measured directly by sensors [5,6] or modeled using physical models [7,8,9] and machine-learning techniques [10,11,12]. However, the accuracy of models can vary due to different sources of noise [1,13,14,15,16,17].
Plant pathosystems’ occurrence and infections are commonly modeled using mechanistic approaches that incorporate well-characterized behaviors of biological agents [18]. Framing weather data as a count of hours under certain conditions is beneficial from the modeling standpoint as it can efficiently compress a large amount of data into a few informative features. For instance, Kang et al. [19] used hourly weather data collected in a set of environments to model rice blast [20] based on temperature and LWD, tracking the disease progression and projecting trends across geographies using interpolation. Similarly, Kassie et al. [21] used a mechanistic model approach to predict early-phase Asian Soybean Rust (ASR) epidemic dynamics in soybeans. The approach included moisture and temperature modeling, producing a system that could lead and contribute to a fungicide-treatment program for ASR in Brazil.
University extension agencies and research groups have proposed correlations between meteorological conditions and crop disease development. These empirical relationships postulate optimal meteorological parameter combinations that must be met before disease expression begins. However, the relationship between weather conditions and crop disease development is complex and variable, since it depends on soil conditions, management practices, and crop stage. This ultimately impacts when and if a curative or preventative fungicide should be applied. Various models incorporating daily meteorological conditions and surface energy balances can be essential for predicting disease conditions and guiding fungicide applications.
Epidemiological models track disease progression based on environmental conditions and host–parasite interactions. These models predict infection efficiency, latent periods, and spore production, which are influenced by factors such as plant nutrition, relative humidity, temperature, and radiation [22]. For example, Kang et al. [19] used hourly weather data to model rice blast progression. El Jarroudi et al. [23,24] iterated on a pre-existing mechanistic model [25], Figure A1, to forecast stripe rust based on relative humidity and temperature combinations. The principles of the epidemiological modeling of plant diseases were formalized by Van der Plank [26]. These models can be utilized for predicting time-to-infection, disease severity, probability of an epidemic, and management actions [27].
Mechanistic approaches are commonly used to model plant pathosystems which incorporate well-characterized behaviors of biological agents [18]. These models frame weather data as a count of hours under certain conditions. Hamer et al. [28] modeled powdery mildew with multiple tree-based machine-learning methods fed with a few pathogen-specific mechanistic features derived from daily summaries of temperature, relative humidity, wind speed, and precipitation collected in specific locations. Skelsey [29] used two main mechanistic features, the daily sum of hours RH ≥ 90% (as LWD proxy) and minimum temperature, on the 28 days prior to infection to model potato blight outbreaks using an ensemble of multiple predictors. As an alternative to mechanistic features, Xu et al. [30] calibrated a fully data-driven model by utilizing weather and soil data to parametrize a spatial–temporal recurrent neural network to forecast yellow rust outbreaks in wheat.
Northern leaf blight (NLB) is a major corn disease caused by Exserohilum turcicum that develops under high humidity and moderate temperatures. The pathogen overwinters on crop debris [31] and has multiple pathotypes, with corn hybrids displaying varying degrees of race-specific resistance [32,33]. Conidial germination and the formation of appressorium on the leaves of susceptible hybrids occur with 3 to 5 consecutive hours of dew at temperatures between 15 and 30 °C, with optimal conditions around 20 °C [34]. Levy and Cohen showed that lesions form within 5 h of dew at 20 °C, and the severity of the disease progression depends on the hybrid’s resistance level. Bowen and Petterson [35], Figure A2, reported that NLB infections occur as a function of dew, relative humidity (60 < RH < 94%), and temperature (10 < T < 35 °C), with an optimal temperature range between 20 and 28 °C. There is no lesion expansion under 15 °C or over 35 °C, and the latency period for new lesions is about 6 days.
Gray leaf spot (GLS) in corn, caused by the fungus Cercospora zeae-maydis, is a major foliar disease that significantly impacts corn yields worldwide. GLS symptoms begin as small, necrotic lesions that expand into rectangular, gray to tan spots following the leaf veins (https://content.ces.ncsu.edu/gray-leaf-spot-in-corn, accessed on 22 January 2025). These lesions reduce the photosynthetic area of the leaves, leading to substantial yield losses, especially when the upper leaves are infected (https://www.extension.purdue.edu/extmedia/bp/BP-56-W.pdf, accessed on 22 January 2025). Studies have shown that environmental conditions such as prolonged periods of high humidity coupled to warm temperatures (22 to 30 °C), favor the development and spread of GLS [36]. Factors such as hybrid susceptibility, weather conditions, crop rotation history, and timely fungicide applications can influence the severity of GLS outbreaks. Paul and Munkvold developed risk assessment models for GLS using preplanting site and maize genotype data for predictors [37]. Research has identified several quantitative trait loci (QTL) in the maize genome that confer resistance to GLS, providing insights into the genetic mechanisms underlying disease resistance [38].
In this paper, we describe a disease-warning system for two major corn diseases, NLB and GLS, which utilizes historical and forecast hourly weather data, as well as a range of soil and field management features. This system leverages pre-existing continuous leaf wetness duration (LWD) conditions (e.g., algorithms when dew is present), coupled with optimal humidity and a range of temperature conditions relative to the specific manifestation of corn pathogens. Modeling these conditions, at scale, in real-time, can be used to predict potential disease onset and provide a recommendation system alerting growers of risk and furthermore guiding their fungicide application timing decisions. This optimal timing system not only improves grower profitability but also contributes to improved sustainability (higher yields with same input) and a reduced risk of pathogen resistance (optimal timing results in better control). We therefore believe that this disease-warning system offers significant advantages for crop disease management and fits very well in integrated pest management practices. By utilizing accurate weather data and advanced modeling techniques, growers can make informed decisions about fungicide applications, improving profitability and reducing environmental impact. This system represents a valuable tool for proactive crop disease management, with potential applications in other regions, crops, and diseases.
2. Materials and Methods
2.1. Weather Information
Hourly weather data utilized for the modeling was sourced from IBM’s weather API (https://www.ibm.com/docs/en/environmental-intel-suite?topic=components-weather-data-apis, accessed on 22 January 2025). Weather stations, including the cabled Vantage Pro2 Plus (www.davisinstruments.com, accessed on 22 January 2025), equipped with leaf wetness sensors (Davis Instruments, Hayward, CA, USA), were installed in 20 sites in 2020 to assess and compare the concordance of locally collected weather data with IBM and GridMet [39]. The alignment among different sources of data is displayed in Figure A3.
2.2. Soil Information
Data for soil parameters used with the machine-learning algorithm were obtained from the Soil Survey Geographic Database (SSURGO) available through the USDA Natural Resources Conservation Service (NRCS) web soil survey (WSS). The parameters under different evaluations included the slope, available water capacity (AWC), cation exchange capacity (CEC-7), soil pH, organic matter, and the texture as a percentage of sand, silt, and clay (Web Soil Survey (usda.gov)). From the described variables, the top-soil fraction, representing the 0–10 cm depth, was utilized for features in the model development.
2.3. Leaf Wetness Determination
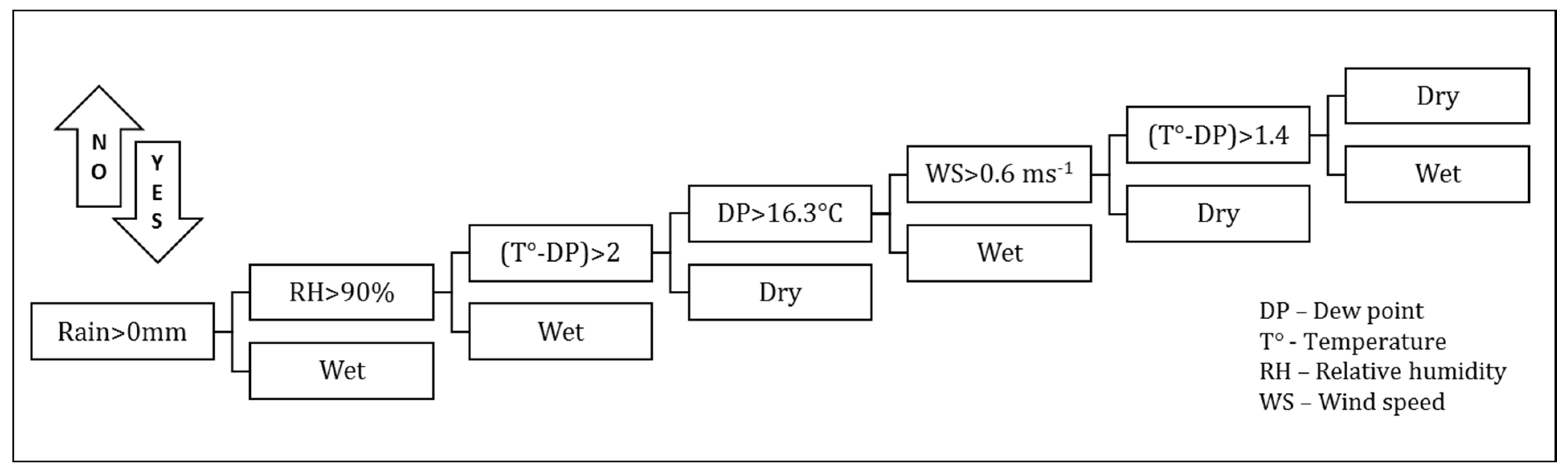
The primary parameter others have used to relate foliar pathogen expression is the leaf wetness duration (LWD) [14]. It is believed that dry periods greater than 5 h retard or eliminate the possibility of disease progression and the process based upon combinations of meteorological variables is reinitiated following a dry period. Dew is defined as the process where moisture condenses on the surface of, for example, a plant leaf during the night. This moisture can come from three sources: the air (dewfall), the soil (dewrise), and plants (guttation) [40]. Dew formation can be simulated using conservation equations for energy and mass (water). LWD was calculated based on precipitation status, dew point, temperature, relative humidity, and wind speed. In this study, we based our estimator of LWD on atmospheric conditions using a condensation model of dew formation in the plant canopy [7,41]. The methodology used herein, from Kim et al. [42], determines whether dew is present from the flowchart decision tree shown below (Figure 1).
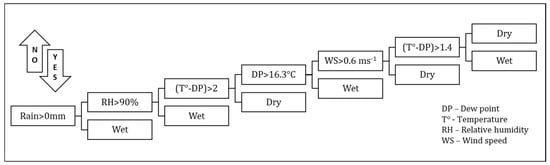
Figure 1.
Schematic decision tree diagram for logic used to determine hourly leaf wetness.
2.4. Mechanistic Features for NLB Model Development
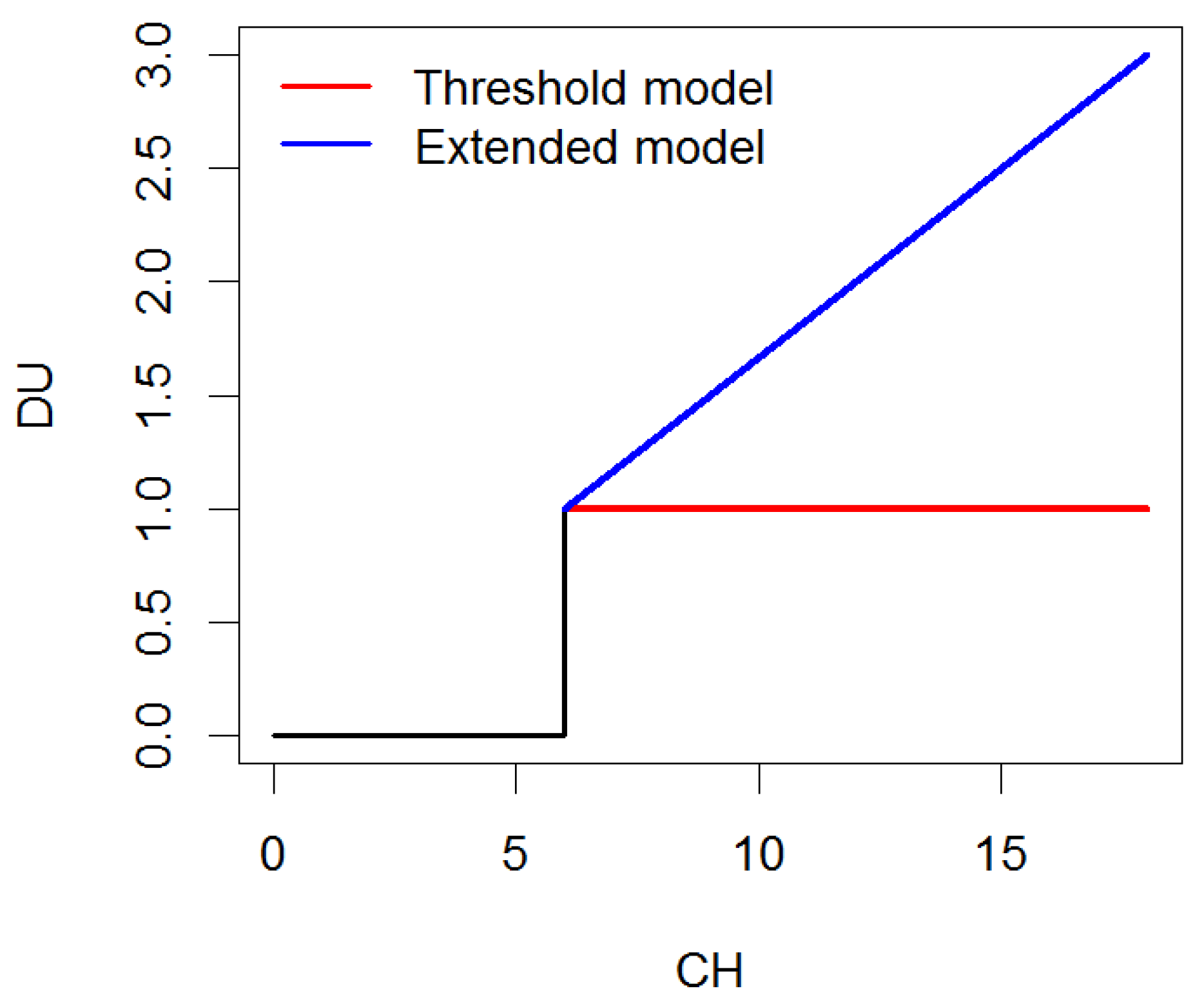
Disease units (DUs) were utilized to parameterize hourly weather data from the planting date to the disease observation date. DUs efficiently compress longitudinal climatic data from multiple meteorological variables and time points and, for the purpose of disease forecasting, enable time-to-event predictions. Disease units were computed by extending the threshold model as follows:
where LWD is the number of consecutive hours under leaf wetness under certain conditions of relative humidity () and temperature (). Such parametrization jointly accounts for leaf wetness duration [43] and the optimal disease development conditions [23,24]. In addition, the extended model described above provides a more quantitative metric of disease units than traditional threshold models, allowing for prolonged periods of time under ideal infection windows to be more favorable than conditions that barely reach the threshold, as illustrated in Figure 2. This mechanistic approach is the way in which the NLB risk DUs were calculated; the optimal humidity, temperature, and LWD ranges were already widely known from the public university pathology websites.
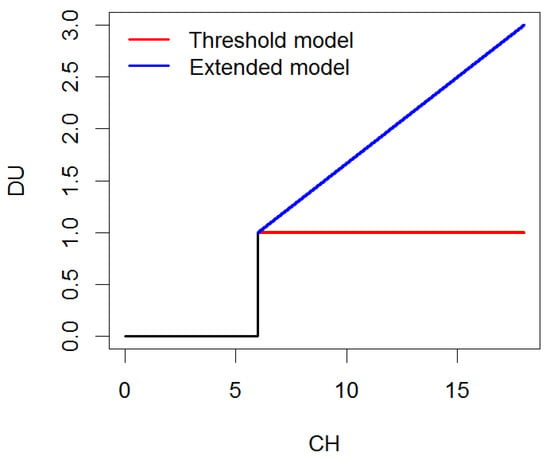
Figure 2.
Disease units (DUs) measured from consecutive hours (LWD) of ideal infection conditions under the threshold model and the extended model.
2.5. Genetic Tolerance Data
The disease tolerance information utilized in this spray timing model was extracted from the Pioneer agronomy catalog (Corn Product Catalog). These data consisted of product scores on a 1–9 scale where 9 represents a highly resistant variety and 1 a highly susceptible one. Most corn hybrids evaluated and utilized in the modeling had product scores between 3 and 7.
2.6. ML Feature Engineering for GLS Model Development
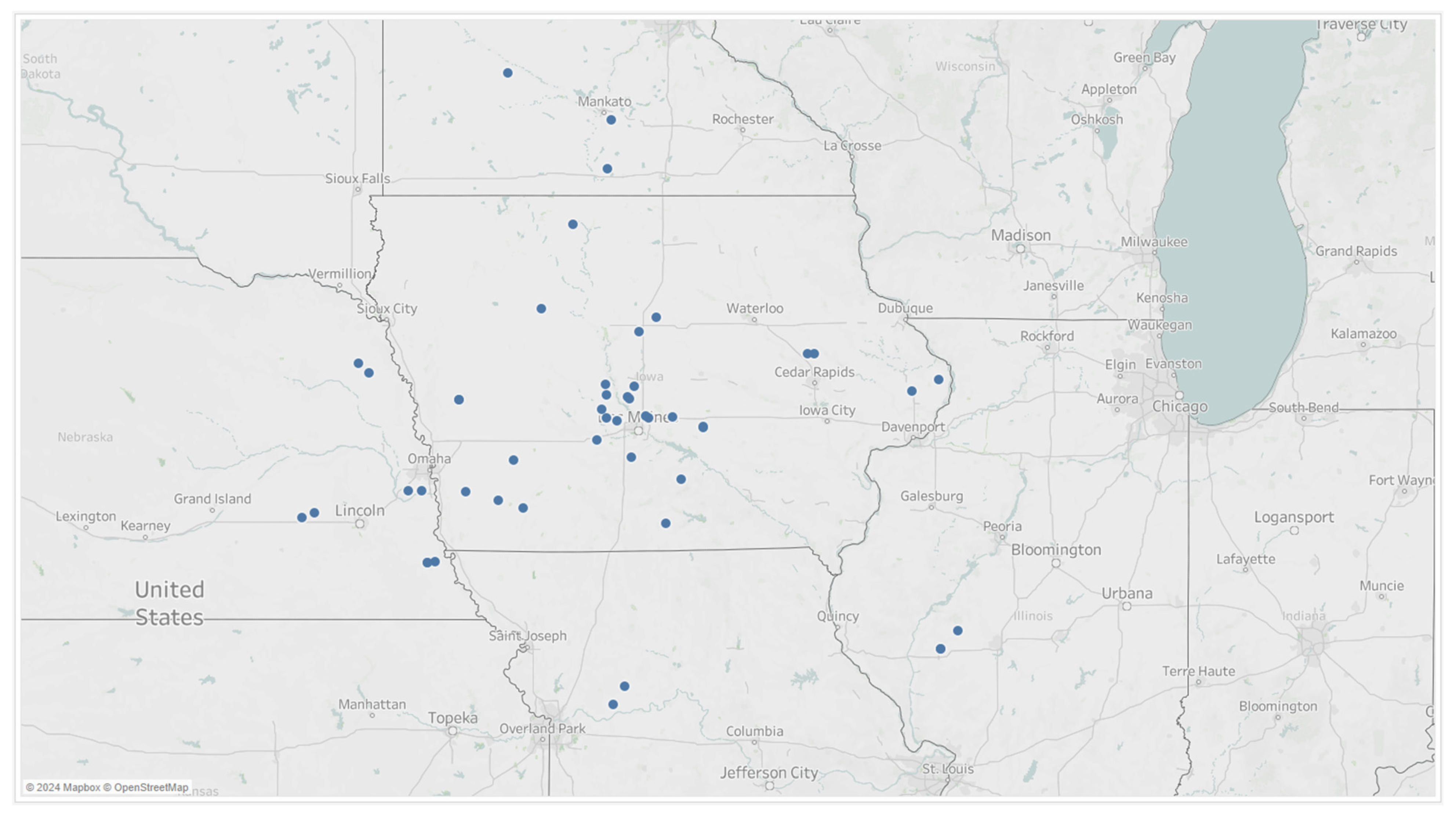
Hourly weather data and disease severity ratings were collected in 2019 across 5 weekly timepoints and across 50 independent trial locations in the Midwest, Figure 3. Natural disease progression was observed and monitored since the trial fields were not treated with a fungicide.
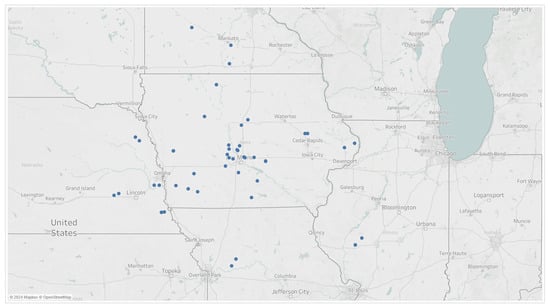
Figure 3.
Trial locations from which 5 observational disease ratings were taken in 2019.
Nine different weather variables were selected as predictors to model GLS onset and progression across the Midwest locations. These chosen weather variables are provided in Table 1. From the hourly weather data, across these observation trial locations, 9 variables were used to generate 216 features from each hour of the day recorded. The weather data were aggregated, by weekly average, starting from the week prior to the first observation date until the week prior to the last observation date. The response variable, utilized in the model training, consisted of the time-series disease observational data. Scores from each timepoint were averaged across hybrids for each of the 5 scoring timepoints.

Table 1.
Set of Inputs utilized for GLS model development.
A classification and regression tree (CART) model algorithm was used to help split the dataset into subsets based on the most significant variables to predict our continuous target outcomes from this data matrix. A CART model was chosen for its simplicity and interpretability while still achieving a strong accuracy in disease prediction; this also avoids the complexity of black-box models. The visible decision points, nodes, and branches of CART models enable easy adjustments and pruning to prevent overfitting as more data are gathered. The model output consists of generating daily GLS risk DUs through accumulated time-series weather data which can help to predict disease onset and progression.
2.7. Disease Severity Ratings
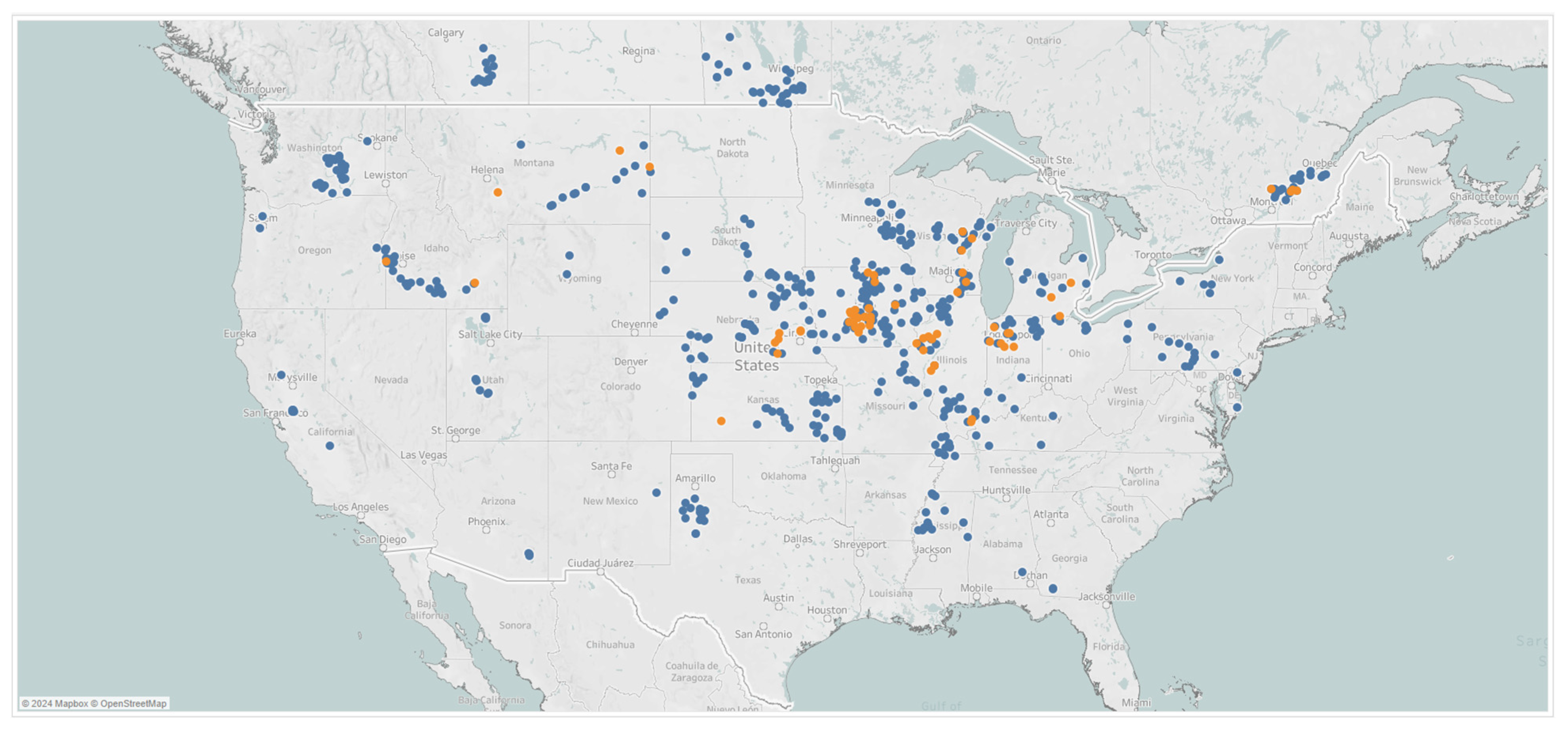
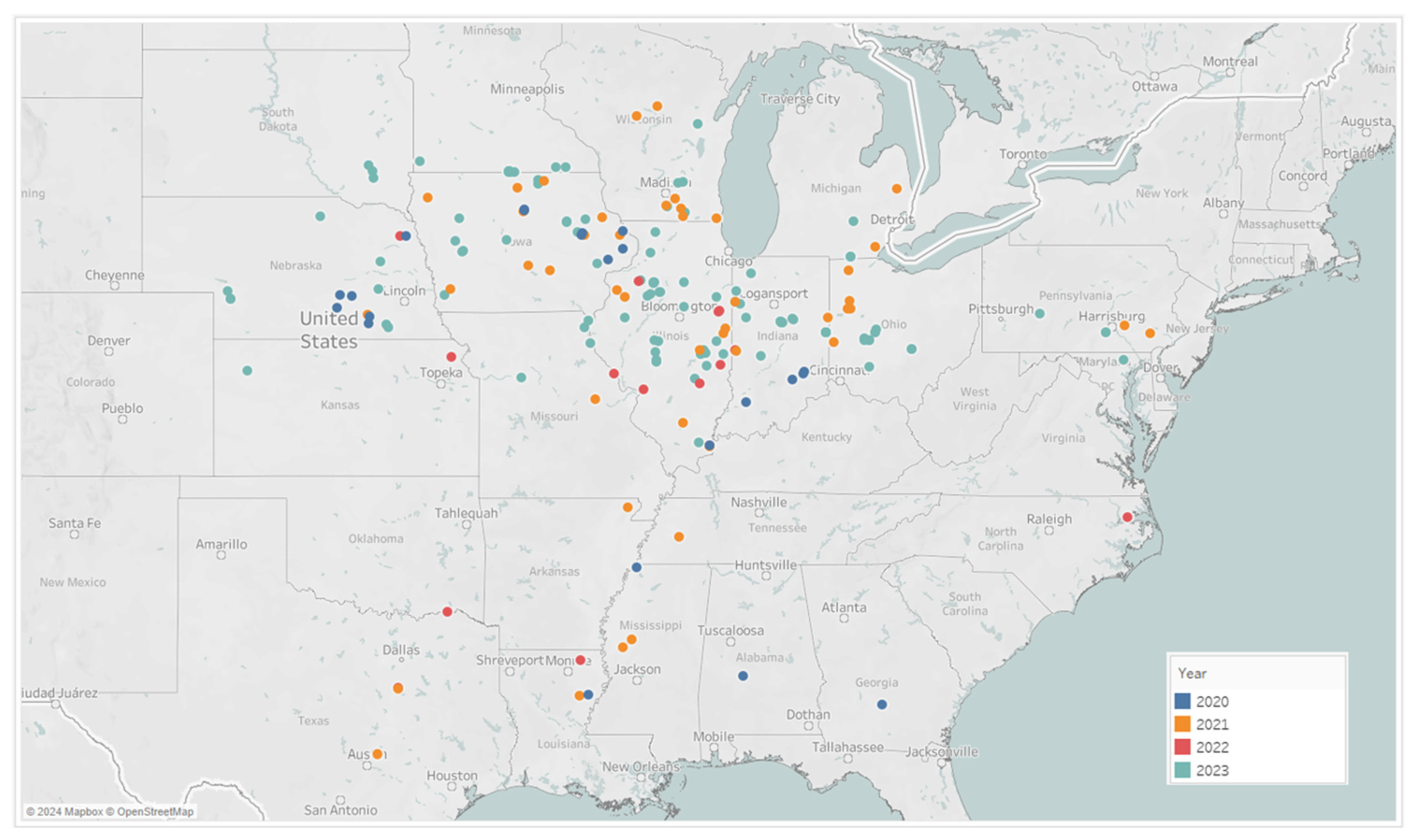
NLB scores representing disease severity were collected on a 1–9 scale in multiple hybrids within each location [44]. The overall severity of a given location was obtained by averaging the score of individual hybrids. An illustration of disease scores is provided in Figure A4 from the Pioneer scoring system. Disease scores, from NLB and GLS, were collected by Corteva field agronomists in 667 locations in the United States and Canada, from which 595 locations were observed in 2019 and 72 locations in 2020, as shown in Figure 4. Some locations exhibited medium-to-high disease pressure; a picture of visible symptoms observed for GLS and NLB, from southern Illinois and eastern Iowa, respectively, is shown in Figure A5.
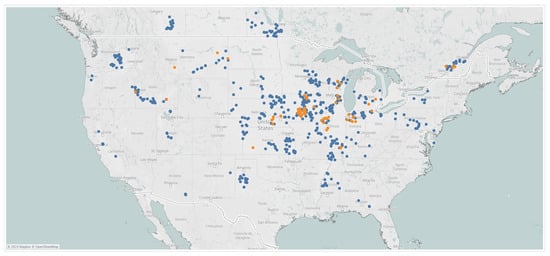
Figure 4.
Geographical distribution of data collected in 2019 (blue) and 2020 (orange).
2.8. Mixed Variable Predictive Modeling for Fungicide Timing
Data were leveraged across the disease-scored locations to develop the fungicide spray timing model; the following features at each location were used: soil features, agronomic management information, and hourly weather data. Agronomic data from each location included the tillage practice (conventional, conservative, none), irrigation (full, limited, none), previous crop (soybean, corn, other), and nitrogen fertilization (lb/a) applied, as well as the row spacing and planting density. These input variables were added to the soil variables already mentioned.
A small set of cumulative weather parameters were used including cumulative LW hours, rainfall, and accumulated growing degree units (GDUs) for corn (Growing Degree Units and Corn Emergence|CropWatch|University of Nebraska–Lincoln). These three continuous variables were combined with the ML-based GLS risk DUs and the mechanistic NLB risk DUs, both described in the previous sections, to serve as the final set of inputs for predicting spray timing in corn. This list of features is shown below in Table 2 across various classes. Hybrid information, such as the disease resistance to select pathogens, was used in the decision tree framework for predicting spray timing across differing tolerance classes. A heuristic set of governance rules were built into a decision tree that included agronomic knowledge, irrigation practice, hybrid information, environmental risk components, and disease models (Figure A6).

Table 2.
Set of variables used in building the spray timing decision model.
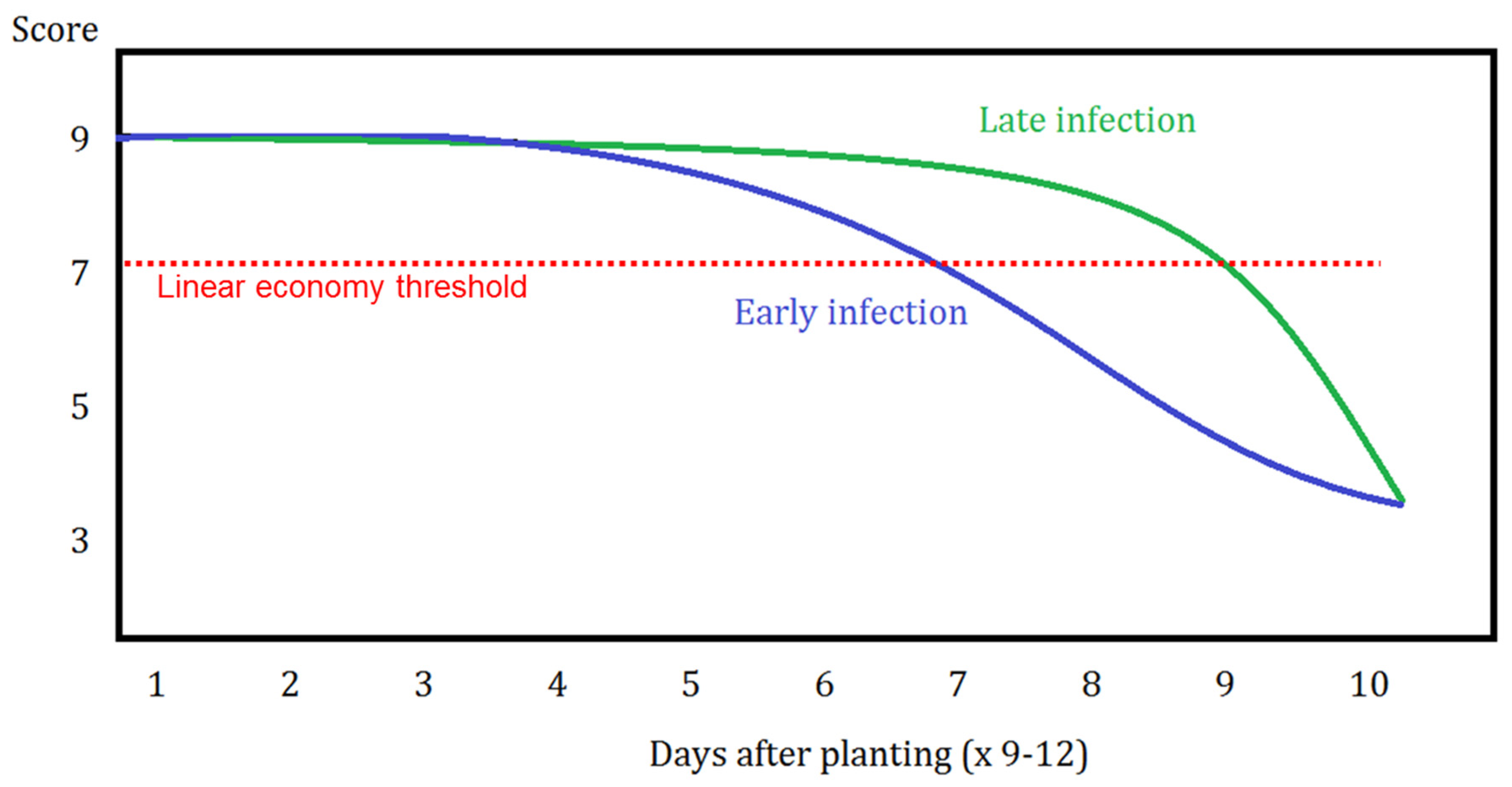
An economic threshold line was drawn to establish and depict differences between early and late infections when predicting the timing to spray fungicides (Figure 5). In this figure, disease score or severity is on the y-axis, whereas time, in days, from planting is on the x-axis. The linear economic threshold line represents some time X after planting where one can expect to observe a “very low” to “low” disease pressure observance in the canopy (ultimately representing a leaf area loss of approximately 10–15%).
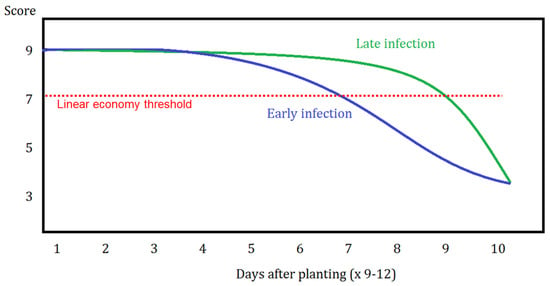
Figure 5.
Framework for spray timing in which an economic threshold line is shown with respect to early or late disease infection possibilities. Illustration of disease severity (y-axis) by days after planting (x-axis) where this can extend to upwards of 90–120 days after planting (×9–12).
2.9. Performance On-Farm Trials and Experimental Design
In testing the spray timing model framework, locations were selected for testing the ROI (return of investment) compared to the standard grower timing and the untreated check (UTC). In total, 190 on-farm trials were carried out from 2020 to 2023 across the US Midwest corn-growing regions, see Figure 6. These locations tested are broken down in Table 3 by state and by year, with totals across years and states also shown. The most-dense corn-growing areas were targeted across Iowa, Illinois, Indiana, Nebraska, Ohio, Wisconsin, and Minnesota, and accounted for 153 of the 190 on-farm trial locations.
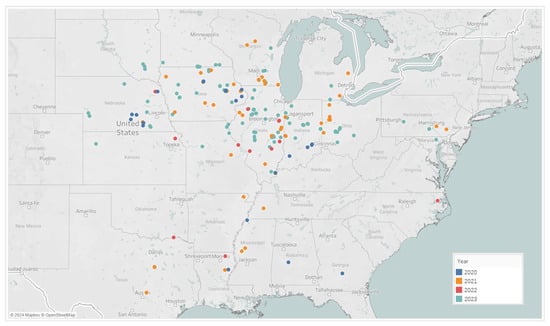
Figure 6.
Performance fungicide ROI trials across the US testing the Corteva model’s timing recommendations versus the farmer’s conventional timing.

Table 3.
Performance on-farm trials broken down by state and year. Totals are shown for each state across the years, as well as the total number of trials conducted within each year.
Most trials were conducted using a simple unrepeated three-treatment strip-plot approach where strips of 5+ acres were leveraged across the field. The three treatment areas consisted of (1) an unsprayed portion or an untreated check (UTC), (2) one area sprayed according to the grower’s standard choice of timing (GT) or conventional timing, and (3) an area treated based on the Corteva model-recommended spray timing (CT). Growers in the Midwest United States commonly use two-way or three-way mode-of-action fungicide products, including strobilurins (group 11), triazoles (group 3), and SDHIs (group 7), to combat fungal diseases in corn and soybeans. Fungicide applications typically occur around the VT/R1 stage, with some growers also scouting fields to determine the presence of disease before spraying.
Experimental layouts for on-farm trials were either designed by the grower or completed by Corteva and given to the grower to reference from. If completed by Corteva, experimental layouts favored uniform placement from previous yield analysis harvest maps, and productivity zones analyzed by satellite imagery, as well as intra-field soil type differences to minimize differences compromising the yield effects between the different strips.
A very low number of trials were not able to keep all 3 treatment blocks fully intact by harvest time; however, partial yield data were still achieved in these fields. This resulted in 35 trials where the UTC block was lost, 15 trials where the CT block was lost, and 24 trials where the GT was lost. The loss of some treatment blocks was due to unforeseen circumstances from logistics, delays, application mishaps, or combine failures at harvest time. Both treated areas leveraged the same chemical fungicide, at the same rate, but differed only in the timing of the application. Aproach PrimaTM (Corteva Agriscience, Indianapolis, IN, USA) fungicide was the preferred chemical used at most all trial sites in this study. Aproach PrimaTM contains picoxystrobin and cyproconazole, at 18% and 7%, respectively. Other common chemicals are frequently used by growers that include, but are not limited to, products such as Trivapro, Quilt Xcel, Veltyma, Headline AMP, Delaro Complete, and Stratego YLD, to name a few.
2.10. Statistical Analyses and Yield Analyses
In creating an ML-based GLS prediction model, a regression decision tree was built by recursively partitioning the data according to relationships between the predictor variables and the response variable (i.e., the GLS disease score). Both the response and predictor variables were continuous data and the decision tree was constructed using JMP®, Version 18.1 SAS Institute Inc., Cary, NC, USA, 1989–2023. A CART model was selected because it is useful for exploring relationships in the data while handling large problems easily with interpretable results. To evaluate the model performance, a 5-fold cross-validation technique was selected since it provides a more robust and reliable performance estimate: it reduces the impact of data variability and minimizes the risk of overfitting. Since the dataset was small to medium in size, this ensured every data point was used in both training and validation for a more comprehensive but still unbiased evaluation. The location-level risk DUs were compared to the ground scores observed from the 2019 and 2020 agronomy trials highlighted from Figure 4 to evaluate the NLB risk model and gauge accuracy. These disease risk units for NLB (calculated from Figure 2) can track and measure potential disease onset and continued progression across time. Scoring data collected from the 2019 and 2020 field seasons were evaluated by viewing the area under the ROC curve for detection accuracies to calculate the disease models’ performance in predicting disease severity.
The risk DUs for the two pathogen models were combined with agronomic data from the field along with soil attributes to serve as a set of input features for the mixed variable model to predict disease observations. Corn hybrid disease ratings from the 50 Midwest observation trials were used to build independent risk thresholds by tolerance class to NLB and GLS; these involved identifying NLB and GLS accumulated risk DUs that accounted for drops in disease pressure observed from the trial observations. For each resistance class, a linear threshold was drawn that predicted a potential disease outbreak for NLB or GLS at very low to low levels of severity. A set of CART-based decision regression trees were constructed using recursive partitioning and the outputs from these trees were averaged for an overall composite stress score prediction.
The results from the on-farm trials to calculate ROI were analyzed by t-tests and p-values were used to assess significance between the different treatment blocks tested. Yield analyses, throughout the different growing seasons, were either analyzed by the growers themselves or completed by Corteva. From the analyses completed by Corteva, the as-planted, as-applied, and as-harvested data layers were gathered and overlayed together to dissect yield cuts from the designed treatment blocks. Some buffering was performed on treatment block lines to avoid contamination from adjacent treatments as well as headland exclusions on the exterior parts of the field edges. This type of bracketed analysis shows more consistent block-to-block differences over a full field breakout analysis which can introduce more unwanted variation due to potential ditches and soil types. Roughly 70% of the fields were analyzed in this fashion versus the other 30% analyzed by the growers themselves.
3. Results
A graph that highlights the likely occurrence of NLB infection, across 2019 and 2020, with increased NLB risk DUs, is shown in Figure A7. This result allowed the establishment of NLB risk DUs to be incorporated into the spray timing decision model. The NLB model was shown to be very predictive of the overall pressure and likely occurrence.
A model selection process was determined to validate that the ML model had the best performance on the GLS data, while the mechanistic model was superior on the NLB data. We can view the model area under the curve (AUC), as in Table 4. The variations in weather and disease pressure between 2019 and 2020 significantly influenced the data quality and quantity, which proportionally affected the accuracy and AUC values of the disease models for NLB and GLS. In 2019, cooler and wetter conditions led to higher NLB pressure, while 2020’s warmer and drier conditions, along with a shift in trial locations, resulted in GLS being more predominant. Despite these differences, both models maintained an average accuracy of around 70% even with the year effect differences observed.

Table 4.
Disease prediction model results shown by area under the ROC curve for detection accuracies.
Disease detection accuracy in our analyses involved distinguishing zero-to-low disease pressure environments from moderate-to-high pressure environments. Receiver operating characteristic (ROC) curves were plotted in viewing the True Positive Rate (TPR) against the False Positive Rate (FPR) for different disease presence versus absence thresholds. In the summarized performance found in Table 4, the area under the ROC curve is measured from 0 to 1, where 1 is best (TPR) and 0 is worst (FPR).
Through feature engineering of extracted hourly weather data, nine variables were selected and leveraged to predict GLS disease onset and progression across the on-farm trials from 2019. A recursive partitioning decision tree was created, using the disease scores (N = 250), from 50 trials (Figure 3) as a y-response variable. The statistical performance metrics of the devised model illustrates the 5k-fold cross-validated accuracies of ~R2 = 0.9, as shown in Table 5. The relative humidity % average at 8 p.m., rainfall (mm) accumulation at 4 a.m., and wind speed (m/s) average at 12 p.m. are the top three contributors to the ML model.

Table 5.
Performance metrics are shown for the GLS featured decision tree ML-model including the R2, RMSE, SSE, N (# of observations), number of splits (N), and the Akaike information criterion (AIC) values.
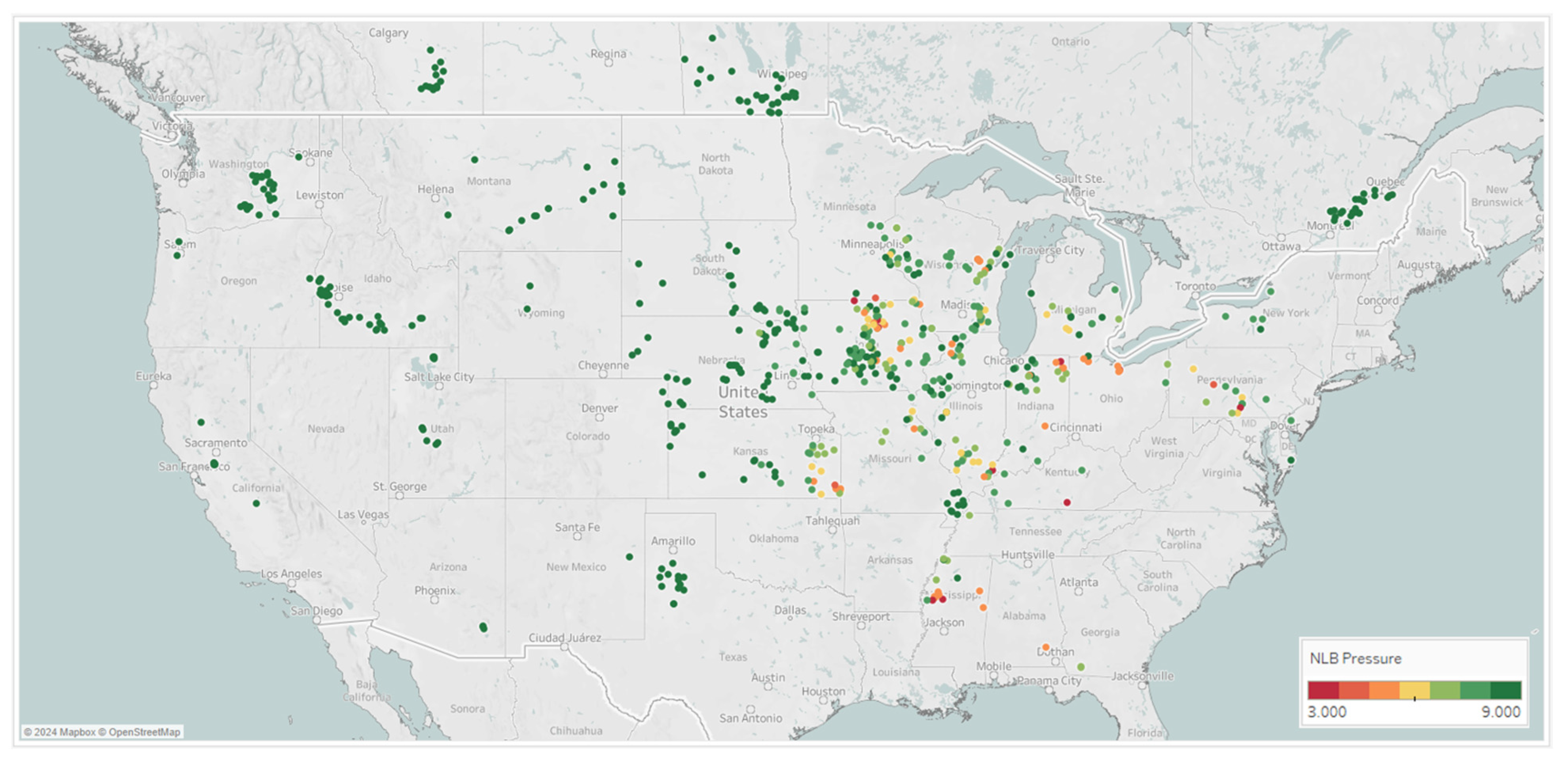
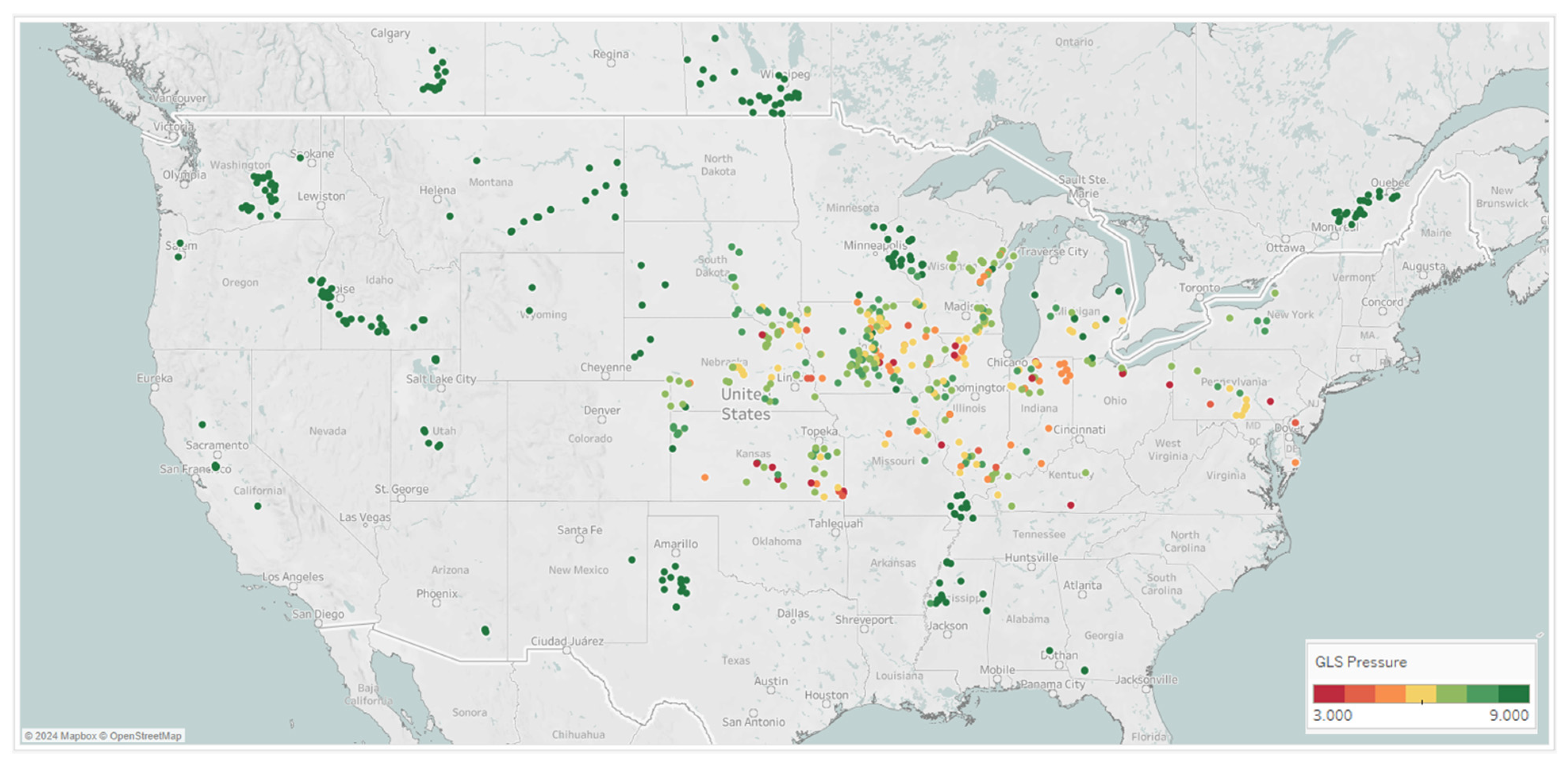
That NLB and GLS average pressure scores collected across the Midwest in 2019 and 2020 helped to build the mixed variable spray timing models, are shown Figure 7. These results show differential GLS and NLB symptoms scored across key parts of the Midwest. This clearly shows that a fungicide spray timing recommendation system would be extremely valuable to growers in this target geography to combat corn diseases and potential yield loss if and when they may manifest.
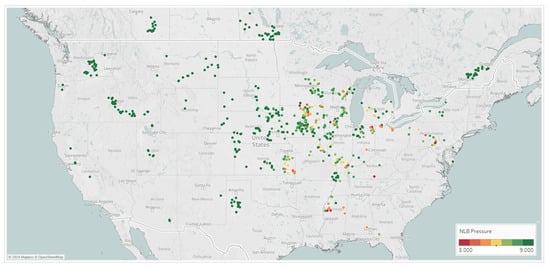
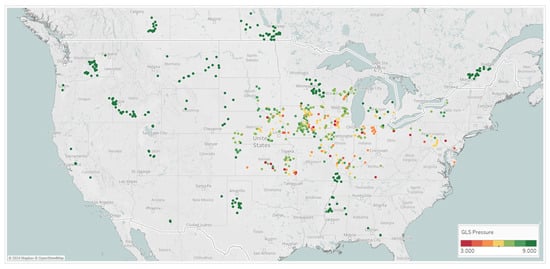
Figure 7.
Locations across 2019 and 2020 from which GLS and NLB disease observations were taken; these Corteva on-farm agronomy trials show the average rating on the 1–9 scoring scale, where 9 indicates no symptoms and 1 indicates very severe symptoms.
A disease prediction model was designed and built to accompany differential field attributes, genetic tolerance, irrigation practice, and environmental risk into a single overall fungicide timing spray decision. Built from the validation data from Figure 4 as a response variable, 18 input variables, shown in Table 2, were utilized within a recursive partitioning decision tree to predict disease stress at the field level. Four different decision tree models were created and used in combination to provide an ensemble disease prediction.
Each of the four models were validated using either a random holdback approach with a set number (N) of locations used for a training and validation set or by using a 5-fold cross-validation method. The statistical performance metrics of the four varying models are shown below in Table 6. These models show R2 values between 0.85 and 0.91.

Table 6.
Four different decision tree models are shown and validated differently between cross-validation and random holdback techniques. The statistical model performance and accuracy metrics and values are shown for the sum of squared error (SSE), R2, AIC, root average square error (RASE), N, and # of splits in the tree.
For CART model 1, the top column contributions were weather features consisting of the NLB risk model, cumulative GDUs, and the soil variables of organic matter, slope%, and pH. These five variables contributed to nearly 77% of the explainable variation. For CART model 2, which did not include the agronomic variables, the top column contributions were a similar set of weather features which included the NLB risk model, cumulative GDUs, and the soil variables of sand%, slope%, and pH; these features contributed to ~78% of the explainable variation seen in the model. For CART model 3, which utilized random holdback validation, the top contributors explained ~73% of the variation which consisted of the weather features of the GLS risk model and NLB risk model, the cumulative GDUs, and the available water capacity and pH from the soil inputs. Lastly, CART model 4, which excluded agronomic variables in the feature set, highlighted weather features such as the cumulative LWD hours, NLB risk model, and cumulative GDUs along with the silt% and pH from the soils; nearly 77% of the variation was explainable by these terms alone.
A set of environmental constraints were established based on the genetic tolerance information for a given hybrid, under a given irrigation management (full, limited, or none) which utilized leaf wetness hours, rainfall (mm), and each of the GLS and NLB DUs. Within a decision tree logical framework, like that shown in Figure A6, values were determined and orchestrated with decision points serving both as logical checkpoints and environmental risk checkmarks to evaluate the season-long accumulated risk. This developed framework also utilized the disease stress prediction models, from Table 6, in developing unique disease threat thresholds by tolerance class; once these thresholds were exceeded, a spray call was then triggered.
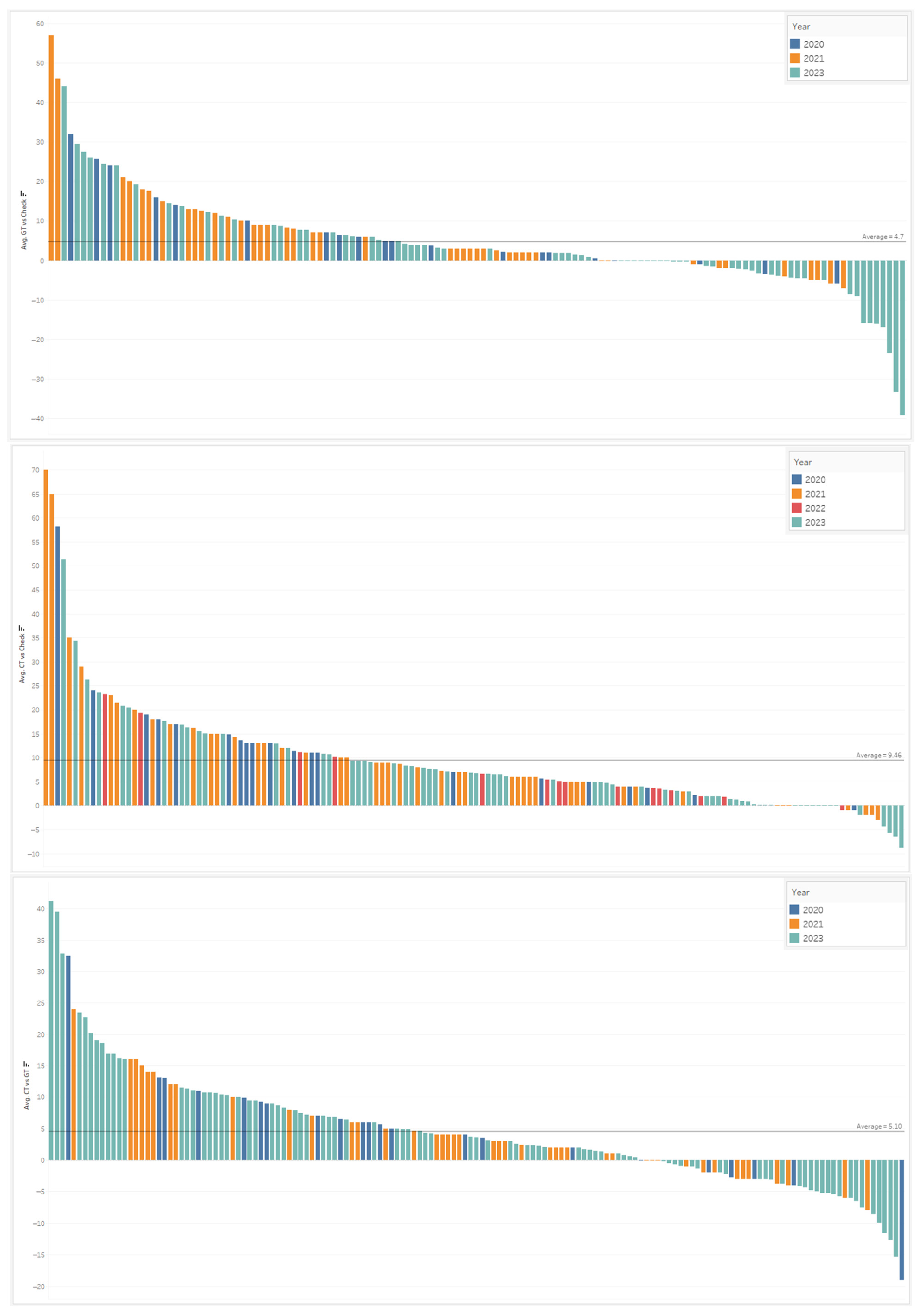
To view the spray timing model performance across the years, from 2020 to 2023, a descending piano bar graph was created showing the differing fungicide treated timings (CT and GT) relative to the UTC. Figure 8 shows these yield advantage breakdowns, by trial, which in turn lead to a positive or negative return of investment (ROI).
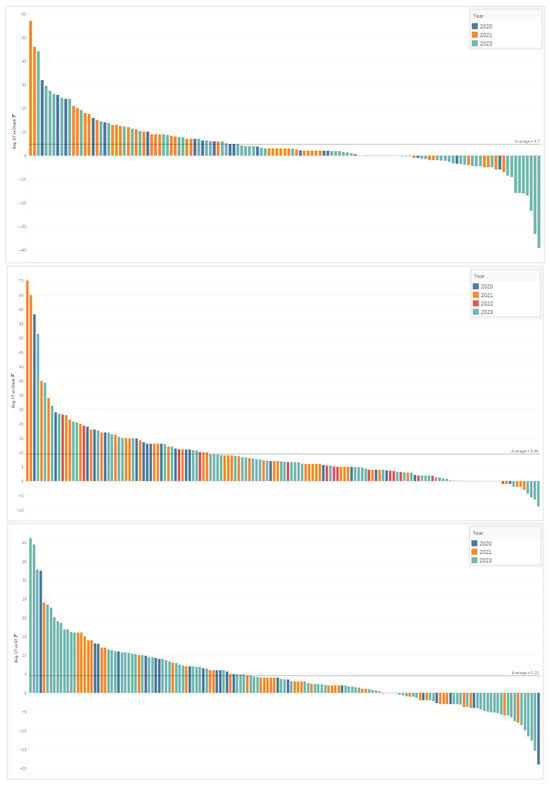
Figure 8.
Performance ROI trials across 2020–2023, which are colored by year, are shown in the piano-style descending bar graphs. The first figure represents the conventional GT compared to the UTC in bu/acre advantage; the second figure shows the CT versus the UTC, and the third figure demonstrates the CT vs. GT advantage.
In the first graph of Figure 8, the yield difference between the conventional timing, chosen by the grower, and the untreated check is shown in descending order. From the results of 3 years of independent trial data collected by our R&D and commercial agronomy teams, the average bu/acre advantage, across 131 trials, for the GT compared to the UTC was 4.7 bu/acre. Our results showed that 55 of 131 trials (42%) surpassed a 4 bu/acre advantage, while only 51 trials (39%) exceeded a 5 bu/acre advantage. Assuming a farmer needs 5 bu/acre to recoup the full cost of the spray, this indicated that around ~60% of the time, farmers do not see a clear ROI when spraying fungicides on their corn crop.
The second graph in Figure 8 shows the yield difference between the Corteva timing compared to the UTC, with an average increase of 9.5 bu/acre. This fungicide spray timing model, developed by Corteva, showed 102/146 trials (70%) exceeded a 4 bu/acre advantage, while 93 trials (64%) exceeded a 5 bu/acre advantage. This fungicide spray timing model shows a 20–30% gain in achieving a positive ROI, per field basis, compared with the grower timing treatments.
Shown in the third graph of Figure 8, the yield difference of the CT was compared to the GT across 151 trials. The yield advantage of the CT averaged +5.1 bu/acre. CT was compared to the GT across 151 trials and averaged a benefit of 5.1 bu/acre. Among these statistics, a win-rate of 62% (94 trials) was achieved for a >2 bu/acre advantage, while 54% of the time (82 trials) there was a >3 bu/acre advantage.
4. Discussion
Agricultural research has recently shifted focus to sustainable farm practices and increasing crop production. The goal of this study was to build and expand pathogen risk models, using predictive ML and mechanistic models, to predict the onset and progression of key corn diseases (NLB and GLS) in real time to better time fungicide applications. Growers benefit from increased corn yield in terms of bushels leading to higher returns of investment from their crop protection inputs through better application timing. This research contributes to guide more sustainable farming practices in opportunities to reduce inputs when crop protection products are not needed to control disease.
The spray timing model incorporates disease risk and LWD models in different ways. Both features serve as checkpoints from which a set of value thresholds can differ across varying tolerance classes. These hybrid tolerance classes were divided up between highly susceptible, moderately susceptible, moderately tolerant, and tolerant. The disease pressure predictor models, from a mixed variable set of inputs, act as triggers when the threshold is reached or exceeded. These models were validated using k-fold cross validation as well as other random holdback techniques.
This research demonstrates a disease model and recommendation system that was developed by Corteva to successfully control and protect against fungal diseases in corn, as evident by the bu/acre advantage seen from the Corteva timing versus the check and grower timings. A 9.46 bu/acre advantage was observed via comparing the CT versus the UTC across 146 trials, while a 5.1 bu/acre advantage was confirmed, across 151 trials, from analyzing the CT versus the GT treated areas.
These results show that by correctly timing the disease onset and progression, there can be a substantial fungicide advantage, not only evident in canopy health, but in total bushels per acre. This disease prediction spray model system builds upon general fungal risk modeling with the addition of the spray timing component. Growers struggle to break even with traditional practices. As shown in the GT vs. UTC comparisons in Figure 8 farmers in the Midwest typically only break even, at >5 bu/acre over the UTC, ~39% of the time. The Corteva timing showed a drastic increase in the win-rate percentage to break even with a 25% advantage over the GT of ~64% of the time.
The variability in disease pressure across different environments in the Midwest significantly impacts the yield potential. Primary diseases like GLS and NLB are major threats during the V14–R1 stages, while tar spot and southern rust become problematic in the later stages (R2–R4) and are very year-dependent. The differences in return on investment (ROI) across trials over four years are mainly due to these environmental and disease variations along with trial placement, not the fungicide chemistry, as Aproach Prima was used consistently.
In conclusion, the disease-warning system described in this document offers a significant advancement in crop disease management by leveraging accurate weather data and advanced modeling techniques. By predicting disease onset and guiding fungicide applications, this system enhances grower profitability and sustainability while reducing pathogen resistance. The integration of historical and forecast weather data, along with soil and field management features, provides a comprehensive approach to managing diseases like NLB and GLS. Field trials have demonstrated the system’s effectiveness, showing substantial yield advantages and a better return on investment compared to conventional methods. Overall, this innovative approach represents a valuable tool for proactive and informed crop disease management.
Empirical models, which rely on observation rather than theory, have extensively been used in academic and industry pest management research. These statistical models such as CART and LWD estimation have been used frequently when modeling for fungal disease risks [15,16,17,37]. Recently, in 2024, new published research has shown improvements in predictions for daily LWD using random forest (RF) models utilizing a multi-step ML approach from the ERA5 reanalysis weather products [45]. Due to the limited availability of leaf wetness sensors and the high cost for physical weather stations, advances in data-based predictions using ML models are emerging as the preferred choice for measuring LWD at scale. Furthermore, with climate change and pest risk modeling becoming major concerns, local daily and hourly LWD predictions are critical to forecast and track pathogen threats.
The strengths of the spray timing framework described herein consist of the mixed variables that are leveraged together as features that combine the field specific agronomic metadata, local hourly historical and forecast weather data, soil information, and product disease tolerance to provide an in-season recommendation. Potential gaps in the feature set that could be added to improve future model predictions could be the use of remote sensing and the utilization of real-time pivot telemetry data from irrigated fields, as well as spore traps which could quantify and identify spore load. The main challenges of practical application are the limited availability of on-farm data capture for non-John Deere (JD) users, which restricts critical input data for field-level recommendations, and the constraints on weather data ingestion, which affect the timeliness and scalability of the model’s daily recommendations. As the project scales, managing increased data processing demands without exceeding computing capacity is also crucial. Along with these model additions, many companies provide comprehensive soil samplings, by field, which can potentially help to identify and determine pathogen load abundance along with other nutrient levels. These agronomic field-level assessments mentioned could be used to tailor the right crop protection chemistries to combat the identified disease targets.
Throughout the trial protocol execution phase, it was critically important for the Corteva field team to overcommunicate with the growers on the field work tasks (data collection, spray timing, etc.) to garner whether the necessary modeling information for trial sites was achieved. Engaging with busy farmers, whose priorities extend far beyond the strip trials you are partnering with them on, need constant reminders and follow-ups. For trial creation and experimental design, it was important to view soil types, satellite imagery, and yield zones (productivity acres) to ensure the size, shape, and placement of the treatment blocks, especially the UTC, were uniform. These consistencies allowed for better execution and better signal-to-noise yield differences for treatment effects, driven by different application timings, and lower error estimates across the bracketed analyses.
Further research and next steps are ongoing to expand the disease model suite to in-corporate spray timing recommendations for tar spot and southern rust. Southern rust, while more common in the southern states of the US, can present difficult and challenging problems for Midwest growers when tropical storms or wind events blow spores from this disease north to infect corn fields (https://www.cropscience.bayer.us/articles/bayer/identifying-and-managing-southern-rust-of-corn) (accessed on 22 January 2025). This devastating fungal disease can cause severe damage to the corn yield as well as spread quickly from field to field. Large fungicide bushel advantages can be observed when protecting from this crucial disease at the right time. Tar spot, which is one of the newer emerging diseases in the upper Midwest and far-east US, thrives in cooler temperatures around 15–22 °C under moderately high humidities >80% (https://www.pioneer.com/us/agronomy/Tar-Spot-of-Corn.html) (accessed on 22 January 2025). While moderate ambient temperatures (18–23 °C) can drive and increase P. maydis stroma, tar spot progression has shown to be hindered by extended periods of higher relative humidity levels of >90% [46].
Expansion into other areas of research can be leveraged from the results of this study to include decisions for two-spray application programs and ROI calculations. From the spray timing model discussed, the first spray timing decision can be initiated. A second application might be warranted using weather forecast information to know whether conditions will potentially progress the disease. As with any spray timing decision, growers are most concerned with the amount of return on investment they potentially might reach after harvest. Genetic tolerance data would need to be incorporated along with the field-level disease risk. These two factors can influence the response a given hybrid has to fungicide as well as project the likely disease pressure impact, or leaf area loss, the disease would have on the crop.
5. Patents
Automated Fungicide Spray Timing
Utility Patent Reference Number: 108467-WO-SEC-1 & 108467-US-UTL-1.
Above is the extension of a provisional patent that was filed on 21 February 2023 (9396-US-PSP).
Author Contributions
L.P. and G.v.d.H. were responsible for the ideation of this research. L.P., A.X., S.C. and J.B. performed the modeling and analysis work. L.P. and A.X. are the primary authors of the manuscript. J.B., G.v.d.H. and L.P. were responsible for the field trial work and data collection. All authors equally supported the writing and editing of the article. All authors have read and agreed to the published version of the manuscript.
Funding
The research work and data collection conducted herein was funded through the internal R&D division of Corteva Agriscience.
Data Availability Statement
The datasets presented in this article are proprietary to Corteva. Requests to access the datasets should be directed to layton.peddicord@corteva.com.
Acknowledgments
The authors would like to thank the Crop Protection Division of Corteva Agriscience for the fungicide chemical cases provided in the multi-year study. They also want to acknowledge the data science teams within Corteva for the treatment block analyses and experimental designs for the fungicide strip trials. Lastly, they want to extend their appreciation to the agronomy, field experimentation, and commercial teams who helped to run the trials in different states and manage the relationships with the customers participating in the ROI trials.
Conflicts of Interest
The work performed in this paper was by salaried employees from Corteva Agriscience, a major American agricultural seed and chemical company. The authors declare no conflicts of interest.
Appendix A
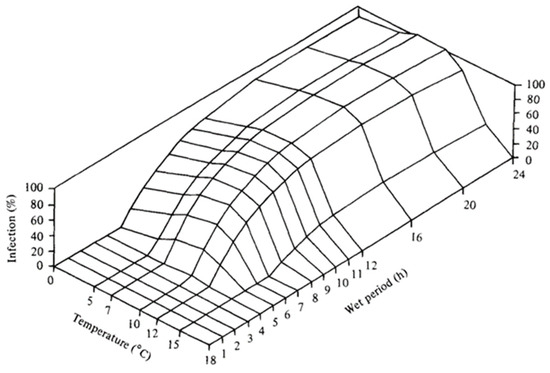
Figure A1.
Dennis (1987) model based on temperature and hours of leaf wetness [25].
Figure A1.
Dennis (1987) model based on temperature and hours of leaf wetness [25].
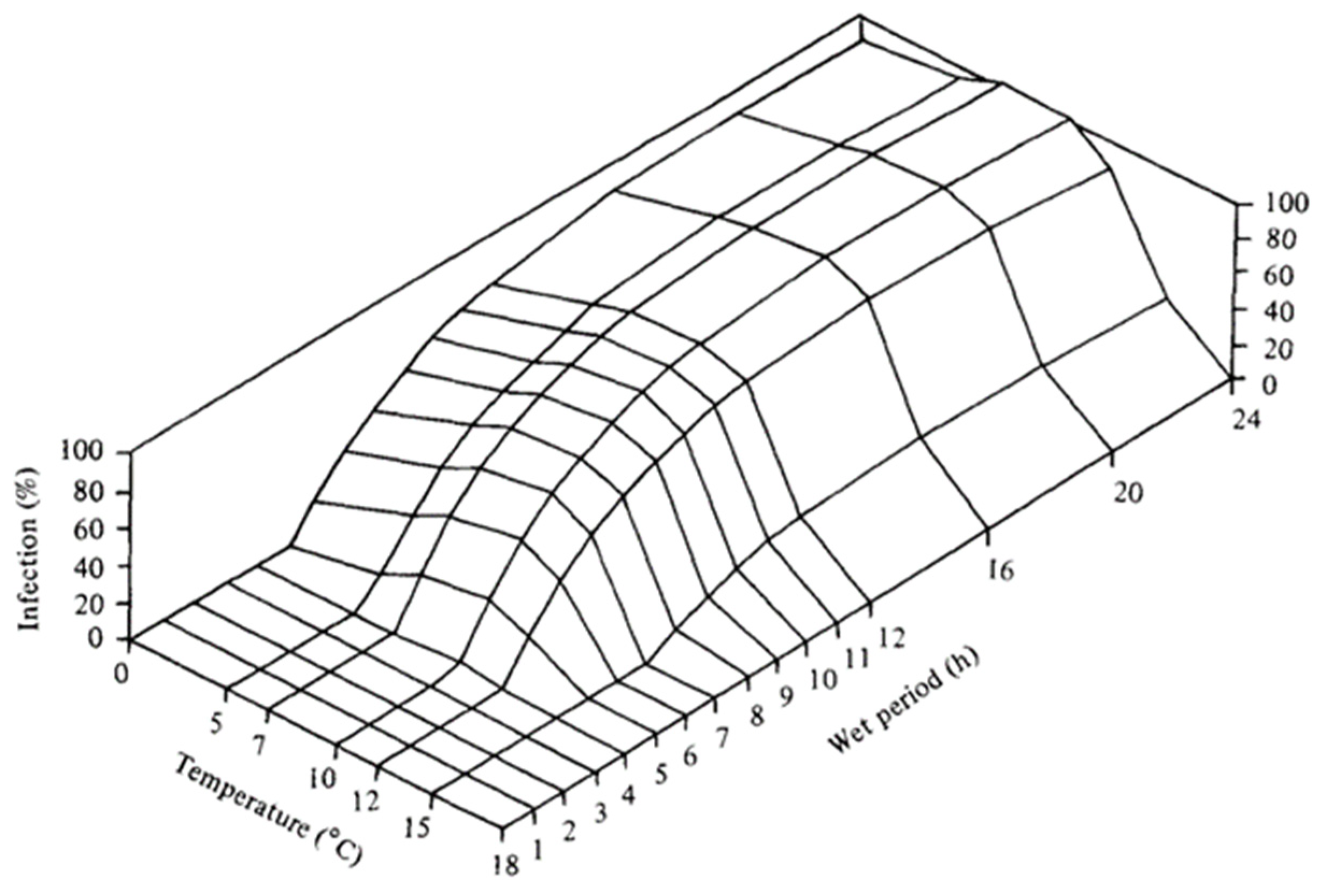
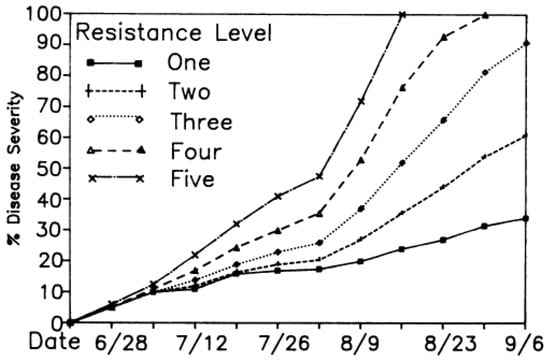
Figure A2.
NLB development from Bowen and Pederson (1989) model Corn-IL1 [35].
Figure A2.
NLB development from Bowen and Pederson (1989) model Corn-IL1 [35].
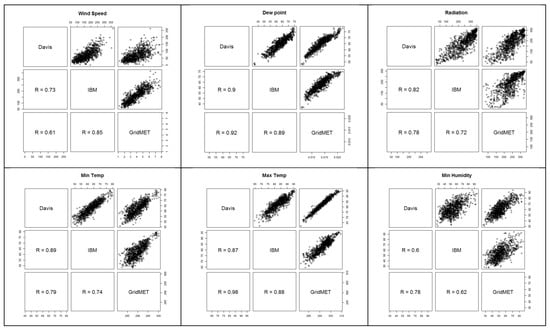
Figure A3.
Correlation among different sources of weather data.
Figure A3.
Correlation among different sources of weather data.
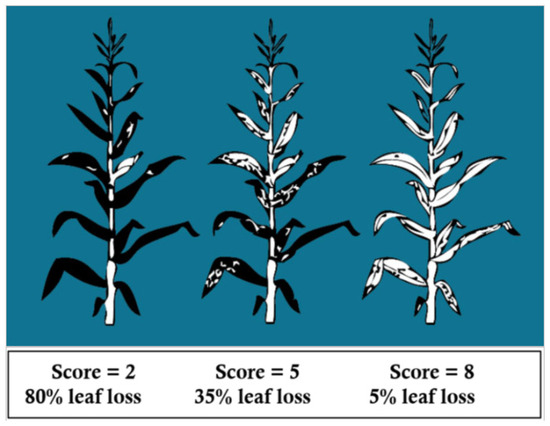
Figure A4.
Illustration of NLB and GLS scores on a 1–9 scale. Lower values represent higher disease severity. Image source example: https://www.pioneer.com/us/agronomy/Managing-Northern-Corn-Leaf-Blight.html (accessed on 22 January 2025).
Figure A4.
Illustration of NLB and GLS scores on a 1–9 scale. Lower values represent higher disease severity. Image source example: https://www.pioneer.com/us/agronomy/Managing-Northern-Corn-Leaf-Blight.html (accessed on 22 January 2025).

Figure A5.
Disease infection levels (medium to high) observed in 2019 from southern Illinois (GLS) and eastern Iowa (NLB). These leaf samples were submitted to the Corteva Agriscience pathology and diagnostics lab located in Johnston, Iowa.
Figure A5.
Disease infection levels (medium to high) observed in 2019 from southern Illinois (GLS) and eastern Iowa (NLB). These leaf samples were submitted to the Corteva Agriscience pathology and diagnostics lab located in Johnston, Iowa.

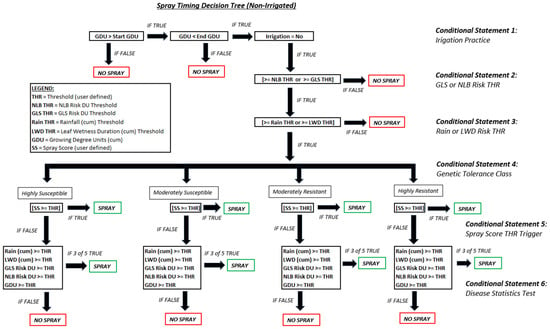
Figure A6.
Spray timing governance decision tree representative of non-irrigated acres.
Figure A6.
Spray timing governance decision tree representative of non-irrigated acres.
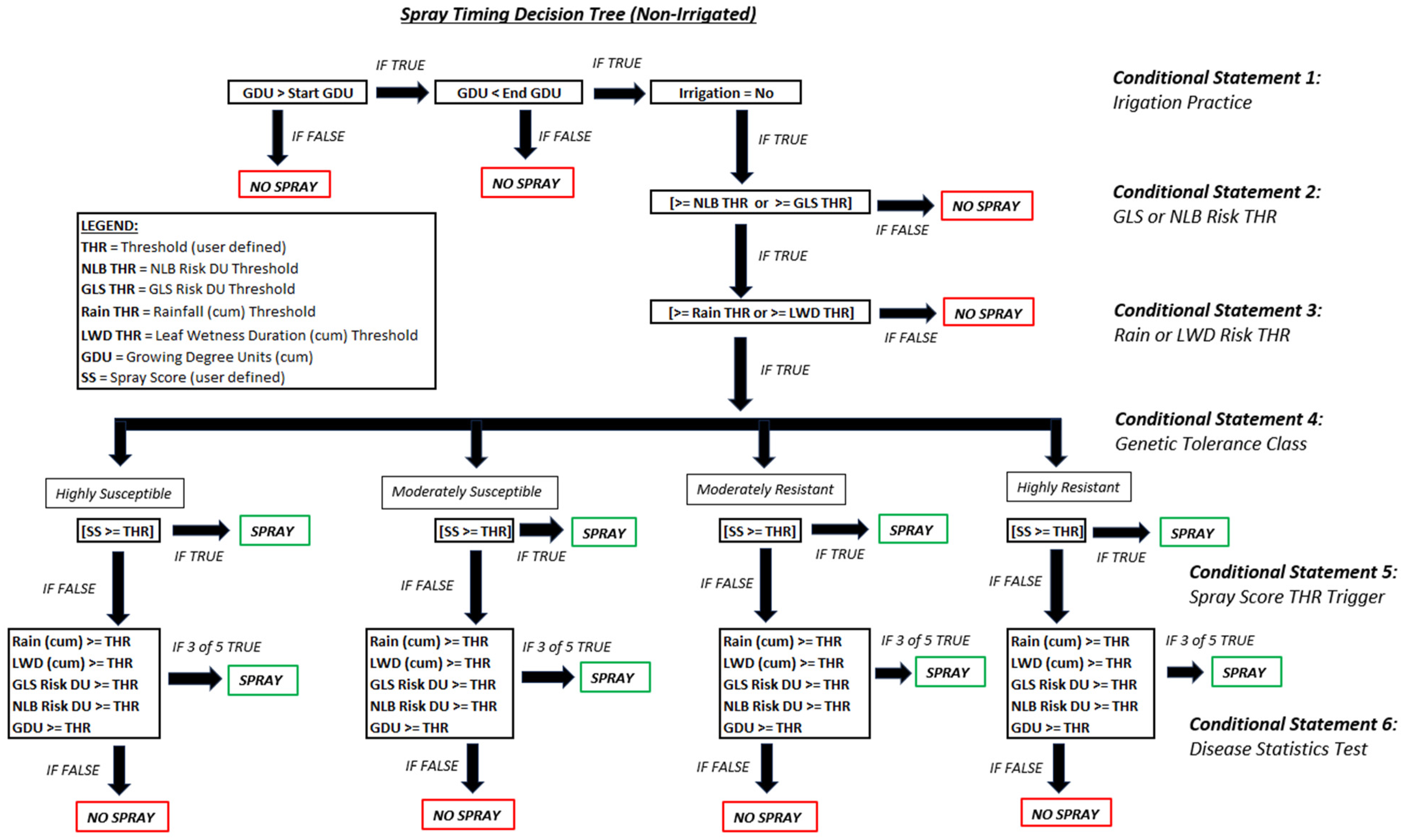
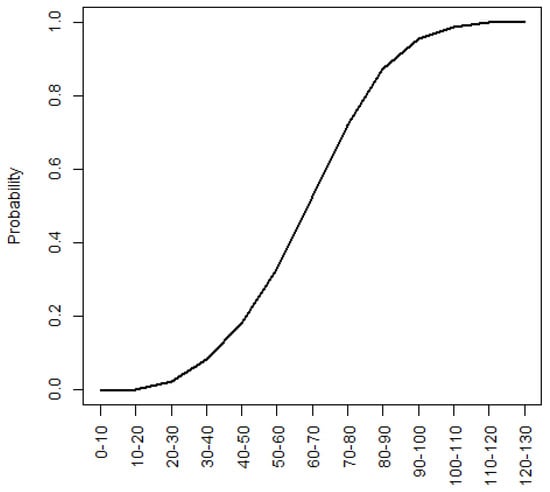
Figure A7.
Probability of NLB infection based on disease units (2019 and 2020 data).
Figure A7.
Probability of NLB infection based on disease units (2019 and 2020 data).
References
- Rowlandson, T.; Gleason, M.; Sentelhas, P.; Gillespie, T.; Thomas, C.; Hornbuckle, B. Reconsidering Leaf Wetness Duration Determination for Plant Disease Management. Plant Dis. 2015, 99, 310–319. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Goudriaan, J. Estimating dew formation in rice, using seasonally averaged diel patterns of weather variables. NJAS Wagening. J. Life Sci. 2004, 51, 391–406. [Google Scholar] [CrossRef]
- Damicone, J.P.; Berggren, G.T.; Snow, J.P. Effect of free moisture on soybean stem canker development. Phytopathology 1987, 77, 1568–1572. [Google Scholar] [CrossRef]
- Grove, G.G.; Madden, L.V.; Ellis, M.A.; Schmitthenner, A.F. Influence of temperature and wetness duration on infection of immature strawberry fruit by Phytophthora cactorum. Phytopathology 1985, 75, 165–169. [Google Scholar] [CrossRef]
- Weiss, A.; Lukens, D.L.; Steadman, J.R. A sensor for the direct measurement of leaf wetness: Construction techniques and testing under controlled conditions. Agric. For. Meteorol. 1988, 43, 241–249. [Google Scholar] [CrossRef]
- Sentelhas, P.C.; Gillespie, T.J.; Batzer, J.C.; Gleason, M.L.; Monteiro, J.E.B.; Pezzopane, J.R.M.; Pedro, M.J. Spatial variability of leaf wetness duration in different crop canopies. Int. J. Biometeorol. 2005, 49, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Pedro, M.J., Jr.; Gillespie, T.J. Estimating dew duration. Agric. Meteorol. 1982, 25, 283–310. [Google Scholar] [CrossRef]
- Magarey, R.D.; Russo, J.M.; Seem, R.C. Simulation of surface wetness with a water budget and energy balance approach. Agric. For. Meteorol. 2006, 139, 373–381. [Google Scholar] [CrossRef]
- Magarey, R.D.; Seem, R.C.; Weiss, A.; Gillespie, T.; Huber, L. Estimating Surface Wetness on Plants. In Micrometeorology in Agricultural Systems; Hatfield, J.L., Baker, J.M., Eds.; American Society of Agronomy: Madison, WI, USA, 2005; Volume 47, pp. 199–226. [Google Scholar]
- Francl, L.J.; Panigrahi, S.; Pahdi, T.; Gillespie, T.J.; Barr, A. Neural network models that predict leaf wetness. Phytopathology 1995, 85, 1128. [Google Scholar]
- Park, J.; Shin, J.Y.; Kim, K.R.; Ha, J.C. Leaf Wetness Duration Models Using Advanced Machine Learning Algorithms: Application to Farms in Gyeonggi Province, South Korea. Water 2019, 11, 1878. [Google Scholar] [CrossRef]
- Wang, H.; Sanchez-Molina, J.A.; Li, M.; Rodríguez Díaz, F. Improving the performance of vegetable leaf wetness duration models in greenhouses using decision tree learning. Water 2019, 11, 158. [Google Scholar] [CrossRef]
- Bregaglio, S.; Donatelli, M.; Confalonieri, R.; Acutis, M.; Orlandini, S. Multi metric evaluation of leaf wetness models for large-area application of plant disease models. Agric. For. Meteorol. 2011, 151, 1163–1172. [Google Scholar] [CrossRef]
- Gleason, M.L.; Duttweiler, K.B.; Batzer, J.C.; Elwynn Taylor, S.; Sentelhas, C.; Boffino, E.; Monteiro, A.; Gillespie, T.J. Obtaining Weather Data for Input to Crop Disease-Warning Systems: Leaf Wetness Duration as a Case Study. Sci. Agric. 2008, 65, 76–87. [Google Scholar] [CrossRef]
- Gleason, M.L.; Taylor, S.E.; Loughin, T.M.; Koehler, K.J. Development and Validation of an Empirical Model to Estimate the Duration of Dew Periods. Plant Dis. 1994, 78, 1011–1016. [Google Scholar] [CrossRef]
- Kim, K.S.; Taylor, S.E.; Gleason, M.L.; Koehler, K.J. Model to enhance site-specific estimation of leaf wetness duration. Plant Dis. 2002, 86, 179–185. [Google Scholar] [CrossRef]
- Kim, K.S.; Taylor, S.E.; Gleason, M.L. Development and validation of a leaf wetness duration model using a fuzzy logic system. Agric. For. Meteorol. 2004, 127, 53–64. [Google Scholar] [CrossRef]
- Krause, R.A.; Massie, L.B. Predictive Systems: Modern Approaches to Disease Control. Annu. Rev. Phytopathol. 1975, 13, 31–47. [Google Scholar] [CrossRef]
- Kang, W.S.; Hong, S.S.; Han, Y.K.; Kim, K.R.; Kim, S.G.; Park, E.W. A web-based information system for plant disease forecast based on weather data at high spatial resolution. Plant Pathol. J. 2010, 26, 37–48. [Google Scholar] [CrossRef]
- Yoshino, R. Ecological studies on the penetration of rice blast fungus, Pyriculariaoryzae into leaf epidermal cells. Bull. Hokuriku Agric. Exp. Stn. 1979, 22, 163–221. [Google Scholar]
- Kassie, B.T.; Onstad, D.W.; Koga, L.; Hart, T.; Clark, R.; van der Heijden, G. Modeling the early phases of epidemics by Phakospora pachyrhizi in Brazilian soybean. Front. Agron. 2023, 5, 1214038. [Google Scholar] [CrossRef]
- de Vallavieille-Pope, C.; Giosue, S.; Munk, L.; Newton, A.C.; Niks, R.E.; Østergård, H.; Pons-Kühnemann, J.; Rossi, V.; Sache, I. Assessment of epidemiological parameters and their use in epidemiological and forecasting models of cereal airborne diseases. Agronomie 2000, 20, 715–727. [Google Scholar] [CrossRef]
- El Jarroudi, M.; Kouadio, L.; Bock, C.H.; El Jarroudi, M.; Junk, J.; Pasquali, M.; Maraite, H.; Delfosse, P. A threshold-based weather model for predicting stripe rust infection in winter wheat. Plant Dis. 2017, 101, 693–703. [Google Scholar] [CrossRef] [PubMed]
- El Jarroudi, M.; Lahlali, R.; Kouadio, L.; Denis, A.; Belleflamme, A.; El Jarroudi, M.; Boulif, M.; Mahyou, H.; Tychon, B. Weather-Based Predictive Modeling of Wheat Stripe Rust Infection in Morocco. Agronomy 2020, 10, 280. [Google Scholar] [CrossRef]
- Dennis, J.I. Temperature and wet-period conditions for infection by Puccinia striiformis f. sp. tritici race 104E137A+. Trans. Br. Mycol. Soc. 1987, 88, 119–121. [Google Scholar] [CrossRef]
- Van der Plank, J.E. Plant Diseases: Epidemics and Control, 3rd ed.; Academic Press: Cambridge, MA, USA; University of Minnesota: Minneapolis, MN, USA, 1963. [Google Scholar]
- Van Maanen, A.; Xu, X.M. Modelling plant disease epidemics. Eur. J. Plant Pathol. 2003, 109, 669–682. [Google Scholar] [CrossRef]
- Hamer, W.B.; Birr, T.; Verreet, J.A.; Duttmann, R.; Klink, H. Spatio-Temporal Prediction of the Epidemic Spread of Dangerous Pathogens Using Machine Learning Methods. ISPRS Int. J. Geo-Inf. 2020, 9, 44. [Google Scholar] [CrossRef]
- Skelsey, P. Forecasting risk of crop disease with anomaly detection algorithms. Phytopathology 2021, 111, 321–332. [Google Scholar] [CrossRef]
- Xu, W.; Wang, Q.; Chen, R. Spatio-temporal prediction of crop disease severity for agricultural emergency management based on recurrent neural networks. GeoInformatica 2018, 22, 363–381. [Google Scholar] [CrossRef]
- Smith, D.R.; White, D.G. Diseases of corn. In Corn and Corn Improvement, 3rd ed.; Sprague, G.F., Dudley, J.W., Eds.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 1988; Volume 18, pp. 687–766. [Google Scholar]
- Lipps, P.E.; Pratt, R.C.; Hakiza, J.J. Interaction of Ht and partial resistance to Exserohilum turcicum in maize. Plant Dis. 1997, 81, 277–282. [Google Scholar] [CrossRef]
- Human, M.P.; Berger, D.K.; Crampton, B.G. Time-Course RNAseq Reveals Exserohilum turcicum Effectors and Pathogenicity Determinants. Front. Microbiol. 2020, 11, 360. [Google Scholar] [CrossRef] [PubMed]
- Levy, Y.; Cohen, Y. Biotic and environmental factors affecting infection of sweet corn with Exserohilum turcicum. Phytopathology 1983, 73, 722–725. [Google Scholar] [CrossRef]
- Bowen, K.L.; Pederson, W. A model of corn growth and disease development for instructional purposes. Plant Dis. 1989, 73, 83–86. [Google Scholar]
- Bhatia, A.; Munkvold, G.P. Relationships of environmental and cultural factors with severity of gray leaf spot on maize. Plant Dis. 2002, 86, 1127–1133. [Google Scholar] [CrossRef]
- Paul, P.A.; Munkvold, G.P. A model-based approach to preplanting risk assessment for gray leaf spot in maize. Phytopathology 2004, 94, 1350–1357. [Google Scholar] [CrossRef]
- Benson, J.M.; Poland, J.A.; Benson, B.M.; Stromberg, E.L.; Nelson, R.J. Resistance to Gray Leaf Spot of Maize: Genetic Architecture and Mechanisms Elucidated through Nested Association Mapping and Near-Isogenic Line Analysis. PLoS Genet. 2015, 11, e1005045. [Google Scholar] [CrossRef] [PubMed]
- Abatzoglou, J.T. Development of gridded surface meteorological data for ecological applications and modelling. Int. J. Climatol. 2013, 33, 121–131. [Google Scholar] [CrossRef]
- Garratt, J.R.; Segal, M. On the contribution of atmospheric moisture to dew formation. Bound.-Layer Meteorol. 1988, 45, 209–236. [Google Scholar] [CrossRef]
- Smith, L.P. Potato blight forecasting by 90 per cent humidity criteria. Plant Pathol. 1956, 5, 83–87. [Google Scholar] [CrossRef]
- Kim, K.R.; Kang, W.S.; Park, E.W. Agricultural Grid Weather Information System based on Digital Weather Forecast in Korea and its Application to Rice Blast Disease Warning. In Proceedings of the International Workshop on the Content, Communication and Use of Weather and Climate Products and Services for Sustainable Agriculture, Queensland, Australia, 18–20 May 2009; National Institute of Meteorological Research, Korea Meteorological Administration, Seoul National University: Seoul, Republic of Korea, 2009. [Google Scholar]
- Huber, L.; Gillespie, T.J. Modeling leaf wetness in relation to plant disease epidemiology. Annu. Rev. Phytopathol. 1992, 30, 553–577. [Google Scholar] [CrossRef]
- Vieira, R.A.; Mesquini, R.M.; Silva, C.N.; Hata, F.T.; Tessmann, D.J.; Scapim, C.A. A new diagrammatic scale for the assessment of northern corn leaf blight. Crop Prot. 2014, 56, 55–57. [Google Scholar] [CrossRef]
- Alsafadi, K.; Alatrach, B.; Sammen, S.S.; Cao, W. Prediction of daily leaf wetness duration using multi-step machine learning. Comput. Electron. Agric. 2024, 224, 109131. [Google Scholar] [CrossRef]
- Webster, R.W.; Nicolli, C.; Allen, T.W.; Bish, M.D.; Bissonnette, K.; Check, J.C.; Chilvers, M.I.; Duffeck, M.R.; Kleczewski, N.; Luis, J.M.; et al. Uncovering the environmental conditions required for Phyllachora maydis infection and tar spot development on corn in the United States for use as predictive models for future epidemics. Sci. Rep. 2023, 13, 17064. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).