Population Structure and Genetic Diversity of Agave Germplasms in China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Isolation
2.2. Marker Development and Amplicon Sequencing Analysis
2.3. Population Structure and Genetic Diversity Analysis
3. Results
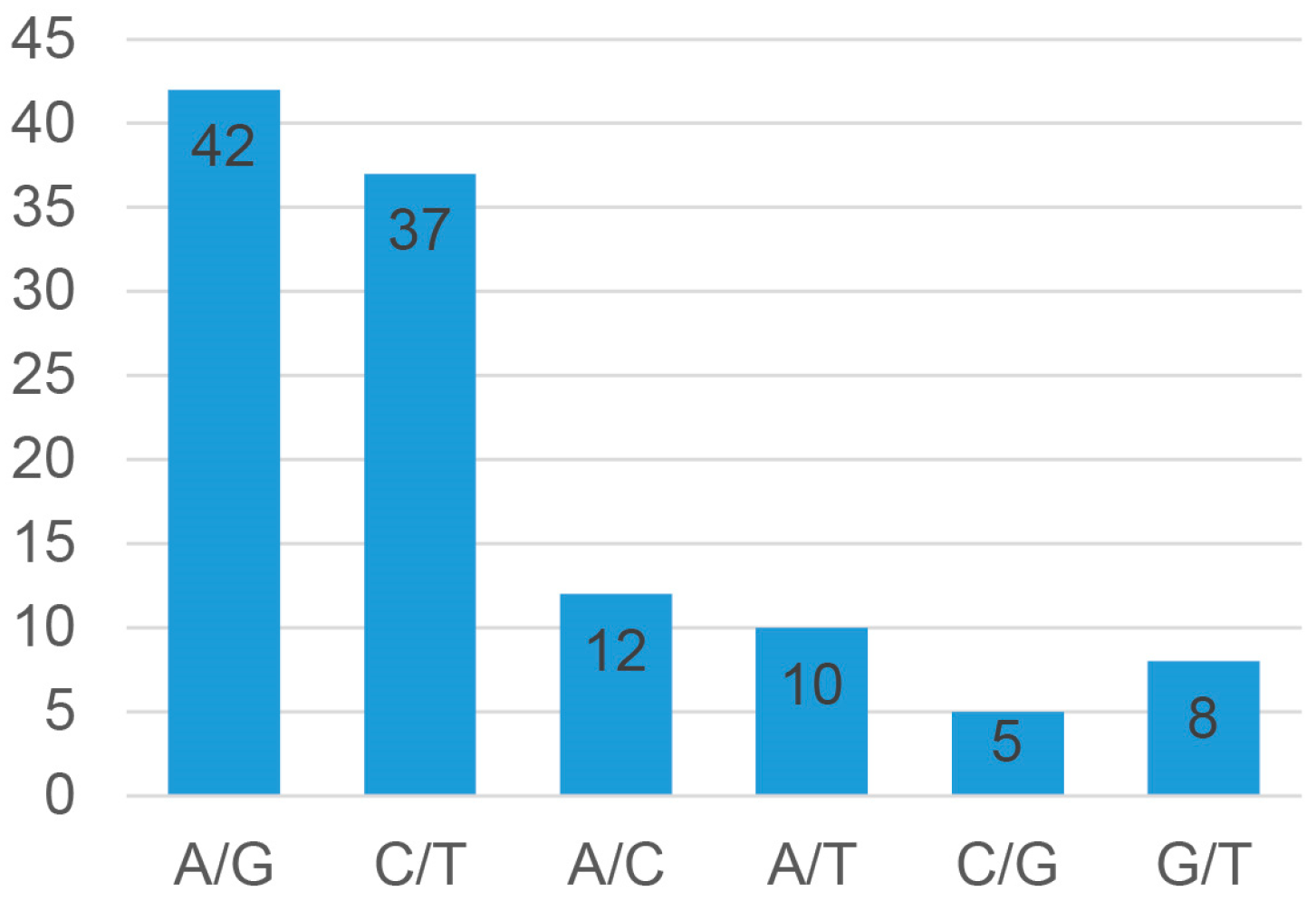
3.1. Genetic Variants by Amplicon Sequencing
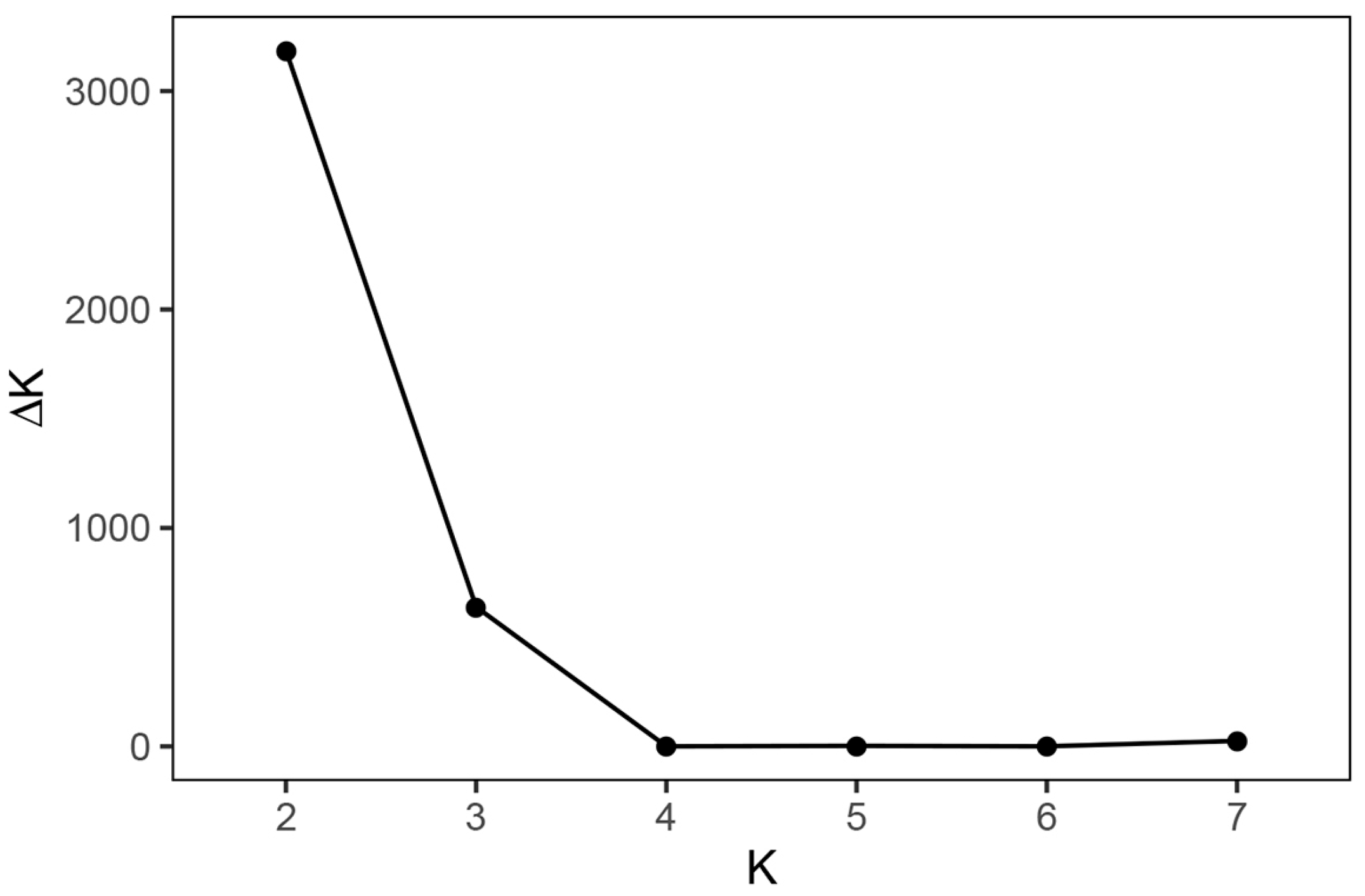
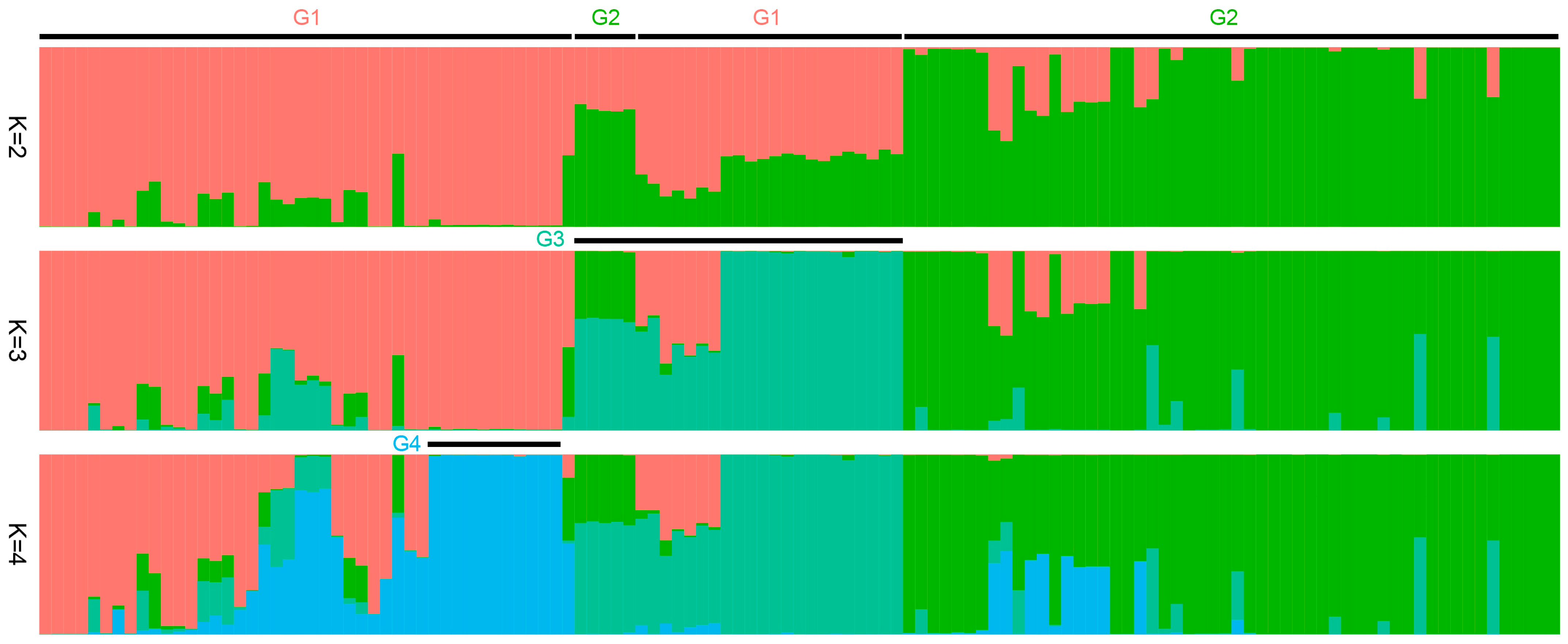
3.2. Population Structure of Agave Germplasms in China
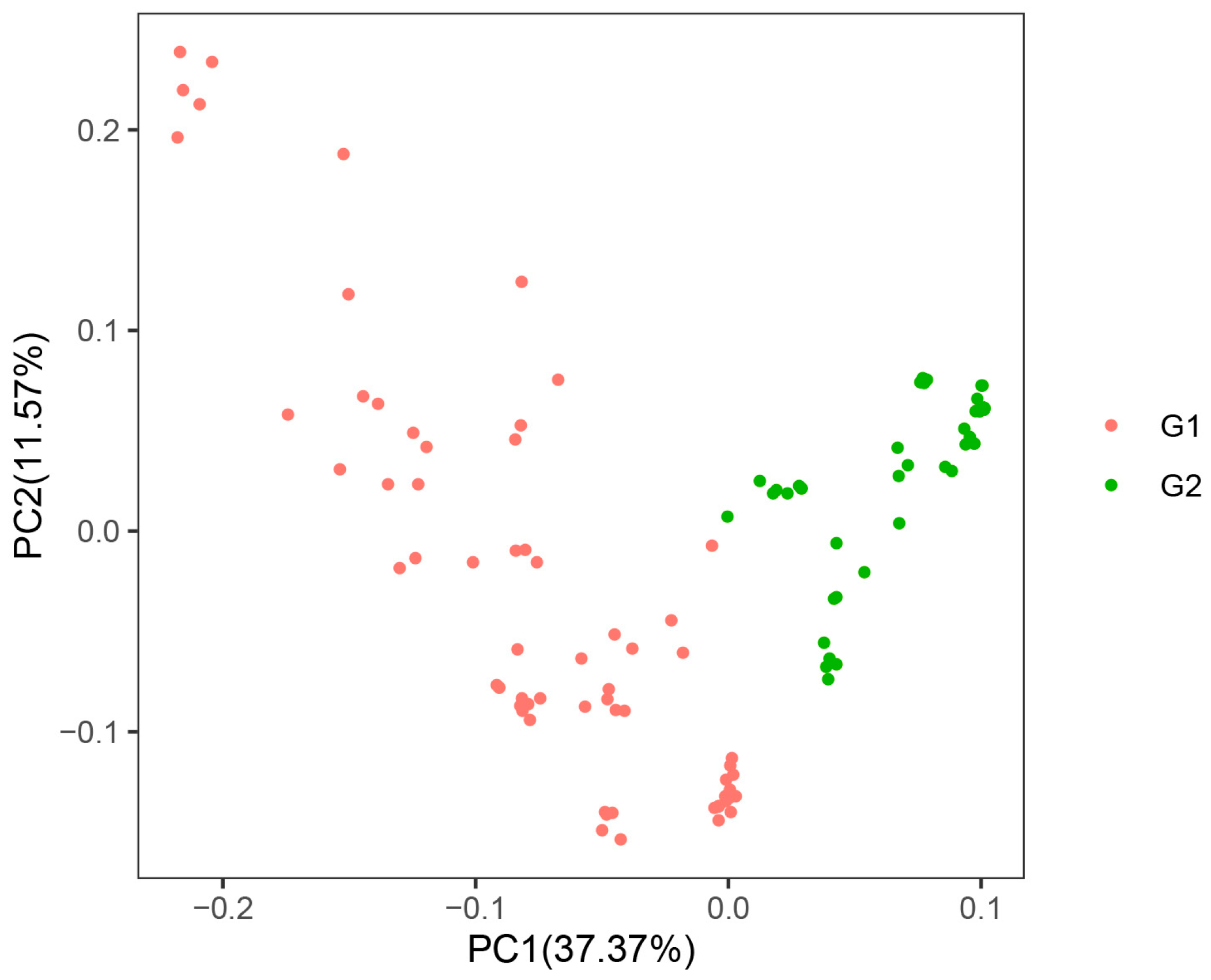
3.3. Genetic Relationships of Agave Germplasms in China
3.4. Genetic Diversity and Differentiation of Subgroups
4. Discussion
4.1. Cost-Effective Characterization of Agave Germplasms
4.2. Agave Breeding in China
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, X.; Xiao, M.; Xi, J.; He, C.; Zheng, J.; Chen, H.; Yi, K. De novo transcriptome assembly of Agave H11648 by Illumina sequencing and identification of cellulose synthase genes in Agave species. Genes 2019, 10, 103. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Zavala, M.L.; Hernández-Arzaba, J.C.; Bideshi, D.K.; Barboza-Corona, J.E. Agave: A natural renewable resource with multiple applications. J. Sci. Food Agric. 2020, 100, 5324–5333. [Google Scholar] [CrossRef] [PubMed]
- Alducin-Martínez, C.; Ruiz Mondragón, K.Y.; Jiménez-Barrón, O.; Aguirre-Planter, E.; Gasca-Pineda, J.; Eguiarte, L.E.; Medellin, R.A. Uses, Knowledge and Extinction Risk Faced by Agave Species in Mexico. Plants 2022, 27, 124. [Google Scholar] [CrossRef] [PubMed]
- Eguiarte, L.E.; Jiménez Barrón, O.A.; Aguirre-Planter, E.; Scheinvar, E.; Gámez, N.; Gasca-Pineda, J.; Castellanos-Morales, G.; Moreno-Letelier, A.; Souza, V. Evolutionary ecology of Agave: Distribution patterns, phylogeny, and coevolution (an homage to Howard S. Gentry). Am. J. Bot. 2021, 108, 216–235. [Google Scholar] [CrossRef]
- Hodgson, W.C.; Jane Rosenthal, E.; Salywon, A.M. Pre-contact Agave domesticates-living legacy plants in Arizona’s landscape. Ann. Bot. 2023, 132, 835–853. [Google Scholar] [CrossRef]
- Colunga-Garcíamarín, P.; Coello-Coello, J.; Eguiarte, L.E.; Piñero, D. Isozymatic variation and phylogenetic relationships between henequen (Agave fourcroydes) and its wild ancestor A. angustifolia (Agavaceae). Am. J. Bot. 1999, 86, 115–123. [Google Scholar] [CrossRef]
- Martínez-Palacios, A.; Eguiarte, L.E.; Furnier, G.R. Genetic diversity of the endangered endemic Agave victoriae-reginae (Agavaceae) in the Chihuahuan Desert. Am. J. Bot. 1999, 86, 1093–1098. [Google Scholar] [CrossRef]
- Silva-Montellano, A.; Eguiarte, L.E. Geographic patterns in the reproductive ecology of Agave lechuguilla (Agavaceae) in the Chihuahuan desert. II. Genetic variation, differentiation, and inbreeding estimates. Am. J. Bot. 2003, 90, 700–706. [Google Scholar] [CrossRef]
- Parker, K.C.; Hamrick, J.L.; Hodgson, W.C.; Trapnell, D.W.; Parker, A.J.; Kuzoff, R.K. Genetic consequences of pre-Columbian cultivation for Agave murpheyi and A. delamateri (Agavaceae). Am. J. Bot. 2007, 94, 1479–1490. [Google Scholar] [CrossRef]
- Figueredo, C.J.; Nassar, J.M. Population genetics of Agave cocui: Evidence for low genetic diversity at the southern geographic limit of genus Agave. J. Hered. 2011, 102, 306–314. [Google Scholar] [CrossRef]
- Parker, K.C.; Trapnell, D.W.; Hamrick, J.L.; Hodgson, W.C. Genetic and morphological contrasts between wild and anthropogenic populations of Agave parryi var. huachucensis in south-eastern Arizona. Ann. Bot. 2014, 113, 939–952. [Google Scholar] [CrossRef]
- Good-Avila, S.V.; Souza, V.; Gaut, B.S.; Eguiarte, L.E. Timing and rate of speciation in Agave (Agavaceae). Proc. Natl. Acad. Sci. USA 2006, 103, 9124–9129. [Google Scholar] [CrossRef] [PubMed]
- Aguirre-Planter, E.; Parra-Leyva, J.G.; Ramírez-Barahona, S.; Scheinvar, E.; Lira-Saade, R.; Eguiarte, L.E. Phylogeography and Genetic Diversity in a Southern North American Desert: Agave kerchovei From the Tehuacán-Cuicatlán Valley, Mexico. Front. Plant Sci. 2020, 11, 863. [Google Scholar] [CrossRef]
- Trejo, L.; Reyes, M.; Cortés-Toto, D.; Romano-Grande, E.; Muñoz-Camacho, L.L. Morphological Diversity and Genetic Relationships in Pulque Production Agaves in Tlaxcala, Mexico, by Means of Unsupervised Learning and Gene Sequencing Analysis. Front. Plant Sci. 2020, 11, 524812. [Google Scholar] [CrossRef] [PubMed]
- Vu, J.P.; Vasquez, M.F.; Feng, Z.; Lombardo, K.; Haagensen, S.; Bozinovic, G. Relative genetic diversity of the rare and endangered Agave shawii ssp. shawii and associated soil microbes within a southern California ecological preserve. Ecol. Evol. 2021, 11, 1829–1842. [Google Scholar]
- Navarro-Quezada, A.; González-Chauvet, R.; Molina-Freaner, F.; Eguiarte, L.E. Genetic differentiation in the Agave deserti (Agavaceae) complex of the Sonoran desert. Heredity 2003, 90, 220–227. [Google Scholar] [CrossRef]
- Vargas-Ponce, O.; Zizumbo-Villarreal, D.; Martínez-Castillo, J.; Coello-Coello, J.; Colunga-Garcíamarín, P. Diversity and structure of landraces of Agave grown for spirits under traditional agriculture: A comparison with wild populations of A. angustifolia (Agavaceae) and commercial plantations of A. tequilana. Am. J. Bot. 2009, 96, 448–457. [Google Scholar] [PubMed]
- Trejo, L.; Alvarado-Cárdenas, L.O.; Scheinvar, E.; Eguiarte, L.E. Population genetic analysis and bioclimatic modeling in Agave striata in the Chihuahuan Desert indicate higher genetic variation and lower differentiation in drier and more variable environments. Am. J. Bot. 2016, 103, 1020–1029. [Google Scholar] [CrossRef]
- Gao, J.; Luoping; Guo, C.; Li, J.; Liu, Q.; Chen, H.; Yi, K. AFLP analysis and zebra disease resistance identification of 40 sisal genotypes in China. Mol. Biol. Rep. 2012, 39, 6379–6385. [Google Scholar] [CrossRef]
- Chávez-Sánchez, C.; Mancilla-Margalli, N.A.; Montero-Cortés, M.I.; Gutiérrez-Miceli, F.A.; Briceño-Félix, G.A.; Simpson Williamson, J.K.; Avila-Miranda, M.E. Asexually propagated Agave tequilana var. azul exhibits variation in genetic markers and defence responses to Fusarium solani. AoB Plants 2022, 14, plac027. [Google Scholar] [CrossRef]
- Lindsay, D.L.; Edwards, C.E.; Jung, M.G.; Bailey, P.; Lance, R.F. Novel microsatellite loci for Agave parryi and cross-amplification in Agave palmeri (Agavaceae). Am. J. Bot. 2012, 99, e295–e297. [Google Scholar] [CrossRef] [PubMed]
- Byers, C.; Maughan, P.J.; Clouse, J.; Stewart, J.R. Microsatellite primers in Agave utahensis (Asparagaceae), a keystone species in the Mojave Desert and Colorado Plateau. Appl. Plant Sci. 2014, 2, apps.1400047. [Google Scholar] [CrossRef]
- Figueredo-Urbina, C.J.; Casas, A.; Torres-García, I. Morphological and genetic divergence between Agave inaequidens, A. cupreata and the domesticated A. hookeri. Analysis of their evolutionary relationships. PLoS ONE 2017, 12, e0187260. [Google Scholar] [CrossRef] [PubMed]
- Cabrera-Toledo, D.; Vargas-Ponce, O.; Ascencio-Ramírez, S.; Valadez-Sandoval, L.M.; Pérez-Alquicira, J.; Morales-Saavedra, J.; Huerta-Galván, O.F. Morphological and Genetic Variation in Monocultures, Forestry Systems and Wild Populations of Agave maximiliana of Western Mexico: Implications for Its Conservation. Front. Plant Sci. 2020, 11, 817. [Google Scholar] [CrossRef]
- Álvarez-Ríos, G.D.; Pacheco-Torres, F.; Figueredo-Urbina, C.J.; Casas, A. Management, morphological and genetic diversity of domesticated agaves in Michoacán, México. J. Ethnobiol. Ethnomed. 2020, 16, 3. [Google Scholar] [CrossRef]
- Figueredo-Urbina, C.J.; Álvarez-Ríos, G.D.; García-Montes, M.A.; Octavio-Aguilar, P. Morphological and genetic diversity of traditional varieties of agave in Hidalgo State, Mexico. PLoS ONE 2021, 16, e0254376. [Google Scholar] [CrossRef]
- Huang, X.; Feng, Q.; Qian, Q.; Zhao, Q.; Wang, L.; Wang, A.; Guan, J.; Fan, D.; Weng, Q.; Huang, T.; et al. High-throughput genotyping by whole-genome resequencing. Genome Res. 2009, 19, 1068–1076. [Google Scholar] [CrossRef]
- Deng, G.; Huang, X.; Xie, L.; Tan, S.; Gbokie, T.J.; Bao, Y.; Yi, K. Identification and expression of SAUR genes in the CAM Plant Agave. Genes 2019, 10, 555. [Google Scholar] [CrossRef]
- Cabrera-Toledo, D.; Mendoza-Galindo, E.; Larranaga, N.; Herrera-Estrella, A.; Vásquez-Cruz, M.; Hernández-Hernández, T. Genomic and Morphological Differentiation of Spirit Producing Agave angustifolia Traditional Landraces Cultivated in Jalisco, Mexico. Plants 2022, 11, 2274. [Google Scholar] [CrossRef]
- Ruiz Mondragon, K.Y.; Aguirre-Planter, E.; Gasca-Pineda, J.; Klimova, A.; Trejo-Salazar, R.E.; Reyes Guerra, M.A.; Medellin, R.A.; Piñero, D.; Lira, R.; Eguiarte, L.E. Conservation genomics of Agave tequilana Weber var. azul: Low genetic differentiation and heterozygote excess in the tequila agave from Jalisco, Mexico. PeerJ 2022, 10, e14398. [Google Scholar] [CrossRef]
- Klimova, A.; Ruiz Mondragón, K.Y.; Aguirre-Planter, E.; Valiente, A.; Lira, R.; Eguiarte, L.E. Genomic analysis unveils reduced genetic variability but increased proportion of heterozygotic genotypes of the intensively managed mezcal agave, Agave angustifolia. Am. J. Bot. 2023, 110, e16216. [Google Scholar] [CrossRef]
- Ruiz Mondragón, K.Y.; Klimova, A.; Aguirre-Planter, E.; Valiente-Banuet, A.; Lira, R.; Sanchez-de la Vega, G.; Eguiarte, L.E. Differences in the genomic diversity, structure, and inbreeding patterns in wild and managed populations of Agave potatorum Zucc. used in the production of Tobalá mezcal in Southern Mexico. PLoS ONE 2023, 18, e0294534. [Google Scholar] [CrossRef] [PubMed]
- Klimova, A.; Gutíerrez-Rivera, J.; Ortega-Rubio, A.; Eguiarte, L.E. Population genomics and distribution modeling revealed the history and suggested a possible future of the endemic Agave aurea (Asparagaceae) complex in the Baja California Peninsula. Ecol. Evol. 2024, 14, e70027. [Google Scholar] [CrossRef]
- Huang, X.; Hu, X.; Liu, Q.; Xie, Z.; Tan, S.; Qin, X.; Yi, K. Full-length agave transcriptome reveals candidate glycosyltransferase genes involved in hemicellulose biosynthesis. Int. J. Biol. Macromol. 2024, 274, 133508. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; He, R.; Tan, S.; Wu, W.; Xi, J.; Liang, Y.; Chen, T.; Huang, X.; Yi, K. Development status, problems and countermeasures of sisal industry in China. Trop. Agric. Eng. 2023, 47, 18–21. (In Chinese) [Google Scholar]
- Huang, X.; Wang, B.; Xi, J.; Zhang, Y.; He, C.; Zheng, J.; Yi, K. Transcriptome comparison reveals distinct selection patterns in domesticated and wild Agave species, the important CAM plants. Int. J. Genom. 2018, 2018, 5716518. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Xu, B.; Tan, S.; Huang, Y.; Xi, J.; Qin, X.; Chen, T.; Chen, H.; Yang, X.; Yi, K. Transcriptome sequencing of Agave angustifolia reveals conservation and diversification in the expression of cinnamyl alcohol dehydrogenase genes in Agave species. Agriculture 2022, 12, 1003. [Google Scholar] [CrossRef]
- Wang, X.; Hu, X.; Lin, C.; Liu, Q.; Li, Y.; Du, D.; Mkapa, D.; Zhang, W.; Huang, X.; Yi, K. Agave schidigera Transcriptome Reveals Stress-Responsive Phenylalanine ammonia-lyase Genes in Agave. Agronomy 2024, 14, 2520. [Google Scholar] [CrossRef]
- Gao, J.; Yang, F.; Zhang, S.; Li, J.; Chen, H.; Liu, Q.; Zheng, J.; Xi, J.; Yi, K. Expression of a hevein-like gene in transgenic Agave hybrid No. 11648 enhances tolerance against zebra stripe disease. Plant Cell Tiss. Organ. Cult. 2014, 119, 579–585. [Google Scholar] [CrossRef]
- Lu, Z.; Hou, X.; Ke, Z.; Zhang, Y.; Yang, Z.; Zhou, W. A newly identified glycosyltransferase AsRCOM provides resistance to purple curl leaf disease in agave. BMC Genom. 2023, 24, 669. [Google Scholar] [CrossRef]
- Campbell, N.R.; Harmon, S.A.; Narum, S.R. Genotyping-in-Thousands by sequencing (GT-seq): A cost effective SNP genotyping method based on custom amplicon sequencing. Mol. Ecol. Resour. 2015, 15, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J.L. A Rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Chen, S. Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. iMeta 2023, 2, e107. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; vonHoldt, B.M. STRUCTUREHARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2011, 4, 359–361. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Xu, S.; Li, L.; Luo, X.; Chen, M.; Tang, W.; Zhan, L.; Dai, Z.; Lam, T.T.; Guan, Y.; Yu, G. Ggtree: A serialized data object for visualization of a phylogenetic tree and annotation data. iMeta 2022, 1, e56. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Grover, A.; Sharma, P.C. Development and use of molecular markers: Past and present. Crit. Rev. Biotechnol. 2016, 36, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Hasan, N.; Choudhary, S.; Naaz, N.; Sharma, N.; Laskar, R.A. Recent advancements in molecular marker-assisted selection and applications in plant breeding programmes. J. Genet. Eng. Biotechnol. 2021, 19, 128. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.J.; Russell, J.; Ramsay, L.; Thomas, W.; Powell, W.; Mackay, I. Overcoming barriers to the registration of new plant varieties under the DUS system. Commun. Biol. 2021, 4, 302. [Google Scholar] [CrossRef]
- Gai, Z.; Zhai, J.; Chen, X.; Jiao, P.; Zhang, S.; Sun, J.; Qin, R.; Liu, H.; Wu, Z.; Li, Z. Phylogeography Reveals Geographic and Environmental Factors Driving Genetic Differentiation of Populus sect. Turanga in Northwest China. Front. Plant Sci. 2021, 12, 705083. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, X.; Li, Y.; Tan, S.; Chen, L.; Mkapa, D.S.; Lin, C.; Liu, Q.; Jin, G.; Chen, T.; Qin, X.; et al. Population Structure and Genetic Diversity of Agave Germplasms in China. Agronomy 2025, 15, 722. https://doi.org/10.3390/agronomy15030722
Hu X, Li Y, Tan S, Chen L, Mkapa DS, Lin C, Liu Q, Jin G, Chen T, Qin X, et al. Population Structure and Genetic Diversity of Agave Germplasms in China. Agronomy. 2025; 15(3):722. https://doi.org/10.3390/agronomy15030722
Chicago/Turabian StyleHu, Xiaoli, Yubo Li, Shibei Tan, Lisha Chen, Dietram Samson Mkapa, Chen Lin, Qingqing Liu, Gang Jin, Tao Chen, Xu Qin, and et al. 2025. "Population Structure and Genetic Diversity of Agave Germplasms in China" Agronomy 15, no. 3: 722. https://doi.org/10.3390/agronomy15030722
APA StyleHu, X., Li, Y., Tan, S., Chen, L., Mkapa, D. S., Lin, C., Liu, Q., Jin, G., Chen, T., Qin, X., Yi, K., & Huang, X. (2025). Population Structure and Genetic Diversity of Agave Germplasms in China. Agronomy, 15(3), 722. https://doi.org/10.3390/agronomy15030722




