Integrative Analysis of the Methylome, Transcriptome, and Proteome Reveals a New Mechanism of Rapeseed Under Freezing Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Samples and Freezing Stress Treatments
2.2. Overwintering Rate, Physiological and Biochemical Determination
2.3. Determination of Photosynthetic Parameters and Observations Using a Transmission Electron Microscope
2.4. DNA Extraction, DNA Methylation Sequencing, and Data Analysis
2.5. RNA Extraction, RNA Sequencing, and Data Filtering
2.6. Protein Extraction, Digestion, Mass Spectrometry Analysis, and Data Analysis
2.7. Functional Annotation and Statistical Analysis
3. Result
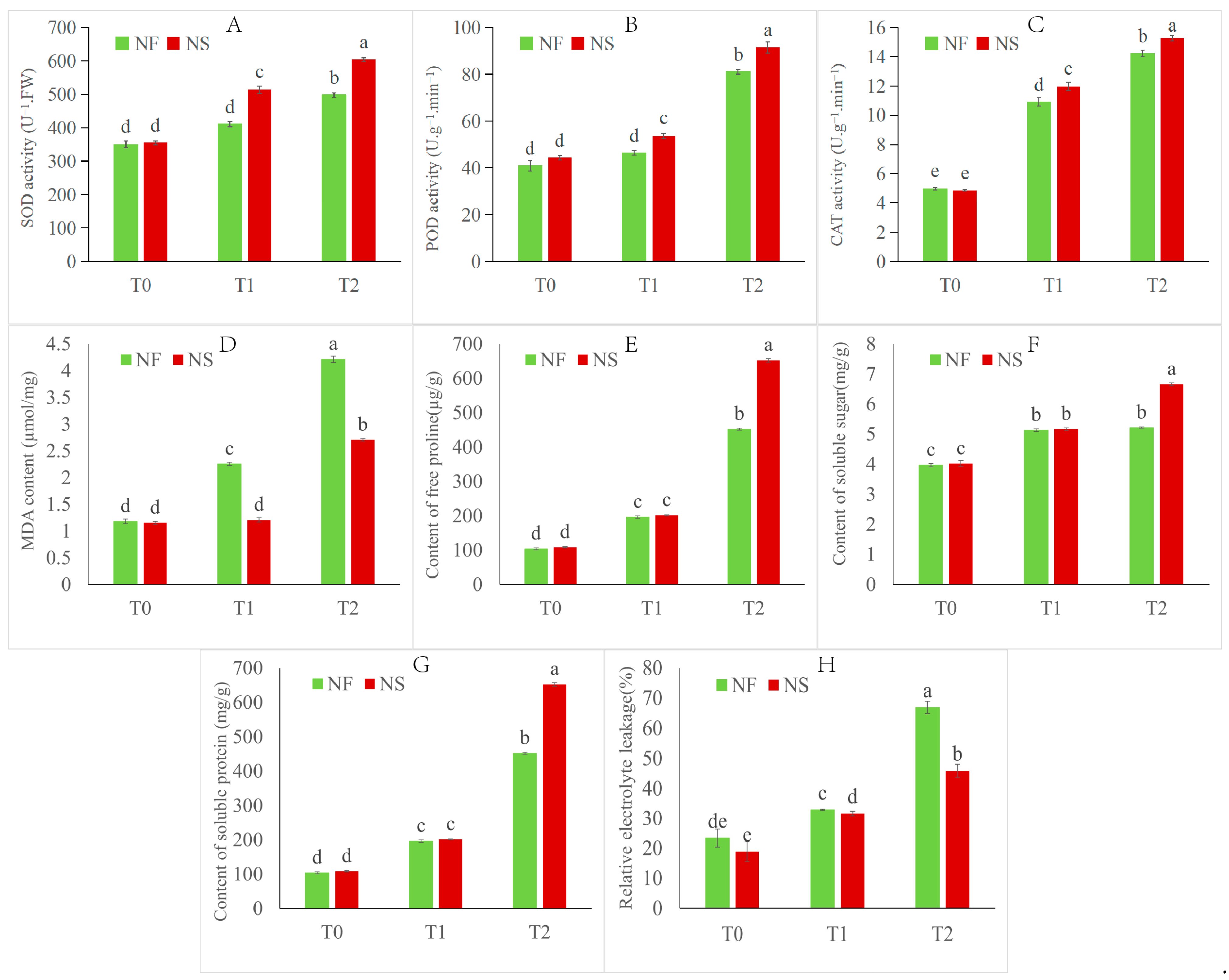
3.1. Morphological, Overwintering Rate, Physiological and Biochemical Responses of Rapeseed to Freezing Stress
3.2. Responses of Photosynthetic Parameters, Microstructure of Rapeseed to Freezing Stress
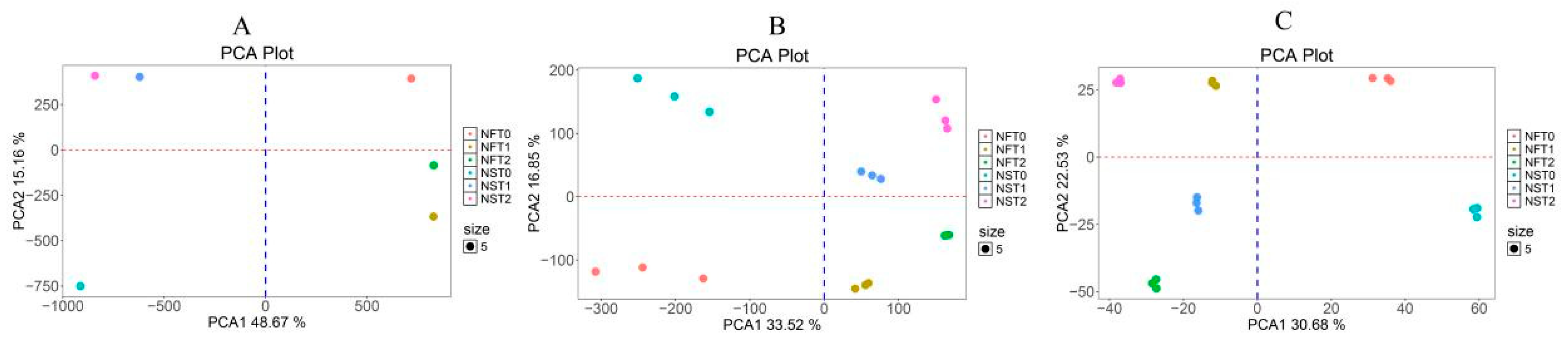
3.3. Quality Control Analysis of the Rapeseed Methylome, Transcriptome, and Proteome
3.4. The DMGs, DEGs, and DAPs Were Identified Between Freezing-Resistant and Freezing-Sensitive Cultivars
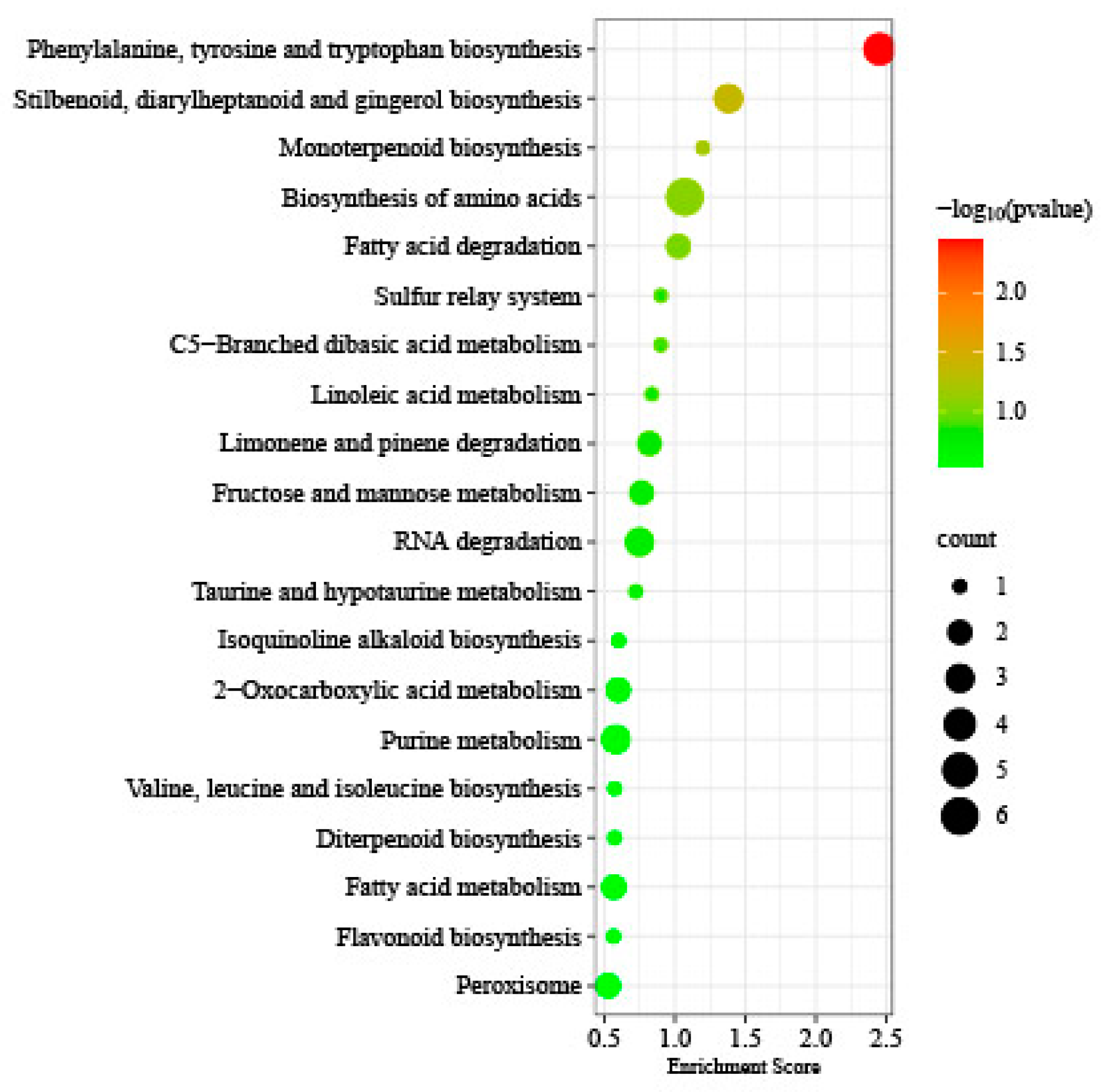
3.5. KEGG Analysis of DMGs, DEGs, and DAPs Under Freezing Stress
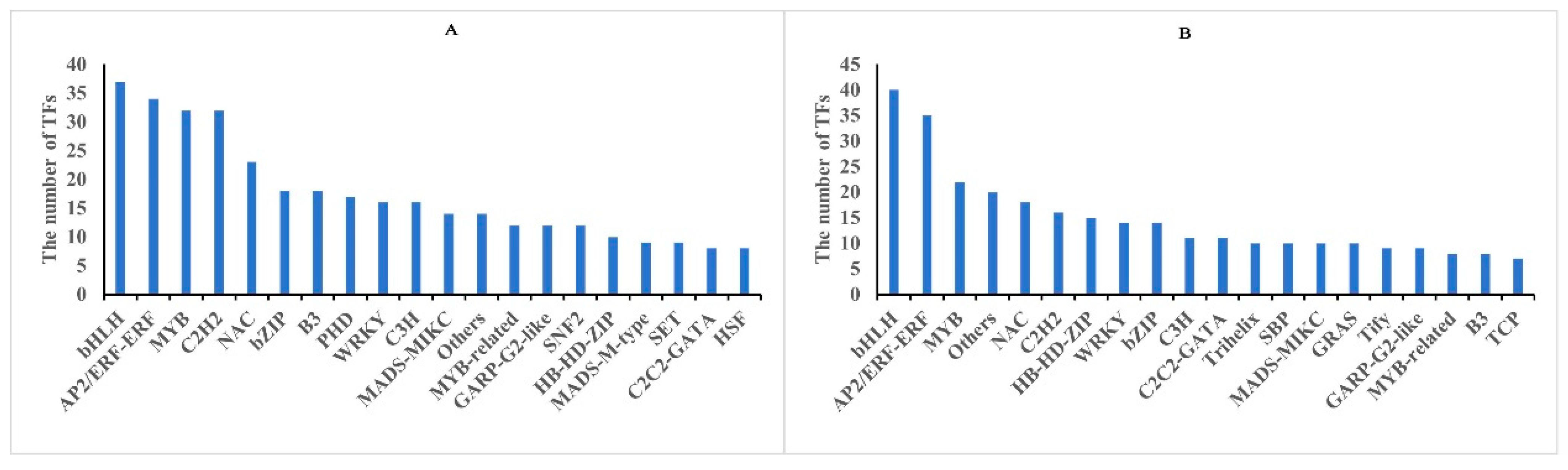
3.6. Identification and Analysis of TFs for Core DMGs, DEGs, and DAPs
3.7. Integrative Analysis of Methylome, Transcriptome, and Proteome
4. Discussion
4.1. Cold Signal Transduction in Rapeseed Under Freezing Stress
4.2. Osmotic Balance Plays an Essential Role in Rapeseed Under Freezing Stress
4.3. Redox and Folate Homeostasis Were Critical Under Freezing Stress in Rapeseed
4.4. The Stability of the Membrane Structure and the Integrity of the Photosynthetic System Are Crucial for the Freezing-Resistant Stress in Rapeseed
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, H. New-demand oriented oilseed rape industry developing strategy. Oil Crop Sci. 2018, 40, 613–617. [Google Scholar]
- Cheng, L.; Jie, H.; Bofeng, L.; Zhongchao, F.; Junpeng, L. Current situation, development difficulties and suggestions of chinese rape industry. J. China Agric. Univ. 2017, 22, 203–210. [Google Scholar]
- Liu, Z.; Dong, X.; Cao, X.; Xu, C.; Wei, J.; Zheng, G.; Wang, J.; Li, H.; Fang, X.; Wang, Y.; et al. QTL mapping for cold tolerance and higher overwintering survival rate in winter rapeseed (Brassica napus). J. Plant Physiol. 2022, 275, 153735. [Google Scholar] [CrossRef]
- Ding, Y.; Shi, Y.; Yang, S. Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 2019, 222, 1690–1704. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Zheng, G.; Yu, X.; Liu, S.; Dong, X.; Cao, X.; Fang, X.; Li, H.; Jin, J.; Mi, W.; et al. Comparative Transcriptomics and Proteomics Analyses of Leaves Reveals a Freezing Stress-Responsive Molecular Network in Winter Rapeseed (Brassica rapa L.). Front. Plant Sci. 2021, 12, 664311. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Liu, Z.; Mi, W.; Xu, C.; Zou, Y.; Xu, M.; Zheng, G.; Fang, X.; Cui, X.; Dong, X.; et al. Analysis on the Adaptability of Northwand Planting of Brassica napus. Sci. Agric. Sin. 2020, 53, 4164–4176. [Google Scholar]
- Chang, Y.; Zhu, C.; Jiang, J.; Zhang, H.; Zhu, J.; Duan, C. Epigenetic regulation in plant abiotic stress responses. J. Integr. Plant Biol. 2020, 62, 563–580. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, X.; Guo, L.; Qi, T.; Liu, G.; Feng, J.; Shahzad, K.; Zhang, B.; Li, X.; Wang, H.; et al. Single-base resolution methylome of cotton cytoplasmic male sterility system reveals epigenomic changes in response to high-temperature stress during anther development. J. Exp. Bot. 2020, 71, 951–969. [Google Scholar] [CrossRef]
- Seymour, D.K.; Becker, C. The causes and consequences of DNA methylome variation in plants. Curr. Opin. Plant Biol. 2017, 36, 56–63. [Google Scholar] [CrossRef]
- Lämke, J.; Bäurle, I. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 2017, 18, 124. [Google Scholar] [CrossRef]
- Ahmad, F.; Farman, K.; Waseem, M.; Rana, R.M.; Nawaz, M.A.; Rehman, H.M.; Abbas, T.; Baloch, F.S.; Akrem, A.; Huang, J.; et al. Genome-wide identification, classification, expression profiling and DNA methylation (5mC) analysis of stress-responsive ZFP transcription factors in rice (Oryza sativa L.). Gene 2019, 718, 144018. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Li, R.; Huang, J.; Zhao, H.; Ge, R.; Wu, Q.; Mallano, A.I.; Wang, Y.; Li, F.; Deng, Y.; et al. Divergent DNA methylation contributes to duplicated gene evolution and chilling response in tea plants. Plant J. 2021, 106, 1312–1327. [Google Scholar] [CrossRef]
- Zheng, G.; Dong, X.; Wei, J.; Liu, Z.; Aslam, A.; Cui, J.; Li, H.; Wang, Y.; Tian, H.; Cao, X. Integrated methylome and transcriptome analysis unravel the cold tolerance mechanism in winter rapeseed (Brassica napus L.). BMC Plant Biol. 2022, 22, 414. [Google Scholar] [CrossRef]
- Wei, J.; Shen, Y.; Dong, X.; Zhu, Y.; Cui, J.; Li, H.; Zheng, G.; Tian, H.; Wang, Y.; Liu, Z. DNA methylation affects freezing tolerance in winter rapeseed by mediating the expression of genes related to JA and CK pathways. Front. Genet. 2022, 13, 968494. [Google Scholar] [CrossRef]
- Hassan, S.A.-Z.; Hesham, F.A.; Shah, F. Antioxidative Defense System, Hormones, and Metabolite Accumulation in Different Plant Parts of Two Contrasting Rice Cultivars as Influenced by Plant Growth Regulators Under Heat Stress. Front. Plant Sci. 2022, 13, 911846. [Google Scholar] [CrossRef]
- Rongmiao, H.; Lizhi, Y.; Tana, W.; Shiyao, C.; Lu, Z. Genes related to osmoregulation and antioxidation play important roles in the response of Trollius chinensis seedlings to saline-alkali stress. Front. Plant Sci. 2023, 14, 1080504. [Google Scholar] [CrossRef]
- Wei, J.; Zheng, G.; Dong, X.; Li, H.; Liu, S.; Wang, Y.; Liu, Z. Integration of transcriptome and proteome analysis reveals the mechanism of freezing tolerance in winter rapeseed. Plant Growth Regul. 2022, 96, 103–118. [Google Scholar] [CrossRef]
- Mi, W.; Liu, Z.; Jin, J.; Dong, X.; Xu, C.; Zou, Y.; Xu, M.; Zheng, G.; Cao, X.; Fang, X.; et al. Comparative proteomics analysis reveals the molecular mechanism of enhanced cold tolerance through ROS scavenging in winter rapeseed (Brassica napus L.). PLoS ONE 2021, 16, e0243292. [Google Scholar] [CrossRef]
- Xu, Y.; Zeng, X.; Wu, J.; Zhang, F.; Li, C.; Jiang, J.; Wang, Y.; Sun, W. iTRAQ-Based Quantitative Proteome Revealed Metabolic Changes in Winter Turnip Rape (Brassica rapa L.) under Cold Stress. Int. J. Mol. Sci. 2018, 19, 3346. [Google Scholar] [CrossRef]
- Gusain, S.; Joshi, S.; Joshi, R. Sensing, signalling, and regulatory mechanism of cold-stress tolerance in plants. Plant Physiol. Biochem. PPB 2023, 197, 107646. [Google Scholar] [CrossRef]
- Zhu, J. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Hwarari, D.; Guan, Y.; Ahmad, B.; Movahedi, A.; Tian, M.; Hao, Z.; Lu, Y.; Chen, J.; Yang, L. ICE-CBF-COR Signaling Cascade and Its Regulation in Plants Responding to Cold Stress. Int. J. Mol. Sci. 2022, 23, 1549. [Google Scholar] [CrossRef]
- Waititu, J.K.; Cai, Q.; Sun, Y.; Sun, Y.; Li, C.; Zhang, C.; Liu, J.; Wang, H. Transcriptome Profiling of Maize (Zea mays L.) Leaves Reveals Key Cold-Responsive Genes, Transcription Factors, and Metabolic Pathways Regulating Cold Stress Tolerance at the Seedling Stage. Genes 2021, 12, 1638. [Google Scholar] [CrossRef]
- Waadt, R.; Seller, C.A.; Hsu, P.K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef]
- Wang, X.; Deng, Y.; Gao, L.; Kong, F.; Shen, G.; Duan, B.; Wang, Z.; Mei, D.; Han, Z. Series-temporal transcriptome profiling of cotton reveals the response mechanism of phosphatidylinositol signaling system in the early stage of drought stress. Genomics 2022, 114, 110465. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Gu, J.; Cui, X.; Fu, H.; Wang, F.; Qi, M.; Sun, Z.; Li, T.; Liu, Y. Genome-wide investigation of the phospholipase C gene family in Solanum lycopersicum and abiotic stress analysis. Environ. Exp. Bot. 2023, 210, 105336. [Google Scholar] [CrossRef]
- Yuan, P.; Yang, T.; Poovaiah, B.W. Calcium Signaling-Mediated Plant Response to Cold Stress. Int. J. Mol. Sci. 2018, 19, 3896. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Zhu, Q.; Yuan, P.; Yan, Y.; Yi, K.; Du, L. Calmodulin and calmodulin-like protein-mediated plant responses to biotic stresses. Plant Cell Environ. 2023, 46, 3680–3703. [Google Scholar] [CrossRef]
- Tarkowski, Ł.P.; Van den Ende, W. Cold tolerance triggered by soluble sugars: A multifaceted countermeasure. Front. Plant Sci. 2015, 6, 203. [Google Scholar] [CrossRef]
- Li, C.; Wan, Y.; Shang, X.; Fang, S. Integration of transcriptomic and metabolomic analysis unveils the response mechanism of sugar metabolism in Cyclocarya paliurus seedlings subjected to PEG-induced drought stress. Plant Physiol. Biochem. PPB 2023, 201, 107856. [Google Scholar] [CrossRef]
- Shen, T.; Li, K.; Yan, R.; Xu, F.; Ni, L.; Jiang, M. The UDP-glucuronic acid decarboxylase OsUXS3 regulates Na + ion toxicity tolerance under salt stress by interacting with OsCATs in rice. Plant Physiol. Biochem. PPB 2023, 196, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Khairudin, N.B.A.; Mazlan, N.S.F. Molecular docking study of Beta-glucosidase with cellobiose, cellotetraose and cellotetriose. Bioinformation 2013, 9, 813–817. [Google Scholar] [CrossRef]
- James, R.K.C.; Karunambigai, A.; Jong-Seong, J.; Jisnuson, S. Functions of rice beta-glucosidases and transglucosidases. Scienceasia 2023, 49, 635. [Google Scholar] [CrossRef]
- Yang, C.; Li, X.; Zhang, Y.; Jiang, H. Transcriptome analysis of Populus × canadensis ’Zhongliao1’ in response to low temperature stress. BMC Genom. 2023, 24, 77. [Google Scholar] [CrossRef]
- Sun, S.; Lin, M.; Qi, X.; Chen, J.; Gu, H.; Zhong, Y.; Sun, L.; Abid, M.; Bai, D.; Hu, C.; et al. Full-length transcriptome profiling reveals insight into the cold response of two kiwifruit genotypes (A. arguta) with contrasting freezing tolerances. BMC Plant Biol. 2021, 21, 365. [Google Scholar] [CrossRef]
- María Elena, A.; Arnould, S.; László, S. Proline metabolism as regulatory hub. Trends Plant Sci. 2022, 27, 39–55. [Google Scholar] [CrossRef]
- Baxter, A.J.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef]
- Xu, D.; Hu, M.; Wang, Y.; Cui, Y. Antioxidant Activities of Quercetin and Its Complexes for Medicinal Application. Molecules 2019, 24, 1123. [Google Scholar] [CrossRef]
- Wei, M.; Xu, L.; Du, Z.; Wang, F.; Zhang, R.; Song, X.; Lam, S.; Shui, G.; Li, Y.; Chye, M.-L. RICE ACYL-COA-BINDING PROTEIN6 Affects Acyl-CoA Homeostasis and Growth in Rice. Rice 2020, 13, 75. [Google Scholar] [CrossRef]
- Zaynab, M.; Peng, J.; Sharif, Y.; Fatima, M.; Albaqami, M.; Al-Yahyai, R.; Raza, A.; Khan, K.A.; Alotaibi, S.S.; Alaraidh, L.A.; et al. Genome-Wide Identification and Expression Profiling of Germin-Like Proteins Reveal Their Role in Regulating Abiotic Stress Response in Potato. Front. Plant Sci. 2021, 12, 831140. [Google Scholar] [CrossRef]
- Anum, J.; O’Shea, C.; Zeeshan Hyder, M.; Farrukh, S.; Skriver, K.; Malik, S.I.; Yasmin, T. Germin like protein genes exhibit modular expression during salt and drought stress in elite rice cultivars. Mol. Biol. Rep. 2022, 49, 293–302. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Tao, M.; Leung, D.W.M.; Yan, X.; Chen, L.; Peng, X.; Liu, E.-E. The rice germin-like protein OsGLP1 participates in acclimation to UV-B radiation. Plant Physiol. 2021, 186, 1254–1268. [Google Scholar] [CrossRef]
- Gangadhar, B.H.; Mishra, R.K.; Kappachery, S.; Venkidasamy, B.; Venkatesh, J.; Nookaraju, A.; Thiruvengadam, M. Enhanced thermo-tolerance in transgenic potato (Solanum tuberosum L.) overexpressing hydrogen peroxide-producing germin-like protein (GLP). Genomics 2021, 113, 3224–3234. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Ge, L.; Ye, X.; Xu, L.; Si, W.; Ding, T. ZmGLP1, a Germin-like Protein from Maize, Plays an Important Role in the Regulation of Pathogen Resistance. Int. J. Mol. Sci. 2022, 23, 14316. [Google Scholar] [CrossRef] [PubMed]
- Tariq, A.A.; Giuseppe, O.; Payam, M.; Aurora, L.N.; Malcolm, J.B.; Jesse, F.G.; Andrew, D.H. A central role for gamma-glutamyl hydrolases in plant folate homeostasis. Plant J. 2010, 64, 256–266. [Google Scholar] [CrossRef]
- Vaishnav, A.; Kumari, S.; Jain, S.; Varma, A.; Choudhary, D.K. Putative bacterial volatile-mediated growth in soybean (Glycine max L. Merrill) and expression of induced proteins under salt stress. J. Appl. Microbiol. 2015, 119, 539–551. [Google Scholar] [CrossRef]
- Salavati, A.; Bushehri, A.A.; Taleei, A.; Hiraga, S.; Komatsu, S. A comparative proteomic analysis of the early response to compatible symbiotic bacteria in the roots of a supernodulating soybean variety. J. Proteom. 2012, 75, 819–832. [Google Scholar] [CrossRef]
- Wei, X.; Tana, W.; Chen, J.; Yu, S.; Zhang, X.; Zhang, L. Responses of Trollius chinensis to drought stress and rehydration: From photosynthetic physiology to gene expression. Plant Physiol. Biochem. PPB 2023, 201, 107841. [Google Scholar] [CrossRef]
- Grabsztunowicz, M.; Rantala, M.; Ivanauskaite, A.; Blomster, T.; Koskela, M.M.; Vuorinen, K.; Tyystjärvi, E.; Burow, M.; Overmyer, K.; Mähönen, A.P.; et al. Root-type ferredoxin-NADP + oxidoreductase isoforms in Arabidopsis thaliana: Expression patterns, location and stress responses. Plant Cell Environ. 2021, 44, 548–558. [Google Scholar] [CrossRef]
- Lin, Y.; Huang, L.; Hase, T.; Huang, H.-T.D.; Feng, T. Expression of plant ferredoxin-like protein (PFLP) enhances tolerance to heat stress in Arabidopsis thaliana. New Biotechnol. 2015, 32, 235–242. [Google Scholar] [CrossRef]
- Shuai, W.; Qiuping, L.; Jianfeng, W.; Yan, Y.; Guoliang, Z.; Huifei, Z.; Jiajie, W.; Feng, C.; Xiaojie, W.; Zhensheng, K.; et al. YR36/WKS1-Mediated Phosphorylation of PsbO, an Extrinsic Member of Photosystem II, Inhibits Photosynthesis and Confers Stripe Rust Resistance in Wheat. Mol. Plant 2019, 12, 1639–1650. [Google Scholar] [CrossRef]
- Gao, L.; Xing, Y.; Li, X.; Guo, G.; Hu, Y.; Ma, W.; Yan, Y. Proteome analysis of wheat leaf under salt stress by two-dimensional difference gel electrophoresis (2D-DIGE). Phytochemistry 2011, 72, 1180–1191. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Bian, B.; Zhang, M.; Wang, C.; Li, C.; Liao, W. The role and proteomic analysis of ethylene in hydrogen gas-induced adventitious rooting development in cucumber (Cucumis sativus L.) explants. PeerJ 2020, 8, e8896. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Zhang, L.; Wu, Y.; Zheng, Y.; Nie, L.; Zhang, S.; Lan, T.; Zhao, Y.; Zhu, S.; Hou, J.; et al. Comparative transcriptome analysis reveals that chlorophyll metabolism contributes to leaf color changes in wucai (Brassica campestris L.) in response to cold. BMC Plant Biol. 2021, 21, 438. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Page, M.T.; Sumida, A.; Tanaka, A.; Terry, M.J.; Tanaka, R. The iron-sulfur cluster biosynthesis protein SUFB is required for chlorophyll synthesis, but not phytochrome signaling. Plant J. 2017, 89, 1184–1194. [Google Scholar] [CrossRef]
- Zhao, X.; Wei, Y.; Zhang, J.; Yang, L.; Liu, X.; Zhang, H.; Shao, W.; He, L.; Li, Z.; Zhang, Y.; et al. Membrane Lipids’ Metabolism and Transcriptional Regulation in Maize Roots Under Cold Stress. Front. Plant Sci. 2021, 12, 639132. [Google Scholar] [CrossRef]
- Mendoza, D.D. Temperature sensing by membranes. Annu. Rev. Microbiol. 2014, 68, 101–116. [Google Scholar] [CrossRef]
- Takahashi, D.; Uemura, M.; Kawamura, Y. Freezing Tolerance of Plant Cells: From the Aspect of Plasma Membrane and Microdomain. Adv. Exp. Med. Biol. 2018, 1081, 61–79. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, G.; Liu, Z.; Wang, J.; Wei, J.; Dong, X.; Li, H.; Wang, Y.; Tian, H.; Wu, Z.; Cui, J. Integrative Analysis of the Methylome, Transcriptome, and Proteome Reveals a New Mechanism of Rapeseed Under Freezing Stress. Agronomy 2025, 15, 739. https://doi.org/10.3390/agronomy15030739
Zheng G, Liu Z, Wang J, Wei J, Dong X, Li H, Wang Y, Tian H, Wu Z, Cui J. Integrative Analysis of the Methylome, Transcriptome, and Proteome Reveals a New Mechanism of Rapeseed Under Freezing Stress. Agronomy. 2025; 15(3):739. https://doi.org/10.3390/agronomy15030739
Chicago/Turabian StyleZheng, Guoqiang, Zigang Liu, Jinxiong Wang, Jiaping Wei, Xiaoyun Dong, Hui Li, Ying Wang, Haiyang Tian, Zefeng Wu, and Junmei Cui. 2025. "Integrative Analysis of the Methylome, Transcriptome, and Proteome Reveals a New Mechanism of Rapeseed Under Freezing Stress" Agronomy 15, no. 3: 739. https://doi.org/10.3390/agronomy15030739
APA StyleZheng, G., Liu, Z., Wang, J., Wei, J., Dong, X., Li, H., Wang, Y., Tian, H., Wu, Z., & Cui, J. (2025). Integrative Analysis of the Methylome, Transcriptome, and Proteome Reveals a New Mechanism of Rapeseed Under Freezing Stress. Agronomy, 15(3), 739. https://doi.org/10.3390/agronomy15030739





