Nitrogen Economy and Nitrogen Environmental Interactions in Conifers
Abstract
:1. Introduction
2. Efficiency of Nitrogen Use, Growth and Biomass Production in Forest Trees
3. Relevant Metabolic Processes Related to N Economy in Conifers
3.1. Arginine Metabolism
3.2. Phenylalanine Metabolism
3.3. Biosynthesis of Lignins and Lignans
4. Other Processes in N Recycling
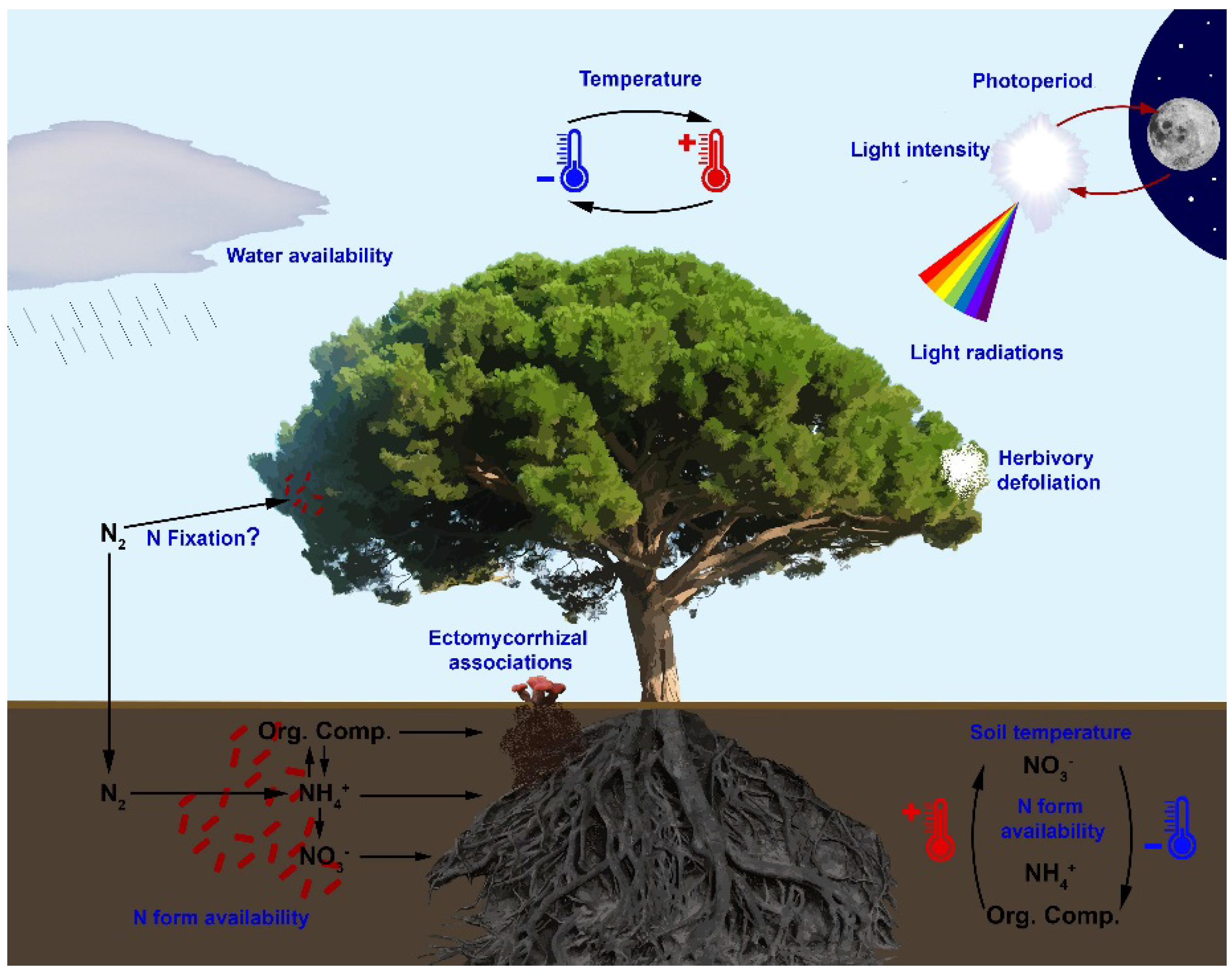
5. Interaction between Nitrogen Metabolism and Environment
5.1. Abiotic Interactions
5.2. Biotic Interactions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hirel, B.; Le Gouis, J.; Ney, B.; Gallais, A. The challenge of improving nitrogen use efficiency in crop plants: Towards a more central role for genetic variability and quantitative genetics within integrated approaches. J. Exp. Bot. 2007, 58, 2369–2387. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Fan, X.; Miller, A.J. Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 2012, 63, 153–182. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.; Kohler, A.; Duplessis, S. Living in harmony in the wood underground: Ectomycorrhizal genomics. Curr. Opin. Plant Biol. 2007, 10, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Tilman, D.; Fargione, J.; Wolff, B.; D´Antonio, C.; Dobson, A.; Howarth, R.; Schindler, D.; Schlesinger, W.H.; Simberloff, D.; Swackhamer, D. Forecasting agriculturally driven global environmental change. Science 2001, 292, 281–284. [Google Scholar] [CrossRef] [PubMed]
- Gruber, N.; Galloway, J.N. An earth-system perspective of the global nitrogen cycle. Nature 2008, 451, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Kronzucker, H.J.; Siddiqi, M.Y.; Glass, A.D.M. Conifer root discrimination against soil nitrate and the ecology of forest succesion. Nature 1997, 385, 59–61. [Google Scholar] [CrossRef]
- Näsholm, T.; Ekblad, A.; Nordin, A.; Glesler, R.; Hogberg, M.; Hogberg, P. Boreal forest plants take up organic nitrogen. Nature 1998, 392, 914–916. [Google Scholar] [CrossRef]
- Lipson, D.; Näsholm, T. The unexpected versatility of plants: Organic nitrogen use and availability in terrestrial ecosystems. Oecologia 2001, 128, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Suddick, E.C.; Whitney, P.; Townsend, A.R.; Davidson, E.A. The role of nitrogen in climate change and the impacts of nitrogen-climate interactions in the United States: Foreword to thematic issue. Biogeochemistry 2013, 114, 1–10. [Google Scholar] [CrossRef]
- Aber, J.; McDowell, W.; Nadelhoffer, K.; Magill, A.; Berntson, G.; Kamakea, M.; McNulty, S.; Currie, W.; Rustad, L.; Fernandez, I. Nitrogen saturation in temperate forest ecosystems. BioScience 1998, 48, 921–934. [Google Scholar] [CrossRef]
- Cooke, J.E.K.; Weih, M. Nitrogen storage and seasonal nitrogen cycling in Populus: Bridging molecular physiology and ecophysiology. New Phytol. 2005, 167, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Farjon, A. A Handbook of the World’s Conifers, 1st ed.; Brill Academic Pub.: Leiden, The Netherlands, 2010. [Google Scholar]
- Bowe, L.M.; Coat, G.; de Pamphilis, C.W. Phylogeny of seed plants based on all three genomic compartments: Extant gymnosperms are monophyletic and Gnetales’ closest relatives are conifers. Proc. Natl. Acad. Sci. USA 2000, 97, 4092–4097. [Google Scholar] [CrossRef] [PubMed]
- Polle, A. The molecular physiology of poplars: Paving the way for knowledge-based biomass production. Plant Biol. 2010, 12, 239–241. [Google Scholar] [CrossRef] [PubMed]
- Stepien, V.; Sauter, J.J.; Martin, F. Vegetative storage proteins in woody plants. Plant Physiol. Biochem. 1994, 32, 185–192. [Google Scholar]
- Good, A.G.; Shrawat, A.K.; Muench, D.G. Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci. 2004, 9, 597–605. [Google Scholar] [CrossRef] [PubMed]
- Finzi, A.C.; Norby, R.J.; Calfapietra, C.; Gallet-Budynek, A.; Gielen, B.; Holmes, W.E.; Hoosbeek, M.R.; Iversen, C.M.; Jackson, R.B.; Kubiske, M.E.; et al. Increases in nitrogen uptake rather than nitrogen-use efficiency support higher rates of temperate forest productivity under elevated CO2. Proc. Natl. Acad. Sci. USA 2007, 104, 14014–14019. [Google Scholar] [CrossRef] [PubMed]
- Birk, E.M.; Vitousek, P.M. Nitrogen availability and nitrogen use efficiency in loblolly pine stands. Ecology 1986, 67, 69–79. [Google Scholar] [CrossRef]
- Nasholm, T.; Nordin, A.; Edfast, A.B.; Hogberg, P. Identification of coniferous forests with incipient nitrogen saturation through analysis of arginine and nitrogen-15 abundance of trees. J. Environ. Qual. 1997, 26, 302–309. [Google Scholar]
- Lawrence, S.D.; Greenwood, J.S.; Korhnak, T.E.; Davis, J.M. A vegetative storage protein homolog is expressed in the growing shoot apex of hybrid poplar. Planta 1997, 203, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Cooke, J.E.K.; Martin, T.A.; Davis, J.M. Short-term physiological and developmental responses to nitrogen availability in hybrid poplar. New Phytol. 2005, 167, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Langenfeld-Heyser, R.; Calfapietra, C.; Polle, A. Influence of free air CO2 enrichment (EUROFACE) and nitrogen fertilisation on the anatomy of juvenile wood of three poplar species after coppicing. Trees 2005, 19, 109–118. [Google Scholar] [CrossRef]
- Cooke, J.E.K.; Brown, K.A.; Wu, R.; Davis, J.M. Gene expression associated with N-induced shifts in resource allocation in poplar. Plant Cell Environ. 2003, 26, 757–770. [Google Scholar] [CrossRef]
- Canales, J.; Flores-Monterrosso, A.; Rueda-López, M.; Avila, C.; Cánovas, F.M. Identification of genes regulated by ammonium availability in the roots of maritime pine trees. Amino Acids 2010, 39, 991–1001. [Google Scholar] [CrossRef] [PubMed]
- MacKay, J.; Dean, J.F.D.; Plomion, C.; Peterson, D.G.; Cánovas, F.M.; Pavy, N.; Ingvarsson, P.K.; Savolainen, O.; Guevara, M.A.; Fluch, S.; et al. Towards decoding the conifer giga-genome. Plant Mol. Biol. 2012, 80, 555–569. [Google Scholar] [CrossRef] [PubMed]
- Plomion, C.; Bastien, C.; Bogeat-Triboulot, M.B.; Bouffier, L.; Déjardin, A.; Duplessis, S.; Fady, B.; Heuertz, M.; Le Gac, A.L.; Le Provost, G.; et al. Forest tree genomics: 10 achievements from the past 10 years and future prospects. Ann. For. Sci. 2015, 73, 77–103. [Google Scholar] [CrossRef]
- Cantón, F.R.; Suárez, M.F.; Cánovas, F.M. Molecular aspects of nitrogen mobilisation and recycling in trees. Photosynth. Res. 2005, 83, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Cánovas, F.M.; Avila, C.; Cantón, F.R.; Cañas, R.A.; de la Torre, F. Ammonium assimilation and amino acid metabolism in conifers. J. Exp. Bot. 2007, 58, 2307–2318. [Google Scholar] [CrossRef] [PubMed]
- Llacer, J.L.; Fita, I.; Rubio, V. Arginine and nitrogen storage. Curr. Opin. Struct. Biol. 2008, 18, 673–681. [Google Scholar] [CrossRef] [PubMed]
- Winter, G.; Todd, C.D.; Trovato, M.; Forlani, G.; Funck, D. Physiological implications of arginine metabolism in plants. Front Plant Sci. 2015, 6, 534. [Google Scholar] [CrossRef] [PubMed]
- Slocum, R.D. Genes, enzymes and regulation of arginine biosynthesis in plants. Plant Physiol. Biochem. 2005, 43, 729–745. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.M.; Ferrar, T.S.; Lohmeir-Vogel, E.; Morrice, N.; Mizuno, Y.; Berenger, B.; Ng, K.K.S.; Muench, D.G.; Moorhead, G.B.G. The PII signal transduction protein of Arabidopsis thaliana forms an arginine-regulated complex with plastid N-acetylglutamate kinase. J. Biol. Chem. 2006, 281, 5726–5733. [Google Scholar] [CrossRef] [PubMed]
- Chellamuthu, V.-R.; Ermilova, E.; Lapina, T.; Lüddecke, J.; Minaeva, E.; Herrmann, C.; Hartmann, M.D.; Forchhammer, K. A widespread glutamine-sensing mechanism in the plant kingdom. Cell 2014, 159, 1188–1199. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.L.; Gifford, D.J. Structural and biochemical changes in loblolly pine (Pinus taeda L.) seeds during germination and early seedling growth. I. Storage protein reserves. Int. J. Plant Sci. 1997, 158, 727–737. [Google Scholar] [CrossRef]
- King, J.E.; Gifford, D.J. Amino acid utilization in seeds of loblolly pine during germination and early seedling growth (I. Arginine and arginase activity). Plant Physiol. 1997, 113, 1125–1135. [Google Scholar] [PubMed]
- Rigault, P.; Boyle, B.; Lepage, P.; Cooke, J.E.K.; Bousquet, J.; MacKay, J.J. A white spruce gene catalog for conifer genome analyses. Plant Physiol. 2011, 157, 14–28. [Google Scholar] [CrossRef] [PubMed]
- Canales, J.; Bautista, R.; Label, P.; Gómez-Maldonado, J.; Lesur, I.; Fernández-Pozo, N.; Rueda-López, M.; Guerrero-Fernández, D.; Castro-Rodriguez, V.; Benzekri, H.; et al. De novo assembly of maritime pine transcriptome: Implications for forest breeding and biotechnology. Plant Biotechnol. J. 2014, 12, 286–299. [Google Scholar] [CrossRef] [PubMed]
- Nystedt, B.; Street, N.R.; Wetterbom, A.; Zuccolo, A.; Lin, Y.C.; Scofield, D.G.; Vezzi, F.; Delhomme, N.; Giacomello, S.; Alexeyenko, A.; et al. The Norway spruce genome sequence and conifer genome evolution. Nature 2013, 497, 579–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimin, A.; Stevens, K.A.; Crepeau, M.W.; Holtz-Morris, A.; Koriabine, M.; Marçais, G.; Puiu, D.; Roberts, M.; Wegrzyn, J.L.; de Jong, P.J.; et al. Sequencing and assembly of the 22-gb loblolly pine genome. Genetics 2014, 196, 875–890. [Google Scholar] [CrossRef] [PubMed]
- Todd, C.D.; Cooke, J.E.; Mullen, R.T.; Gifford, D.J. Regulation of loblolly pine (Pinus taeda L.) arginase in developing seedling tissue during germination and post-germinative growth. Plant Mol. Biol. 2001, 45, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Suárez, M.F.; Ávila, C.; Gallardo, F.; Cantón, F.R.; García-Gutiérrez, A.; Claros, M.G.; Cánovas, F.M. Molecular and enzymatic analysis of ammonium assimilation in woody plants. J. Exp. Bot. 2002, 53, 891–904. [Google Scholar] [CrossRef] [PubMed]
- Cañas, R.A.; Villalobos, D.P.; Díaz-Moreno, S.M.; Cánovas, F.M.; Cantón, F.R. Molecular and functional analyses support a role of Ornithine-δ-aminotransferase in the provision of glutamate for glutamine biosynthesis during pine germination. Plant Physiol. 2008, 148, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Dudareva, N. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.; Widhalm, J.R.; Qian, Y.; Maeda, H.; Cooper, B.R.; Jannasch, A.S.; Gonda, I.; Lewinsohn, E.; Rhodes, D.; Dudareva, N. An alternative pathway contributes to phenylalanine biosynthesis in plants via a cytosolic tyrosine: Phenylpyruvate aminotransferase. Nat. Commun. 2013, 4, 2833. [Google Scholar] [CrossRef] [PubMed]
- De la Torre, F.; El-Azaz, J.; Ávila, C.; Cánovas, F.M. Deciphering the role of aspartate and prephenate aminotransferase activities in plastid nitrogen metabolism. Plant Physiol. 2014, 164, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Van Heerden, P.S.; Towers, G.H.; Lewis, N.G. Nitrogen metabolism in Lignifying Pinus taeda cell cultures. J. Biol. Chem. 1996, 271, 12350–12355. [Google Scholar] [CrossRef] [PubMed]
- Claros, M.G.; Aguilar, M.L.; Cánovas, F.M. Evidence for an operative glutamine translocator in chloroplasts from maritime pine (Pinus pinaster Ait.) cotyledons. Plant Biol. 2010, 12, 717–723. [Google Scholar] [CrossRef] [PubMed]
- Craven-Bartle, B.; Pascual, M.B.; Cánovas, F.M.; Avila, C. A Myb transcription factor regulates genes of the phenylalanine pathway in maritime pine. Plant J. 2013, 74, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Canales, J.; Rueda-López, M.; Craven-Bartle, B.; Avila, C.; Cánovas, F.M. Novel insights into regulation of asparagine synthetase in conifers. Front Plant Sci. 2012, 3, 100. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.; Donaldson, L.; Ralph, J. Lignification and lignin manipulation in conifers. Adv. Bot. Res. 2012, 61, 37–76. [Google Scholar]
- Villalobos, D.P.; Díaz-Moreno, S.M.; El-Sayed, S.S.; Cañas, R.A.; Osuna, D.; van Kerckhoven, S.H.; Bautista, R.; Claros, M.G.; Cánovas, F.M.; Cantón, F.R. Reprogramming of gene expression during compression wood formation in pine: Coordinated modulation of S-adenosylmethionine, lignin and lignan related genes. BMC Plant Biol. 2012, 12, 100. [Google Scholar] [CrossRef] [PubMed]
- García-Gutiérrez, A.; Cantón, F.R.; Gallardo, F.; Sánchez-Jiménez, F.; Cánovas, F.M. Expression of ferredoxin-dependent glutamate synthase in dark-grown pine seedlings. Plant Mol. Biol. 1995, 27, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Avila-Ospina, L.; Moison, M.; Yoshimoto, K.; Masclaux-Daubresse, C. Autophagy, plant senescence, and nutrient recycling. J. Exp. Bot. 2014, 65, 3799–3811. [Google Scholar] [CrossRef] [PubMed]
- Minina, E.A.; Filonova, L.H.; Fukada, K.; Savenkov, E.I.; Gogvadze, V.; Clapham, D.; Sanchez-Vera, V.; Suarez, M.F.; Zhivotovsky, B.; Daniel, G.; et al. Autophagy and metacaspase determine the mode of cell death in plants. J. Cell Biol. 2013, 203, 917–927. [Google Scholar] [CrossRef] [PubMed]
- Vuosku, J.; Sutela, S.; Kestila, J.; Sarjala, T.; Haggman, H. Expression of catalase and retinoblastoma-related protein genes associates with cell death processes in Scots pine zygotic embryogenesis. BMC Plant Biol. 2015, 15, 88. [Google Scholar] [CrossRef] [PubMed]
- Harms, U.; Sauter, J.J. Localization of a storage proteinin the wood ray parenchyma cells of Taxodium distichum (L.), L.C. Rich, by immunogold labeling. Trees 1992, 6, 37–40. [Google Scholar] [CrossRef]
- Galindo-González, L.M.; El Kayal, W.; Ju, C.J.T.; Allen, C.C.G.; King-Jones, S.; Cooke, J.E.K. Integrated transcriptomic and proteomic profiling of white spruce stems during the transition from active growth to dormancy. Plant Cell Environ. 2012, 35, 682–701. [Google Scholar] [CrossRef] [PubMed]
- Millard, P. A review of internal cycling of nitrogen within trees in relation to soil fertility. Optim. Plant Nutr. 1993, 53, 623–628. [Google Scholar]
- Pettengill, E.A.; Pettengill, J.B.; Coleman, G.D. Elucidating the evolutionary history and expression patterns of nucleoside phosphorylase paralogs (vegetative storage proteins) in Populus and the plant kingdom. BMC Plant Biol. 2013, 13, 118. [Google Scholar] [CrossRef] [PubMed]
- Malaguti, D.; Millard, P.; Wendler, R.; Hepburn, A.; Tagliavini, M. Translocation of amino acids in the xylem of apple (Malus domestica Borkh.) trees in spring as a consequence of both N remobilization and root uptake. J. Exp. Bot. 2001, 52, 1665–1671. [Google Scholar] [CrossRef] [PubMed]
- Prunier, J.; Verta, J.P.; MacKay, J.J. Conifer genomics and adaptation: At the crossroads of genetic diversity and genome function. New Phytol. 2015, 209, 44–62. [Google Scholar] [CrossRef] [PubMed]
- Giordano, M. Homeostasis: An underestimated focal point of ecology and evolution. Plant Sci. 2013, 211, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Gillooly, J.F.; Brown, J.H.; West, G.B.; Savage, V.M.; Charnov, E.L. Effects of size and temperature on metabolic rate. Science 2001, 293, 2248–2251. [Google Scholar] [CrossRef] [PubMed]
- Tjoelker, M.G.; Reich, P.B.; Oleksyn, J. Changes in leaf nitrogen and carbohydrates underlie temperature and CO2 acclimation of dark respiration in five boreal tree species. Plant Cell Environ. 1999, 22, 767–778. [Google Scholar] [CrossRef]
- Angelcheva, L.; Mishra, Y.; Antti, H.; Kjellsen, T.D.; Funk, C.; Strimbeck, R.G.; Schröder, W.P. Metabolomic analysis of extreme freezing tolerance in Siberian spruce (Picea obovata). New Phytol. 2014, 204, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Junttila, O.; Palva, E.P. Environmental regulation and physiological basis of freezing tolerance in woody plants. Acta Physiol. Plant 2004, 26, 213–222. [Google Scholar] [CrossRef]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef] [PubMed]
- Odlum, K.; Blake, T.; Kim, Y.; Glerum, C. Influence of photoperiod and temperature on frost hardiness and free amino acid concentrations in black spruce seedlings. Tree Physiol. 1993, 13, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.T.; Glerum, C. Seasonal free amino-acid fluctuations in red pine and white spruce needles. Can. J. For. Res. 1995, 25, 697–703. [Google Scholar] [CrossRef]
- Mano, Y.; Nemoto, K. The pathway of auxin biosynthesis in plants. J. Exp. Bot. 2012, 63, 2853–2872. [Google Scholar] [CrossRef] [PubMed]
- Peer, W.A. From perception to attenuation: Auxin signalling and responses. Curr. Opin. Plant Biol. 2013, 16, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A. Auxin: A regulator of cold stress response. Physiol. Plant 2013, 147, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Cañas, R.A.; Canales, J.; Muñoz-Hernández, C.; Granados, J.M.; Ávila, C.; García-Martín, M.L.; Cánovas, F.M. Understanding developmental and adaptive cues in pine through metabolite profiling and co-expression network analysis. J. Exp. Bot. 2015, 66, 3113–3127. [Google Scholar] [CrossRef] [PubMed]
- Mayne, M.; Coleman, J.; Blumwald, E. Differential expression during drought conditioning of a root-specifi S-adenosylmethionine synthetase from jack pine (Pinus banksiana Lamb.) seedlings. Plant Cell Environ. 1996, 19, 958–966. [Google Scholar] [CrossRef]
- Jaglo-Ottosen, K.R.; Gilmour, S.J.; Zarka, D.G.; Schabenberger, O.; Thomashow, M.F. Arabidopsis CBF1 overexpression induces cor genes and enhances freezing tolerance. Science 1998, 280, 104–106. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, M.; Webb, R.; Balsamo, R.; Close, T.J.; Yu, X.M.; Griffith, M. Purification, immunolocalization, cryoprotective, and antifreeze activity of PCA60: A dehydrin from peach (Prunus persica). Physiol. Plant 1999, 105, 600–608. [Google Scholar] [CrossRef]
- Kjellsen, T.D.; Shiryaeva, L.; Schröder, W.P.; Strimbeck, G.R. Proteomics of extreme freezing tolerance in Siberian spruce (Picea obovata). J. Proteom. 2010, 73, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Kjellsen, T.D.; Yakovlev, I.A.; Fossdal, C.G.; Strimbeck, G.R. Dehydrin accumulation and extreme low-temperature tolerance in Siberian spruce (Picea obovata). Tree Physiol. 2013, 33, 1354–1366. [Google Scholar] [CrossRef] [PubMed]
- Warren, C.R. Why does temperature affect relative uptake rates of nitrate, ammonium and glycine: A test with Eucalyptus pauciflora. Soil Biol. Biochem. 2009, 41, 778–784. [Google Scholar] [CrossRef]
- Boczulak, S.A.; Hawkins, B.J.; Roy, R. Temperature effects on nitrogen form uptake by seedling roots of three contrasting conifers. Tree Physiol. 2014, 34, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Bassisirad, H. Kinetics of nutrient uptake by roots: Responses to global change. New Phytol. 2000, 147, 155–169. [Google Scholar] [CrossRef]
- Krywult, M.; Smykla, J.; Kinnunen, H.; Martz, F.; Sutinen, M.L.; Lakkala, K.; Turunen, M. Influence of solar UV radiation on the nitrogen metabolism in needles of Scots pine (Pinus sylvestris L.). Environ. Pollut. 2008, 156, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Cantón, F.R.; Suárez, M.F.; Josè-Estanyol, M.; Cánovas, F.M. Expression analysis of a cytosolic glutamine synthetase gene in cotyledons of Scots pine seedlings: Developmental, light regulation and spatial distribution of specific transcripts. Plant Mol. Biol. 1999, 40, 623–634. [Google Scholar] [CrossRef] [PubMed]
- Cañas, R.A.; de la Torre, F.; Cánovas, F.M.; Cantón, F.R. High levels of asparagine synthetase in hypocotyls of pine seedlings suggest a role of the enzyme in re-allocation of seed-stored nitrogen. Planta 2006, 224, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Takashima, T.; Hikosaka, K.; Hirose, T. Photosynthesis or persistence: nitrogen allocation in leaves of evergreen and deciduous Quercus species. Plant Cell Environ. 2004, 27, 1047–1054. [Google Scholar] [CrossRef]
- Yoshimura, K. Irradiance heterogeneity within crown affects photosynthetic capacity and nitrogen distribution of leaves in Cedrela sinensis. Plant Cell Environ. 2010, 33, 750–758. [Google Scholar] [PubMed]
- Benomar, L.; Lamhamedi, M.S.; Villeneuve, I.; Rainville, A.; Beaulieu, J.; Bousquet, J.; Margolis, H.A. Fine-scale geographic variation in photosynthetic-related traits of Picea glauca seedlings indicates local adaptation to climate. Tree Physiol. 2015, 35, 864–878. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Källman, T.; Ma, X.; Gyllenstrand, N.; Zaina, G.; Morgante, M.; Bousquet, J.; Eckert, A.; Wegrzyn, J.; Neale, D.; et al. Disentangling the roles of history and local selection in shaping clinal variation of allele frequencies and gene expression in Norway spruce (Picea abies). Genetics 2012, 191, 865–881. [Google Scholar] [CrossRef] [PubMed]
- Kujala, S.T.; Savolainen, O. Sequence variatio n patterns along a latitudinal cline in Scots pine (Pinus sylvestris): Signs of clinal adaptation? Tree Genet. Genom. 2012, 8, 1451–1467. [Google Scholar] [CrossRef]
- Avia, K.; Kärkkäinen, K.; Lagercrantz, U.; Savolainen, O. Association of FLOWERING LOCUS T/TERMINAL FLOWER 1-like gene FTL2 expression with growth rhythm in Scots pine (Pinus sylvestris). New Phytol. 2014, 204, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Zhang, Y.; Zhang, Y.; Zhang, S. Continuous planting under a high density enhances the competition for nutrients among young Cunninghamia lanceolata saplings. Ann. For. Sci. 2015. [Google Scholar] [CrossRef]
- Feng, Y.; Lei, Y.; Wang, R.; Callaway, R.M.; Valiente-Banuet, A.; Li, Y.; Zheng, Y. Evolutionary tradeoffs for nitrogen allocation to photosynthesis versus cell walls in an invasive plant. Proc. Natl. Acad. Sci. USA 2009, 106, 1853–1856. [Google Scholar] [CrossRef] [PubMed]
- Duursma, R.A.; Marshall, J.D.; Nippert, J.B.; Chambers, C.C.; Robinson, A.P. Estimating leaf-level parameters for ecosystem process models: A study in mixed conifer canopies on complex terrain. Tree Physiol. 2005, 25, 1347–1359. [Google Scholar] [CrossRef] [PubMed]
- Lilles, E.B.; Astrup, R.; Lefrançois, M.L.; David Coates, K. Sapling leaf trait responses to light, tree height and soil nutrients for three conifer species of contrasting shade tolerance. Tree Physiol. 2014, 34, 1334–1347. [Google Scholar] [CrossRef] [PubMed]
- Zepp, R.G.; Erickson, D.J.; Paul, N.D.; Sulzberger, B. Interactive effects of solar UV radiation and climate change on biogeochemical cycling. Photochem. Photobiol. Sci. 2007, 6, 286–300. [Google Scholar] [CrossRef] [PubMed]
- Bornman, J.F.; Barnes, P.W.; Robinson, S.A.; Ballaré, C.L.; Flint, S.D.; Caldwell, M.M. Solar ultraviolet radiation and ozone depletion-driven climate change: Effects on terrestrial ecosystems. Photochem. Photobiol. Sci. 2015, 14, 88–107. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Liu, Q. Responses in growth, physiology and nitrogen nutrition of dragon spruce (Picea asperata) seedlings of different ages to enhanced ultraviolet-B. Acta Physiol. Plant. 2007, 29, 217–224. [Google Scholar] [CrossRef]
- Schnitzler, J.P.; Jungblut, T.P.; Feicht, C.; Köfferlein, M.; Langebartels, C.; Heller, W.; Sandermann, H. UV-B induction of flavonoid biosynthesis in Scots pine (Pinus sylvestris L.) seedlings. Trees 1997, 11, 162–168. [Google Scholar] [CrossRef]
- Martz, F.; Sutinen, M.L.; Derome, K.; Wingsle, G.; Julkunen-Tiitto, R.; Turunen, M. Effects of ultraviolet (UV) exclusion on the seasonal concentration of photosynthetic and UV-screening pigments in Scots pine needles. Glob. Change Biol. 2007, 13, 252–265. [Google Scholar] [CrossRef]
- Mouradov, A.; Spangenberg, G. Flavonoids: a metabolic network mediating plants adaptation to their real estate. Front. Plant Sci. 2014, 5, 620. [Google Scholar] [CrossRef] [PubMed]
- Du, S.; Yamamoto, F. An Overview of the biology of reaction wood formation. J. Integr. Plant Biol. 2007, 49, 131–143. [Google Scholar] [CrossRef]
- Palacio, S.; Hernández, R.; Maestro-Martínez, M.; Camarero, J.J. Fast replenishment of inicial carbon stores alter defoliation by the pine processionary moth and its relationship to the re-growth ability of tress. Tress 2012, 26, 1627–1640. [Google Scholar] [CrossRef] [Green Version]
- Asiegbu, F.O.; Choi, W.; Li, G.; Nahalkova, J.; Dean, R.A. Isolation of a novel antimicrobial peptide gene (Sp-AMP) homologue from Pinus sylvestris (Scots pine) following infection with the root rot fungus Heterobasidion annosum. FEMS Microbiol. Lett. 2003, 228, 27–31. [Google Scholar] [CrossRef]
- Ekramoddoullah, A.K.M.; Liu, J.J.; Zamani, A. Cloning and characterisation of a putative antifungal peptide gene (Pm-AMP1) in Pinus monticola. Phytopatholy 2006, 96, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Ekramoddoullah, A.K.M. Molecular tools in the study of the white pine blister rust (Cronartium ribicola) pathosystem. Can. J. Plant Pathol. 2005, 27, 510–520. [Google Scholar] [CrossRef]
- Canales, J.; Avila, C.; Cánovas, F.M. A maritime pine antimicrobial peptide involved in ammonium nutrition. Plant Cell Environ. 2011, 34, 1443–1453. [Google Scholar] [CrossRef] [PubMed]
- Brundett, M.C. Coevolution of roots and mycorrhizas of land plants. New Phytol. 2002, 134, 275–304. [Google Scholar] [CrossRef]
- Chalot, M.; Plassard, C. Ectomycorrhiza and nitrogen provision to the host tree. In Ecological Aspects of Nitrogen Metabolism in Plants, 1st ed.; Polacco, J.C., Todd, C.D., Eds.; John Wiley and Sons: Oxford, UK, 2011; pp. 69–94. [Google Scholar]
- Averill, C.; Turner, B.L.; Finzi, A.C. Mycorrhiza-mediated competition between plants and decomposers drives soil carbon storage. Nature 2014, 505, 543–545. [Google Scholar] [CrossRef] [PubMed]
- Hobbie, E.A.; Colpaert, J.V.; White, M.W.; Ouimette, A.P.; Macko, S.A. Nitrogen form, availability, and mycorrhizal colonization affect biomass and nitrogen isotope patterns in Pinus sylvestris. Plant Soil 2008, 310, 121–136. [Google Scholar] [CrossRef]
- Klironomos, J.N.; Hart, M.M. Animal nitrogen swap for plant carbon. Nature 2001, 410, 651–652. [Google Scholar] [CrossRef] [PubMed]
- Kranabetter, J.M.; Dawson, C.R.; Dunn, D.E. Indices of dissolved organic nitrogen, ammonium and nitrate across productivity gradients of boreal forests. Soil Biol. Biochem. 2007, 39, 3147–3158. [Google Scholar] [CrossRef]
- Chalot, M.; Blaudez, D.; Brun, A. Ammonia: A candidate for nitrogen transfer at the mycorrhizal interface. Trends Plant Sci. 2006, 11, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Gruffman, L.; Jämtgård, S.; Näsholm, T. Plant nitrogen status and co-occurrence of organic and inorganic nitrogen sources influence root uptake by Scots pine seedlings. Tree Physiol. 2014, 34, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Flores-Monterroso, A.; Canales, J.; de la Torre, F.; Ávila, C.; Cánovas, F.M. Identification of genes differentially expressed in ectomycorrhizal roots during the Pinus pinaster-Laccaria bicolor interaction. Planta 2013, 237, 1637–1650. [Google Scholar] [CrossRef] [PubMed]
- Warren, C.R.; Adams, M.A. Possible causes of slow growth of nitrate-supplied Pinus pinaster. Can. J. For. Res. 2002, 32, 569–580. [Google Scholar] [CrossRef]
- Carrell, A.A.; Frank, A.C. Pinus flexilis and Picea engelmannii share a simple and consistent needle endophyte microbiota with a potential role in nitrogen fixation. Front Microbiol. 2014, 5, 333. [Google Scholar] [CrossRef] [PubMed]


© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cañas, R.A.; De la Torre, F.; Pascual, M.B.; Avila, C.; Cánovas, F.M. Nitrogen Economy and Nitrogen Environmental Interactions in Conifers. Agronomy 2016, 6, 26. https://doi.org/10.3390/agronomy6020026
Cañas RA, De la Torre F, Pascual MB, Avila C, Cánovas FM. Nitrogen Economy and Nitrogen Environmental Interactions in Conifers. Agronomy. 2016; 6(2):26. https://doi.org/10.3390/agronomy6020026
Chicago/Turabian StyleCañas, Rafael A., Fernando De la Torre, Maria Belén Pascual, Concepción Avila, and Francisco M. Cánovas. 2016. "Nitrogen Economy and Nitrogen Environmental Interactions in Conifers" Agronomy 6, no. 2: 26. https://doi.org/10.3390/agronomy6020026
APA StyleCañas, R. A., De la Torre, F., Pascual, M. B., Avila, C., & Cánovas, F. M. (2016). Nitrogen Economy and Nitrogen Environmental Interactions in Conifers. Agronomy, 6(2), 26. https://doi.org/10.3390/agronomy6020026









